Abstract
In this paper, we introduce a Python library called mad-GP that enables users to more easily explore the design space of Gaussian process (GP) surrogate models for modeling potential energy surfaces (PESs). A user of mad-GP only needs to write down the functional form of the prior mean function (i.e., a prior guess for the PES) and kernel function (i.e., a constraint on the class of PESs), and the library handles all required derivative implementations via automatic differentiation (AD). We validate the design of mad-GP by applying it to perform geometry optimization of small molecules. In particular, we test the effectiveness of fitting GP surrogates to energies and/or forces, and perform a preliminary study on the use of non-constant priors and hierarchical kernels in GP PES surrogates. We find that GPs that fit forces perform comparably with GPs that fit both energies and forces, although force-only GPs are more robust for optimization because they do not require an additional step to be applied during optimization. We also confirm that constant mean functions and Matérn kernels work well as reported in the literature, although our tests also identify several other promising candidates (e.g., Coulomb matrices with three-times differentiable Matérn kernels). Our tests validate that AD is a viable method for performing geometry optimization with GP surrogate models on small molecules.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Potential energy surfaces (PESs) are fundamental to theoretical chemistry. The PES of a system of \(N_{\text {A}}\) atoms takes the form of a \((3N_{\text {A}}- 6)\)-dimensional hypersurface and is the solution to the multi-electron Schrödinger equation under a fixed-nuclei approximation. Crucially, one can study the chemical properties of a system (e.g., thermodynamics, activation energies, reaction mechanisms, and reaction rates) by identifying the local extrema on its PES, a process known as geometry optimization. In particular, local minima correspond to reactants, products, and reactive intermediates, while first-order saddle points correspond to transition-state structures.
There is a rich line of work on developing electronic-structure methods (e.g., density-functional theory) that approximately evaluate the electronic-structure energy of a system for a given geometry (i.e., positions of nuclei). These methods can be applied to perform, e.g., single-point energy (SPE) calculations to evaluate the electronic-structure energy of a given point on a PES. However, calculating a system’s energy at even a single geometry with relatively high accuracy (i.e., to an error under 1 kcal/mol) can be computationally expensive. In particular, the computational cost of high-level electronic-structure methods scales unfavorably with respect to system size (i.e., number of basis functions). Consequently, it is often best practice to keep to a minimum the number of SPE calculations required for convergence in geometry optimization. (Alternatively, one can attempt to accelerate the SPE computation itself.)
One promising approach for managing the number of SPE calculations is to use a surrogate for a PES with the following properties: (1) it is computationally inexpensive to evaluate the surrogate at any point, and (2) the surrogate can incorporate information from an electronic-structure calculation (e.g., energy and force) to refine its approximation of the PES. The first property allows for rapid but potentially inaccurate evaluations, while the second enables us to refine the surrogate’s accuracy in a controlled manner by determining when and where to pay the cost of an electronic-structure calculation.
Researchers have demonstrated that Gaussian processes (GPs) form an effective surrogate for representing the PESs of molecular systems [1,2,3,4,5], and can reduce the number of SPE calculations required for geometry optimization (e.g., finding local extrema and minimum-energy reaction paths [6,7,8,9]). One particularly favorable property of GPs for modeling PESs is that they can encode both the PES and its gradient in a consistent manner. As a result, GP surrogates naturally express the physical relationship between energy and forces, and can exploit electronic-structure methods providing information on both.
The encouraging results on GPs as PES surrogates (e.g., see [1, 2, 5, 10]) warrant further study, especially since GPs form a large and flexible class of models parameterized by prior mean and kernel functions. The prior mean function for a surrogate GP encodes prior beliefs about the functional form of a PES (e.g., the asymptotic behavior of stretching a bond from equilibrium length to the dissociation limit) while the kernel function constrains its shape (e.g., it should be “smooth”).
A primary barrier to exploring the GP design space, particularly when incorporating force information, is that the user must manually implement the first and second-order derivatives for a GP’s mean and kernel functions. Prior work [6, 7, 11, 12] has largely examined GPs with mean and kernel functions that have simple derivatives (e.g., constant mean functions with constant 0 derivative or Matérn kernels with well-known derivatives). As researchers begin to explore more complex mean and kernel functions that model more of the chemistry and physics, the burden of deriving and implementing derivatives will only increase.
