Abstract
Lung cancer is one of the types of cancer with highest mortality rate in the world. In case of early detection and diagnosis, the survival rate of patients significantly increases. In this study, a novel method and system that provides automatic detection of juxtapleural nodule pattern have been developed from cross-sectional images of lung CT (Computerized Tomography). Shape-based and both shape and texture based 7 features are contributed to the literature for lung nodules. System that we developed consists of six main stages called preprocessing, lung segmentation, detection of nodule candidate regions, feature extraction, feature selection (with five feature ranking criteria) and classification. LIDC dataset containing cross-sectional images of lung CT has been utilized, 1410 nodule candidate regions and 40 features have been extracted from 138 cross-sectional images for 24 patients. Experimental results for 10 classifiers are obtained and presented. Adding our derived features to known 33 features has increased nodule recognition performance from 0.9639 to 0.9679 AUC value on generalized linear model regression (GLMR) for 22 selected features and being reached one of the most successful results in the literature.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lung cancer, is one of the types of cancer, cells in lung tissue can invade other organs by covering the surrounding tissue cells, has highest mortality rate in the world. According to report of the World Health Organization (WHO), lung cancer is type of cancer that causes to death in men ranked first and in woman ranked second among cancer types in the world [1]. Treatment of lung cancer and the survival rate of patient depend on early detection of the cancer and patient not to smoke.
One of the most important signs of lung cancer in initial stage is detection of cancerous (malignant) lung nodules. Medical imaging techniques such as LDCT or low dose CT (Computerized Tomography), PET (Positron Emission Tomography), chest radiography, ultrasonography and X-ray are extensively used for early detection of lung cancer and imaging of nodules. CT medical imaging method has highest sensitivity and lower error rate. CT enables visualization of small or low-contrast nodules that could hardly be screened in conventional radiograms [2].
Development of computer aided lung nodule region detection method and system on CT images is so important in terms of being useful and helpful to expert radiologists in the process of early diagnosis and treatment of lung cancer, increasing of the survival rate of patients, reduction and minimization of error rate of nodule detection and interpretation of radiologists, reducing the time required for these operations, facilitating detection of cancerous nodules which can be overlooked. These and many other benefits of the system which can be used in various fields, are motivation of the project. So, these factors have led to this work.
In this study, a method and software that provides automatic detection of juxtapleural nodule region have been developed from cross-sectional images of lung CT using classifiers and image processing techniques. Juxtapleural nodules which have a significant degree of connection to the pleural surface, emerge segmentation problems in the stage of lung segmentation due to the fact that nodules have Hounsfield Units near to pleura. In the scope of this study, lungs have been segmented using image processing techniques by separating from complex background, juxtapleural nodule candidate regions have been identified using α-hull method [3], then morphological, statistical and texture based features have been extracted from these regions and feature selection has been applied, original nodule region has been detected by means of classification models. LIDC dataset containing cross-sectional images of lung CT has been utilized.
Main contribution to the literature is to gain novel seven features (five shape-based and two both shape and texture based) extracted from nodule candidates which identified by means of α-hull method. In the developed system, distinctiveness factor between nodule candidates has been increased by providing wealth of features.
Related work
Numerous studies have been made for computer aided detection and diagnosis of lung nodules [4, 5]. While some studies consist of scientific publications, another portion of studies consist of patents of CAD (Computer Aided Detection) systems having commercial value used extensively in hospitals nowadays. CAD systems used in lung cancer generally consist of five main stages. These stages are image acquisition, preprocessing, lung segmentation, detection of nodule candidate regions, reduction of false positives.
Image acquisition stage is based on obtaining images which contain lung nodules from medical imaging methods. For example, Lung Image Database Consortium (LIDC) [6], Public Lung Database to Address Drug Response (ELCAP) [7] data sets. Image pre-processing refers to the process of improving both the quality and interpretability of the acquired lung images. The pre-processing component reduces noise and artefacts in the lung image slices [2]. Ezoe et al. [8] applied N-Quoit (Q) filter based on mathematical morphology to enhance candidate nodules, Frangi et al. [9] applied blob-enhancement filter based on Hessian Matrix to enhance sphere-like objects [10], Ochs et al. [11] and Paik et al. [12] applied sphericity structure enhancement filter to enhance the nodule like structure in CT images, Gaussian smoothing was employed by Retico et al. [13] to eliminate the image artefacts and Li et al. [14] applied multi-scale enhancement filter to suppress blood vessels and highlight nodule like structures in their studies [2].
Lung segmentation stage consists of some operations for separating lung regions and removing other parts from image. Healthy lung tissues constitute darker region than structures located in and around the lungs. This feature of lung tissues is one of the basic methods for segmentation of lungs. While isolated nodules don’t have segmentation problem, juxtapleural nodules have some segmentation problems because of the fact that these nodules have similar color, Hounsfield Unit near to pleural surface. In these situations, different segmentation approaches are needed to solve this problem. Gray level thresholding based approaches were used by Armato et al. [15], Hu et al. [16]. Apart from the gray level thresholding and pixel based approaches, different approaches such as active shape model (ASM) (snakes), active appearance model (AAM), deformable model, level set are also used [17]. Adaptive border marching method proposed in [18], alpha hull method proposed in [19, 20] were developed for detection of juxtapleural nodule regions.