In this paper, we introduce a Python library called mad-GP—molecular/material and autodiff GPs—that enables researchers to more systematically explore GPs as PES surrogates (Sect. 2). Users of mad-GP need only specify the functional form of the prior mean and kernel functions parameterizing a GP as Python code, and the library implements the first and second-order derivatives. As proof of concept, we have written non-constant mean functions (e.g., Leonard-Jones potential) and kernel functions based on chemical descriptors (e.g., Coulomb matrices [13], Smooth Overlap of Atomic Positions (SOAP) [14]) in mad-GP.
mad-GP handles differentiation of mean and kernel functions by applying automatic differentiation (AD, or autodiff), a technique from computer science which has played a significant role in the rise of deep learning. The hope is that researchers, unencumbered from the task of manually implementing derivatives, will be able to experiment with functions that better capture the chemical structure (e.g., by design or by learning from data).
We apply mad-GP to the task of geometry optimization for small molecules to demonstrate its use (Sect. 3). Notably, mad-GP enables users to fit GP surrogates to energies and/or forces. We use this functionality to compare the efficacy of two popular approaches to GP surrogates for geometry optimization: (1) those that fit energies and forces (e.g., [6, 7, 11, 12]) and (2) those that fit forces exclusively (e.g., [3, 10, 15]). We also evaluate a third approach, namely, the energy-only approach. To the best of our knowledge, there has been relatively little work comparing these approaches. We show that both the first two approaches are comparable at reducing the number of SPE calculations, but that force-only GPs are more robust for optimization because they do not require an additional step (i.e., set the mean function to a maximum energy) to be applied during optimization. We also perform a preliminary study on the use of non-constant priors and hierarchical kernels in GP PES surrogates. We confirm that constant mean functions and Matérn kernels work well as reported in the literature, although our studies also identify several other promising combinations (e.g., Coulomb matrices with three-times differentiable Matérn kernels). Our tests validate that AD is a viable method for performing geometry optimization with GP surrogate models on small molecules.
2 Methods
In this section, we review PESs and PES surrogate models (2.1). In the pursuit of self-containment, we also give a brief introduction of Gaussian processes (Sect. 2.2); likewise, we later discuss AD, which is central to the implementation of mad-GP (Sect. 2.4).
2.1 Models of potential energy surfaces
PESs are complex hypersurfaces and there is a rich line of work on characterizing their properties and their connections to chemistry (e.g., see [16] for an earlier account). Notably, Fukui et al. [17] introduces the formal concept of an intrinsic reaction coordinate for a PES which enables us to trace the “path" of a chemical reaction on a PES between products and/or reactants through transition states (e.g., see [18, 19]), and consequently, study chemical reactions (e.g., see [20,21,22,23,24,25] for PES models used to study chemical reactions). Mezey in a series of works [26,27,28,29,30,31,32,33,34,35] studies PESs from a topological perspective. One important result shows that a PES can be formally partitioned into catachment regions (i.e., basins of attractions around local minima), thus justifying the view that PESs do indeed encode the information necessary for studying chemical reactions (i.e., reactants, products, transition states). Crucially, the topological perspective enables a description of a PES in a coordinate-free manner, which provides insight into the design of computational representations of PESs that are invariant to physical symmetries (e.g., by working with a unique representation of an equivalence class given by a quotient space).
The mathematical analysis of PESs inspires computational representations of PESs for modeling and simulation purposes (e.g., see the software POTLIB [36]), including many-mode representations [37,38,39,40,41,42,43] as well as sum-of-product representations [44,45,46,47]. Recently, many ML models have been explored as candidates for modeling PESs. ML models take a statistical point-of-view and attempt to model the PES directly from data in line with earlier work that uses simpler models (e.g., see [48,49,50]). These models thus have to pay more attention to capturing important physical properties of PESs with the hope of having better computational scaling potential. These ML models include neural networks [51,52,53,54,55,56,57,58,59,60,61,62], kernel methods [1,2,3,4], and Gaussian processes [6, 7, 11, 12]. Some ML models are designed specifically for modeling PESs (e.g., Gaussian Approximation Potentials [1, 2], Behler-Parrinello Neural Networks [55,56,57], and q-Spectral Neighbor Analysis Potentials [63]).
The choice of PES representation is informed by the task at hand. Neural network surrogates have demonstrated success in regression settings where we would like to directly predict a quantity (e.g., energy, force, dipole moments) from a geometry. However, it is expensive to train a neural network and so their use case in geometry optimization is limited. GP surrogates, on the other hand, have demonstrated success in geometry optimization because they are trainable in an online manner and also provide a closed form solution that is searchable by gradient descent (Sect. 2.2). Moreover, GPs can also express conservation of energy. However, GPs are hard to train when the number of examples is large (cubic scaling in the number of examples).
2.2 Gaussian processes
A Gaussian process (GP) defines a probability distribution on a class of real-valued functions as specified by a mean function \(\mu : \mathbb {R}^D \rightarrow \mathbb {R}^K\) and a (symmetric and positive definite) kernel function \(k: \mathbb {R}^D \times \mathbb {R}^D \rightarrow \mathbb {R}^{(K \times K)}\). The notation
indicates that \(f: \mathbb {R}^D \rightarrow \mathbb {R}^K\) is a function drawn from a GP with mean \(\mu\) and kernel k. The connection with multivariate Gaussian distributions is that
where the notation \(f(\mathbf {X})\) is shorthand for
for \(\varvec{x}_1, \dots , \varvec{x}_M \in \mathbb {R}^D\) (similarly, for \(\mu (\mathbf {X})\)) and the covariance matrix \(K(\mathbf {X}, \mathbf {Y})\) is defined as
with \(\mathbf {X}^{(D \times M)} = \begin{pmatrix} \varvec{x}_1&\hdots&\varvec{x}_M \end{pmatrix}\) (similarly, \(\mathbf {Y}^{(D \times L)} = \begin{pmatrix} \varvec{y}_1&\hdots&\varvec{y}_L \end{pmatrix}\)).
We will review two properties of GPs that make them useful as PES surrogates: (1) GP surrogates support gradient-based optimization which can be used in application such as geometry optimization (Sect. 2.2.1) and (2) GP surrogates support fitting force information (Sect. 2.2.3). Before we do this, we start by highlighting aspects of GPs that are usually left abstract from a mathematical point-of-view that are important for their application to PESs. For more background on GPs, we refer the reader to standard references (e.g., see Williams et al. [64]).
2.2.1 Kernels for atomistic systems
As a reminder, GPs are parameterized by a kernel function \(k: \mathbb {R}^D \times \mathbb {R}^D \rightarrow \mathbb {R}^{(K \times K)}\). Intuitively, the kernel function can be thought of as a measure of similarity between two vectors \(\varvec{x}, \varvec{y}\in \mathbb {R}^D\). For the use case of modeling PESs, the vectors \(\varvec{x}\) and \(\varvec{y}\) are then vector encodings of two molecular structures \(m_{\varvec{x}}\) and \(m_{\varvec{y}}\). A molecular structure \(m_{\varvec{x}}\) describes a molecule with \(N_{\text {A}}\) atoms by giving (1) the atomic nuclear charges \((Z_1 \, \dots \, Z_{N_{\text {A}}})\), (2) the masses \((W_1 \, \dots \, W_{N_{\text {A}}})\), and nuclear positions \((x^1 \dots x^{3N_{\text {A}}})^\text{T}\) in Cartesian (xyz) coordinates. Consequently, we will need to convert molecular structures into vectors so that kernel functions can operate on them.