Lung nodule detection refers to the process of determining whether nodule patterns are present in the image, and identifying the location of the nodules [2]. Lung nodule detection is classified into intensity and model based detection methods. It can be achieved using template-based, segmentation-based, and classification-based methods. An ensemble classification aided by clustering (CAC) method was proposed for classification based method [21]. In intensity based detection methods, thresholding [22], clustering algorithms [23], artificial neural network (ANN) [24], mathematical morphology and ANN [25], 3D region growing approach [14] were developed. In model based detection methods, template matching based on genetic algorithm [26], deformable template [27], object based deformation [28], anatomy based generic model [29] were proposed [30]. Studies using combination of intensity based and model based methods are also available. Surface normals and gradient-based method to capture the concentration of normals by calculating derivatives of intensity images was proposed by [12], [30].
False positives reduction refers to the process of further eliminating the false positives from the output of nodule detection or lung segmentation components [2]. Appearance, shape, spectral and textural and contextual features are used to describe the content of image. Some fundamental feature types, representing the appearance of medical images in the most discriminatory way eases the classification procedure and detection and understanding disease process [31]. Retico et al. [13] used voxel based neural approach by extracting features from voxel neighborhood to reduce the amount of false positive findings in the lists of nodule candidates in their study. Nunzio et al. [20] applied threshold values on features extracted from juxtapleural nodule candidate regions identified by α-hull method and used a supervised two-layer, 13-input, 20-hidden-neuron, 1-output feed-forward neural network, trained with gradient descent learning rule with momentum as the classifier system. Choi and Choi [32] developed three dimensional shape based feature descriptor to detect nodules and iterative wall elimination method for refinement of feature descriptor. Ozekes and Osman [33] introduced a CAD system based on three-dimensional (3D) feature extraction to detect lung nodules. Kuruvilla and Gunavathi [34] extracted features and used different ANNs for classification. Daliri [35] proposed a hybrid automatic system based on genetic algorithm and fuzzy extreme learning machines. Other studies and detailed experimental results in this area can be achieved from [2] and [17].
Materials and methods
Image acquisition (materials)
Lung CTs have been obtained from LIDC-IDRI data set [36]. This dataset is publicly available in the National Biomedical Imaging Archive (NBIA) and it provides a reference for the medical imaging research community.
The characteristics of LIDC-IDRI dataset is shown in Table 1. In the initial blinded-read phase, each radiologist independently reviewed each CT scan and marked lesions belonging to one of three categories (“nodule ≥3 mm,” “nodule < 3 mm,” and “non-nodule ≥ 3 mm”) [6]. In the subsequent unblinded-read phase, each radiologist independently reviewed their own marks along with the anonymized marks of the three other radiologists to render a final opinion [6]. Radiologists used the computer interface to construct outlines of around the nodules in each CT section [6].
These images have been obtained using several CT scanners and CT protocols in helical mode. These nodules have at least 3 mm and less than 30 mm diameter in lung CTs. In the scope of this study, only lung CT slices containing juxtapleural lung nodules have been utilized.
System methodology
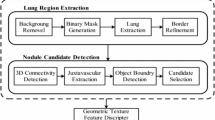
System architecture and stages of the project are illustrated in Fig. 1. Details of these stages will be explained in the following subsections, methods and approaches used for every stage will be discussed.
Preprocessing
Preprocessing consists of initial operations which select appropriate images to be suitable for processing on the computer. These operations used in this stage are shown in Fig. 2.
Firstly, slices containing juxtapleural lung nodule are selected from LIDC dataset. Images obtained according to medical imaging methods have DICOM image format. Representation of nodule regions that outlined by expert radiologists is implemented on these lung CT slices (Fig. 3). The ground truth regions are stored in xml file containing all information of every patient about all slice images for each case. Then, pixel values are converted into double format in terms of accuracy of applied operations.
Representation of ground truth region and information of nodule [36]
Image information stored in DICOM consists of pixel values in form of binary data. In order to identify window level and width appropriate for lung region, pixel values must be converted into Hounsfield Unit (HU). Linear transformation is made according to Rescale Slope and Rescale Intercept tags in DICOM file. One is used for Rescale Slope and −1024 is used for Rescale Intercept.
Window width is equal to 1500 and window level is equal to −400 for lung CTs. HU values having [−1150, 350] can be shown effectively for this window values. While HU values that exceeds upper limit of this range are shown in white, HU values that exceeds lower limit of this range are shown in black. Besides, so as to apply image processing operations appropriately, 16-bits image is converted into 8-bits image (Fig. 4).
Identifying of window variables a original lung CT b setting the window level and width [36]
In the next step, some morphological operations are applied for obtaining thorax by removing other undesirable structures over 8-bit lung slice image. First of all, the inside of chest region is filled with gaps. By this process, lungs are covered with color of portion fall outside the lung region of the thorax. Then, mask of thorax and line at the bottom of the thorax are extracted by using Otsu thresholding method [37] on CT slices. Otsu method selects the optimal threshold automatically to maximize the seperability of resultant classes in gray levels [37]. In order to remove the pieces outside of the thorax, disk shaped structural element with 10 units of radius is constructed and erosion operation is applied on image. After the erosion operation, thorax mask is obtained, areas that outside of thorax mask are covered in white color on original slice and thorax is obtained. An example of this process is illustrated in Fig. 5.
Lung segmentation
In this stage, lungs are segmented by applying Otsu thresholding method on thorax region. As a result of segmentation, some parts where outside of the lungs may occur. So as to solve this problem, morphological opening operation is carried out with disk shaped structural element four units in radius on segmented lung images. Besides, lung boundaries are obtained by filling the gaps in lung regions and applying Otsu thresholding method on lung mask. As a result of these operations, lung segmentation process is carried out by identifying two regions with highest area in regions with borderline. An example of lung segmentation process is illustrated in Fig. 6.
Detection of nodule candidates
Juxtapleural nodules have a significant degree of connection to the pleural surface. While tissue densities of these nodules have Hounsfield Units near to pleura and thorax surrounding lung region, their tissue densities are different from lung tissue. For this reason, segmentation operation depending on tissue density is not suitable for detection of nodule candidates.
Alpha shape is an approach that generalizes convex hull for given finite set of points. Alpha shapes are also subgraph of delaunay triangulation. Alpha parameter that identifies level of detail is used for generalization operation. This approach has been firstly emerged by Edelsbrunner, Kirkpatrick and Seidel [3]. Given a set S of n points in the plane, the α-hull of S is the intersection of all closed disks with radius 1/α that contain all the points of S [3]. For α-hull method, there is not any point in all disks, points locate on only boundary lines of disks. α values are inversely proportional to level of detail. If α value is decreased then level of detail increases or vice versa.
In the scope of this study, juxtapleural lung nodule candidate regions are detected by using α-hull method based on generalizing of convex hull. Code example and algorithm used in this study for α-hull method can be accessed from [38]. Therefore, radius values that change between 45 units and 80 units with an interval of five units are separately tried on images of left and right lungs in different slice images and nodule candidate regions are extracted. The best result of radius values for every lung CT is identified as parameter. In order to select radius values, coverage of nodule boundaries of lines occurred as a result of α-hull method is taken into account.
Nodule candidate regions are defined as a region between lines obtained as a result of α-hull method and boundaries obtained as a result of lung segmentation. New boundary points that contact with boundary of segmented lung image are accepted as starting and finishing point of nodule candidate region. Due to the fact that locations between these boundary points must be integer, Bresenham line drawing algorithm has been carried out. So as to discard small regions, minimum distance between starting and finishing point of nodule candidates is set to 10 units.
In the developed system, only one real lung nodule is available among lung nodule candidate regions for each slice. So as to identify and enumerate real lung nodule as a true positive, central point of every nodule candidate region and ground truth region is computed. One region having minimum Euclidean distance between central point of the nodule candidate region and ground truth region is defined as a real nodule and it is enumerated by red color on image. An example of this process is shown in Fig. 7.
Feature extraction
Feature extraction stage has been carried out for reduction of false positives by identifying nodule candidates that do not contain ground truth region from convex nodule candidate regions obtained by using α-hull method in the previous step. Threshold values on features are identified by means of classifiers. In the scope of this project, 40 features based on shape information, texture information, both texture and shape information are extracted from nodule candidates (Fig. 8). Total seven novel features including five of them are based on shape information, two of them are based on both shape and texture information are extracted and presented to the literature. Some portion of these 40 features is available in [19, 20]. Information about these features is provided on following subsections.
Shape based features
Shape based features are geometrical features that color and texture information are not considered. There are 16 features based on shape information in this study. These shape based features are span, depth, boundary length, depth over span, radius, area, circularity ratio, eccentricity, solidity, convex area, extent, bound over span, major axis over boundary, minor axis over boundary, span over major axis and Mj_MnAxis over boundary.
The span of concave lung nodule candidate is the length of the segment that joins its extremal points, the depth is the greatest perpendicular distance between the boundary points of concave nodule candidate and the segment defining the span, the boundary length is the number of boundary points composing concave nodule candidate, depth over span is equal to depth / span, radius is the mean distance of boundary points from the centroid of concave nodule candidate, the area of region candidate is defined by the closed boundary formed from the concavity boundary points and span segment connecting the first and last boundary points, circularity ratio is the ratio of area of concavity region candidate and that of the circle having the same perimeter [19, 20], eccentricity is the aspect ratio of concavity region, solidity is the ratio of the area of nodule region candidate (As) to the convex hull area of the shape (Ahull) [39], convex area is the total number of pixels in the convex hull of the shape [40], extent is the ratio of the number of pixels in the lung nodule candidate to pixels in the minimum bounding box of the shape [40]. Representation of lung nodule candidate and some shape based features are illustrated in Fig. 9.
Texture based features
Texture information consists of color, brightness, shape and size information about the surface of image. First order statistical features considering only original tissue density values and second order statistical features considering neighbor tissue density pair with distance d and relative positions of gray levels are extracted in this study. Besides, total number of texture based features is 22.
There are 15 first order statistical features including arithmetic, geometric and harmonic mean, standard deviation, skewness, kurtosis, entropy, interquartile range, seven Hu geometric moments.
Gray level co-occurrence matrix (Gray Level Spatial Dependence Matrix) (GLCM) is used for second order statistical features. Then, gray level nodule candidate regions are scaled into eight different texture levels. While GLCM is independent from translation operation, it is sensitive to rotation operation. So as to eliminate this problem, GLCMs with distance one in four different directions are constructed according to 0, 45, 90 and 135°. Then, the sum of these matrices is computed. As a result of this operation, GLCM is independent from rotation operation.
After obtaining GLCM stage, second order statistical features are extracted by normalizing GLCM so that sum of the matrix elements is equal to 1. There are seven GLCM features including autocorrelation, contrast, cluster prominence, cluster shade, energy, entropy and homogeneity.
Novel features proposed to the literature
In this study, seven novel features extracted from lung nodule candidates are proposed to the literature. Five of these features are based on shape information and two of them are based on shape and texture information of lung nodule candidates. These features are explained as follows:
-
Shape Based Novel Features
-
Bound over Span: It is the ratio of the value of boundary length to the value of span. This feature is closely related with nodule candidate size and noise. Boundary length provides identification of lung nodule caused by noise by setting minimum threshold and span is based on nodule candidate size [19]. It is denoted as:
$$ \mathrm{B}\mathrm{o}\mathrm{S}=\frac{\mathrm{BL}}{\mathrm{S}} $$(1)where BoS is value of bound over span, BL is boundary length and S is the span of nodule candidate.
-
Major Axis over Boundary: It is the ratio of the length of major axis of nodule candidate (Dmax) to boundary length. Major axis length is longest axis length of the nodule candidate. This feature is closely related with nodule candidate size and noise. It is denoted as:
$$ \mathrm{MjoB}=\frac{{\mathrm{D}}_{\max }}{\mathrm{BL}} $$(2)where MjoB is value of major axis over boundary, Dmax is major axis length and BL is boundary length of nodule candidate.
-
Minor Axis over Boundary: It is the ratio of the length of minor axis of nodule candidate (Dmin) to boundary length. Minor axis is shortest axis of the nodule candidate. This feature is closely related with nodule candidate size and noise. It is denoted as:
$$ \mathrm{MnoB}=\frac{{\mathrm{D}}_{\min }}{\mathrm{BL}} $$(3)where MnoB is value of minor axis over boundary, Dmin is minor axis length and BL is boundary length of nodule candidate.
-
Span over Major Axis: It is the ratio of the value of span to length of major axis. It is denoted as:
$$ \mathrm{SoMj}=\frac{\mathrm{S}}{{\mathrm{D}}_{\max }} $$(4)where SoMj is value of span over major axis, S is the span and Dmax is major axis length of nodule candidate.
-
Mj_MnAxis over Boundary: It is the ratio of the product of length of major axis and minor axis to boundary length. It is denoted as:
$$ \mathrm{MjMnoB}=\frac{{\mathrm{D}}_{\max}\times {\mathrm{D}}_{\min }}{\mathrm{BL}} $$(5)where MjMnoB is value of Mj_MnAxis axis over boundary, Dmax is major axis length, Dmin is minor axis length and BL is boundary length of nodule candidate.
-
-
Shape and Texture Based Novel Features
-
Span over Variance: It is the ratio of span to variance of tissue density of nodule region candidate. It is denoted as:
$$ \mathrm{S}\mathrm{o}\mathrm{V}=\frac{\mathrm{S}}{{\mathrm{Var}}_{\mathrm{ROI}}} $$(6)where SoV is value of span over variance, S is the span and VarROI is variance of tissue density of nodule candidate.
-
F1: It is ratio of the product of major axis length of nodule region candidate and minor axis length of nodule region candidate to variance of tissue density of shape. It is denoted as:
$$ \mathrm{F}1=\frac{{\mathrm{D}}_{\max}\times {\mathrm{D}}_{\min }}{{\mathrm{Var}}_{\mathrm{ROI}}} $$(7)where Dmax is major axis length, Dmin is minor axis length and VarROI is variance of tissue density of nodule candidate.
-
Feature Selection
This stage is carried out for selecting features having high level distinctive information and discarding features having low level information in extracted 40 features. Feature selection is important to increase prediction performance and improve generalization ability of classifier by reducing data size in classifier design.
Filter method that ranks features independent of the choice of classifiers has been utilized in this study. Statistical t test, Kullback—Leibler Divergence (Relative Entropy), Bhattacharyya Distance, Wilcoxon Test and ROC (Receiver Operating Characteristic) value have been used as feature ranking criteria [41, 42]. The number of features has been identified by evaluating classifier performance on top five classifiers. Every classifier has been run 10 times and mean of results has been computed. The number of features is based on area of the best classifier under the ROC curve (AUC) (Area Under Curve) according to every feature selection criteria. When success rate decreases and there is not significant degree of improvement, increasing of feature number is stopped and it is identified as a terminating condition. As a result of feature selection stage, GLMR is identified as a classifier, ROC is identified as a feature selection criterion and the number of features is 22 for classification operation.
When feature selection stage is applied, 22 features are obtained. These features are DepthOverSpan, MjoB, M2, harmonic mean, eccentricity, geometric mean, BoS, MnoB, M1, Circularity, Mean, M3, M4, autocorrelation, depth, homogeneity, extent, entropy, skewness, F1, MjMnoB, Convex Area, respectively.
In addition to this stage, importance degree of features is determined by measuring correlation with each other and target classes. It is expected that features must have high correlation with output values of target class and must have low correlation among them. Pair linear relationship between feature values is computed with Pearson correlation coefficient used frequently in statistics. Heat Map data visualization tool has been used for representation of matrix values. It is shown in Fig. 10.
Classification
This stage is nodule recognition stage that identifies whether region of interest is a real nodule or not by means of classifiers using extracted features from lung nodule candidates. In the scope of this project, performance comparison has been made on 10 different classifiers according to AUC values and GLMR has been identified as the best classifier in the developed system. Experimental results have been implemented for top of five classifiers. These 10 classifiers consist of linear discriminant classifier (LDC) [43], quadratic discriminant classifier (QDC) [44], k nearest neighbor classifier (K-NN) [45], multi-layer perceptron (MLP) [46], probabilistic neural network (PNN) [47], support vector machines (SVM) [48], classification tree (CT) [49], Naïve Bayes Classifier (NBC) [50], generalized linear model regression (GLMR) [51] and ensemble classifiers (ECT) [52]. As a result of the trials, k value is identified as one in K-NN, spread parameter is taken as 0.02 in PNN, the number of neurons of hidden layer in MLP is assigned to 5, the ratio of training data to test data is identified as 80–20 or 4, classification trees is made 20 learning cycle by bagging method in ensemble classifiers. Binomial distribution is used for GLMR. For NBC and CT classifier, default parameter values in Matlab are used. Dot product kernel function is used for SVM. GLMR, QDC, LDC, K-NN and PNN are top five classifiers in the developed system.
Besides, k fold cross validation technique is used for the selection of classification model. k value is identified as 10 for balancing variance and error value. The ratio of training data to test data is identified as 90–10 or 9 in the developed system. There are 1269 training data and 141 test data. In each of 10 runs, 127 of 1269 data are used as a validation data and other parts of 1269 data are used as a training data by using 10 fold cross validation. Real nodule depends on the fact that lung nodule candidate comprises of ground truth region. If nodule candidate is a real nodule, the output value will become 1, otherwise it will become 0. Additionally, GLMR classifier has been evaluated by using AUC and classification accuracy on test data according to different feature selection criteria and the number of features.
Results
Using of all data and software has been implemented on Matlab R2011a environment and notebook with Intel i7-2670 QM 2.2 Ghz processor in this study. In the scope of this project, LIDC dataset containing only slice images with juxtapleural lung nodule is used. Every CT image can contain many slices. Besides, every slice image has only one nodule. Information about the dataset is illustrated in Table 2.
The suitability of performance criteria used in classification depends on features of training data. The number of real nodule region is 124 of 1269 regions in training data are real nodule region. The number of non-nodule region is 1145. Due to the fact that class skewness of training set (the ratio of the number of nodule regions to the number of non-nodule regions) is not equal to 1, AUC (Area Under the Curve) that is not sensitive to skewness is used in this study. Additionally, classification accuracy is also used for measuring overall performance of system.
Computation time of the proposed system is shown in Table 3. Nodule detection stage consists of preprocessing, lung segmentation, detection of nodule candidates and feature extraction. Classification stage consists of feature selection and classification. Computation time is computed for data set (design and test set) and only one nodule candidate.
In the next subsection, experimental results of nodule recognition without derived seven novel features based on 33 features on design set is presented. After that, effect of derived novel features on design set is shown. Next, nodule recognition system and effect of derived features using test set is presented. Finally, effect of feature selection stage on design set is shown.
Nodule Recognition (without derived features)
Experimental results based on total 33 features which do not contain seven novel features that we developed, ROC feature selection criterion and 22 selected features are shown in Table 4. The best result has been obtained on GLMR classifier with 0.9640 AUC value for 33 features. In this case, the newly formed five features are boundary length, area, radius, contrast (GLCM), cshade (GLCM), respectively. Novel seven features in feature extraction stage have increased performance of the nodule recognition system by 0.41 % according to AUC and by 0.25 % according to classification accuracy performance criteria for GLMR classifier.
Nodule Recognition System and Effect of Derived Features
When feature selection stage is applied and ROC feature selection criteria is used, experimental results based on 40 features for 10 classifiers are illustrated in Table 5 and Fig. 11. The best result is obtained with GLMR classifier and %95.88 classification accuracy. As a result of these experimental results, it is shown that feature selection stage has increased the success rate of nodule recognition and classification stage and has decreased training time of classifiers. Additionally, five of seven novel features presented literature including MjoB, BoS, MnoB, MjMnoB, F1 have entered into first 22 features of system according to ROC feature selection criterion and have increased nodule recognition performance of the system.
Nodule Recognition System and Effect of Derived Features Using Test Set
GLMR is selected as best classifier using ROC performance criterion and training set. So as to determine generalization performance of selected model on the new data, experimental results are applied on the test data. When feature selection stage is applied and seven novel features presented the system, experimental results are illustrated in Table 6. When derived features are not used in the system, experimental results based on 33 features are illustrated in Table 7. Feature selection stage uses only 22 features and five of seven novel features including MjoB, BoS, MnoB, MjMnoB, F1 have entered to system. Novel features have increased performance of the nodule recognition system by 0.45 % according to AUC value for test set (Fig. 12).
The Effect of Feature Selection Stage
When feature selection stage is not applied in the system, experimental results based on 40 features for top five classifiers are illustrated in Table 8. While feature selection stage is done, 0.9679 mean AUC value is obtained with 22 different features, 0.9551 mean AUC value is obtained when it is not applied. In both cases, the best result has been provided with GLMR classifier. As a result of feature selection stage, other three classifiers except K-NN and PNN classifiers have performance improvement with AUC value. Besides, classification performance has been measured by increasing the number of features from 1 to 25 on GLMR classifier step by step. This process is repeated for each feature selection criterion and other four classifiers.
Conclusion
Computer aided detection of lung cancer with highest mortality rate is so important in terms of increasing of the survival rate of patients and providing early detection and diagnosis. In this study, automatic juxtapleural lung nodule region detection method and related software is developed. α-hull method is used for detection of nodule candidates. Total seven novel features based on shape, texture information are presented as a new method to the literature. Besides, ranking and selection of all features using statistical criteria made a contribution in terms of performance improvement. It is shown that feature selection stage increased the overall performance of the system by increasing AUC value from 0.9551 to 0.9679. Feature extraction and classification stages are important for reduction of false positives.
In the future studies, the scope of this study may be extended by developing the system which considers other types of lung nodules. Parameter tuning of all classifiers is out of scope of this paper and may be considered as another study. Additionally, the comparison of methods used with alternatives in literature can be made by applying different techniques and approaches on developed system for every stage.
References
World Health Organization, World Health Report. http://www.who.int/whr/2004/en/. Accessed 12 Feb 2014, 2004.
Lee, S. L. A., Kouzani, A. Z., and Hu, E. J., Automated detection of lung nodules in computed tomography images: A review. Mach. Vis. Appl. 23(1):151–163, 2012. doi:10.1007/s00138-010-0271-2.
Edelsbrunner, H., Kirkpatrick, D. G., and Seidel, R., On the shape of a set of points in the plane. IEEE Trans. Inf. Theory 29(4):551–559, 1983.
Wang, Q., Zhu, W., and Wang, B., Three-dimensional SVM with latent variable: Application for detection of lung lesions in CT images. J. Med. Syst. 39(1):171, 2015. doi:10.1007/s10916-014-0171-5.
Avci, E., A new expert system for diagnosis of lung cancer: GDA—LS_SVM. J. Med. Syst. 36(3):2005–2009, 2012. doi:10.1007/s10916-011-9660-y.
Armato, S. G., 3rd, McLennan, G., Bidaut, L., McNitt-Gray, M. F., Meyer, C. R., Reeves, A. P., Zhao, B., Aberle, D. R., Henschke, C. I., Hoffman, E. A., Kazerooni, E. A., MacMahon, H., van Beek, E. J. R., Yankelevitz, D., et al., The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys. 38(2):915–931, 2011.
Cornell University, Public lung database to address drug response. Vision and Image Analysis Group (VIA) and International Early Lung Cancer Action Program (I-ELCAP) Labs. http://www.via.cornell.edu/crpf.html, Accessed 18 Feb 2014, 2008
Ezoe, T., Takizawa, H., Yamamoto, S., Shimuzu, A., Matsumoto, T., Tateno, Y., Iimura, T., Matsumoto, M., An automatic detection method of lung cancers including ground glass opacities from chest X-ray CT images. In: Proc. of SPIE 4684:1672–1680, 2002.
Frangi, A. F., Niessen, W. J., Hoogeveen, R. M., Walsum, T. V., and Viergever, M. A., Model-based quantitation of 3-D magnetic resonance angiographic images. IEEE Trans. Med. Imaging 18(10):946–956, 1999.
Suzuki, K., Supervised “lesion-enhancement” filter by use of a Massive-Training Artificial Neural Network (MTANN) in Computer-Aided Diagnosis (CAD). Phys. Med. Biol. 54(18):31–45, 2009.
Ochs, R. A., Goldin, J. G., Abtin, F., Kim, H. J., Brown, K., Batra, P., Roback, D., McNitt-Gray, M. F., and Brown, M. S., Automated classification of lung bronchovascular anatomy in CT using Adaboost. Med. Image Anal. 11(3):315–324, 2007.
Paik, D. S., Beaulieu, C. F., Rubin, G. D., Acar, B., Jeffrey, R. B., Yee, J., Dey, J., and Napel, S., Surface normal overlap: A computer-aided detection algorithm with application. IEEE Trans. Med. Imaging 23(6):661–675, 2004.
Retico, A., Delogu, P., Fantacci, M. E., Gori, I., and Martinez, A. P., Lung nodule detection in low-dose and thin-slice computed tomography. Comput. Biol. Biomed. 38(4):525–534, 2008.
Li, Q., Li, F., and Doi, K., Computerized detection of lung nodules in thin-section CT images by use of selective enhancement filters and an automated rule-based classifier. Acad. Radiol. 15(2):165–175, 2008.
Armato, S. G., 3rd, Giger, M. I., Moran, C. J., Blackburn, J. T., Doi, K., and Macmahon, H., Computerized detection of pulmonary nodules on CT scans. Radiographics 19(5):1303–1311, 1999.
Hu, S., Hoffman, E. A., and Reinhardt, J. M., Automatic lung segmentation for accurate quantitation of volumetric X-ray CT images. IEEE Trans. Med. Imaging 20(6):490–498, 2001.
El-Baz, A., Beache, G. M., Gimel’farb, G., Suzuki, K., Okada, K., Elnakib, A., Soliman, A., Abdollahi, B., Computer-aided diagnosis systems for lung cancer: challenges and methodologies. Int. J. Biomed. Imaging Article ID 942353, 46 pages, 2013
Pu, J., Roos, J., Yi, C. A., Napel, S., Rubin, G. D., and Paik, D. S., Adaptive border marching algorithm: Automatic lung segmentation on chest CT images. Comput. Med. Imaging Graph. 32(6):452–462, 2008.
Sensakovic, W. F., Starkey, A., Armato, S. G. 3rd, A general method for the identification and repair of concavities in segmented medical images. IEEE Nuclear Science Symposium Conference Record 5320–5326, 2008
Nunzio, G. De, Massafra, A., Cataldo, R., Mitri I. De, Peccarisi, M., Fantacci, M. E., Gargano, G., Torres, E. L., Approaches to juxta-pleural nodule detection in CT images within the MAGIC-5 Collaboration. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 648(1):103–106, 2011.
Lee, S. L. A., Kouzani, A. Z., and Hu, E. J., Random forest based lung nodule classification aided by clustering. Comput. Med. Imaging Graph. 34(7):535–542, 2010.
Armato, S. G., 3rd, Giger, M. L., and MacMahon, H., Automated detection of lung nodules in CT scans: Preliminary results. Med. Phys. 28(8):1552–1561, 2001.
Gurcan, M,, Sahiner, B,, Petrick, N,, Chan, H, P., Kazerooni, E. A., Cascade, P. N., Hadjiiski, L., Lung nodule detection on thoracic computed tomography images: preliminary evaluation of a computer-aided diagnosis system. Med. Phys. 29(11):2552–2558.
Suzuki, K., Armato, S. G., 3rd, Li, F., Sone, S., and Doi, K., Massive training artificial neural network (MTANN) for reduction of false positives in computerized detection of lung nodules in low-dose computed tomography. Med. Phys. 30(7):1602–1617, 2003.
Awai, K., Murao, K., Ozawa, A., Komi, M., Hayakawa, H., Hori, S., and Nishimura, Y., Pulmonary nodules at chest CT: Effect of computer-aided diagnosis on radiologists’ detection performance. Radiology 230(2):347–352, 2004.
Lee, Y., Hara, T., Fujita, H., Itoh, S., and Ishigaki, T., Automated detection of pulmonary nodules in helical CT images based on an improved template-matching technique. IEEE Trans. Med. Imaging 20(7):595–604, 2001.
Farag, A., El-Baz, A., Gimel’farb, G. G., Falk, R., Hushek, S. G. Automatic detection and recognition of lung abnormalities in helical CT images using deformable templates. Lecture Notes in Computer Science, Springer-Verlag, Medical Image Computing and Computer-Assisted Intervention 3217:856–864, 2004.
Ge, Z. Y., Sahiner, B., Chan, H. P., Hadjiiski, L. M., Cascade, P. N., Bogot, N., Kazerooni, E. A., Wei, J., and Zhou, C., Computer-aided detection of lung nodules: False positive reduction using a 3D gradient field method and 3D ellipsoid fitting. Med. Phys. 32(8):2443–2454, 2005.
Brown, M. S., McNitt-Cray, M. F., Golldin, J. G., Suh, R. D., Sayre, J. W., and Aberle, D. R., Patient-specific models for lung nodule detection and surveillance in CT images. IEEE Trans. Med. Imaging 20(12):1242–1250, 2001.
Ye, X., Lin, X., Dehmeshki, J., Slabaugh, G., and Beddoe, G., Shape-based computer-aided detection of lung nodules in thoracic CT images. IEEE Trans.Biomed. Eng. 56(7):1810–1820, 2009.
Bağci, U., Bray, M., Caban, J., Yao, J., and Mollura, D. J., Computer-assisted detection of infectious lung diseases: A review. Comput. Med. Imaging Graph. 36(1):72–84, 2012.
Choi, W. J., and Choi, T. S., Automated pulmonary nodule detection based on three-dimensional shape-based feature descriptor. Comput. Methods Prog. Biomed. 113(1):37–54, 2014.
Ozekes, S., and Osman, O., Computerized lung nodule detection using 3D feature extraction and learning based algorithms. J. Med. Syst. 34(2):185–194, 2010. doi:10.1007/s10916-008-9230-0.
Kuruvilla, J., and Gunavathi, K., Lung cancer classification using neural networks for CT images. Comput. Methods Prog. Biomed. 113(1):202–209, 2014.
Daliri, M. R., A hybrid automatic system for the diagnosis of lung cancer based on genetic algorithm and fuzzy extreme learning machines. J. Med. Syst. 36(2):1001–1005, 2012. doi:10.1007/s10916-011-9806-y.
Cancer Imaging Archive (2014) LIDC-IDRI. https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI. Accessed 9 Oct 2014.
Otsu, N., A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9(1):62–66, 1979.
Lundrgen J. Alpha Shapes http://www.mathworks.com/matlabcentral/fileexchange/28851-alpha-shapes/content/alphavol.m. Accessed 12 Feb 2014, 2010
Mingqiang, Y., Kidiyo, K., Joseph, R. A survey of shape feature extraction techniques. Pattern Recognition Techniques, Technology and Applications 43–90, 2008
Math Works Inc, Matlab R2011a documentation. http://www.mathworks.com/help/index.html. Accessed 12 Feb 2014, 2014
Theodoridis S., Koutroumbas K. (1999) Pattern recognition. Academic Press.
Liu, H., and Motoda, H., Feature selection for knowledge discovery and data mining. Kluwer Academic Publishers, Boston, 1998.
Fisher, R. A., The use of multiple measurements in taxonomic problems. Ann. Eugen. 7(2):179–188, 1936. doi:10.1111/j.1469-1809.1936.tb02137.x.
Cover, T. M., Geometrical and statistical properties of systems of linear inequalities with applications in pattern recognition. IEEE Trans. Electron. Comput. EC-14(3):326–334, 1965. doi:10.1109/pgec.1965.264137.
Cover, T. M., and Hart, P. E., Nearest neighbor pattern classification. IEEE Trans. Inf. Theory IT-13(1):21–27, 1967. doi:10.1109/TIT.1967.1053964.
Rumelhart D. E., Geoffrey, E. H., Williams, R. J., Learning internal representations by error propagation. Parallel distributed processing: explorations in the microstructure of cognition 1:318–362. MIT Press, Cambridge, 1986
Specht, D. F., Probabilistic neural networks. Neural Netw. 3(1):109–118, 1990. doi:10.1016/0893-6080(90)90049-Q.
Vapnik, V., Estimation of dependences based on empirical data. Springer Verlag, New York, 1982.
Breiman, L., Friedman, J. H., Olshen, R. A., and Stone, C. J., Classification and regression trees. CRC Press LLC, Florida, 1984.
Good, I. J., Probability and the weighing of evidence. Charles Griffin, London, 1950.
Dobson, A. J., An introduction to generalized linear models. Chapman & Hall, New York, 1990.
Breiman, L., Bagging predictors. Mach. Learn. 24(3):123–140, 1996. doi:10.1023/A:1018054314350.
Acknowledgments
This study has been supported by Scientific and Technological Research Council of Turkey (TÜBİTAK) 2211 National Graduate Scholarship Program.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Systems-Level Quality Improvement.
Rights and permissions
About this article
Cite this article
Taşcı, E., Uğur, A. Shape and Texture Based Novel Features for Automated Juxtapleural Nodule Detection in Lung CTs. J Med Syst 39, 46 (2015). https://doi.org/10.1007/s10916-015-0231-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-015-0231-5