For example, if molecular structures are described in Cartesian coordinates, then D would be \(3N_{\text {A}}\) where \(N_{\text {A}}\) is the number of atoms in the system. The atomic charges and masses would be ignored. The encoding would then be an identity function on the nuclear positions, and \(\user2{x} = (x^{1} \ldots x^{{3N_{A} }} )^\text{T} = (\user2{x}^{1} \ldots \user2{x}^{{N_{A} }} )^\text{T}\) would then correspond to a particular geometry in Cartesian coordinates represented by a column vector consisting of coordinates of all atoms. We could then use a standard GP kernel such as a (twice-differentiable, or \(p=2\)) Matérn kernel defined as
where \(\rho = \Vert \varvec{x}- \varvec{y} \Vert\), and \(\sigma _{\text {M}}\) and l are hyper-parameters, to measure the geometric similarity between two structures. In full detail, the kernel function would be
where \(\text {xyz}\) converts a molecular structure into its xyz coordinates (an identity function on the nuclear positions in this case). This kernel is the one used in many state-of-the-art works on GP surrogates (e.g., see [11]). This approach, while effective, leaves at least two directions of potential improvement.
First, the description of a molecule in terms of xyz coordinates does not respect basic physical invariances such as permutation invariance. To solve these problems, researchers have developed chemical descriptors (e.g., global descriptors such as Coulomb matrices [13] and local descriptors such as SOAP [14] that describe a molecule as a set of local environments around its atoms) that describe molecules in a way such that these physical invariances are respected. We can take advantage of these more sophisticated (and physically accurate) descriptors by defining a GP kernel
where \(d: \mathcal {M}\rightarrow \mathcal {D}\) is some descriptor, \(\mathcal {M}\) is the space of molecular structures, and \(\mathcal {D}\) is the target space of a descriptor (e.g., \(\mathcal {D}= \mathbb {R}^D\)). Different descriptors may be better for different kinds of molecules and chemical systems.
Second, once we generalize from xyz coordinates to descriptors, the Matérn kernel \(k_m\) may no longer be a sensible choice as a measure of similarity. For example, local descriptors such as SOAP descriptors produce a set of variable-sized local environments that describe a neighborhood of a system from the perspective of each atom in the system. Consequently, we may also be interested in kernels \(k_d: \mathcal {D}\times \mathcal {D}\rightarrow \mathbb {R}^{(K \times K)}\) that compare descriptors directly, resulting in a GP kernel of the form
Note that descriptors of different molecules may return encodings of different sizes (e.g., the xyz encoding of molecules with different number of atoms has different sizes), and so these kernel functions \(k_d\) are not necessarily the standard ones defined on a fixed-size vector space. For instance, in the case of local descriptors, we may need to perform structure matching (e.g., see [65]).
In short, while we can leave aspects of a GP such as the kernel function k abstract, the use case of PESs highlights a hierarchical and compositional structure. The choice of kernel depends on factors such as (1) what invariants we hope to capture (e.g., rotational and permutational invariance), (2) whether we are working with molecules or materials, and (3) do we need to compare molecules with a different number of atoms and atomic types. Constructing the appropriate kernels that capture the relevant chemistry and physics can lead to improved GP PES surrogates.
2.2.2 Geometry optimization with GPs
Previously, we saw that we could define hierarchical kernels with descriptors for PES modeling. Although this may be interesting as a theoretical exercise, it is not useful from a practical perspective if we cannot effectively compute with a GP surrogate defined in this manner. For example, in an application such as using a GP surrogate for geometry optimization, we would like to be able to extract a physical geometry out. In this section, we will see that defining hierarchical kernels interacts nicely with gradient-based surrogate optimization. Before we explain this, we will first review how to update a GP surrogate with data obtained from an electronic-structure calculation. This produces a posterior surrogate which will more closely approximate a PES.
Fitting a GP We can fit GPs to observed data, called Gaussian Process Regression (GPR), to refine the class of functions defined by a GP in a principled manner (e.g., maximum a posteriori or MAP inference) by solving a linear system of equations. In the context of modeling a PES of a molecule, the observed data would be a set of observations \(\{ (\varvec{x}_1, E_1), \dots , (\varvec{x}_N, E_N) \}\) (i.e., training set) where each \(\varvec{x}_i\) is a vector encoding of a molecule’s geometry and \(E_i\) the corresponding energy as obtained from an electronic-structure method. (When forces are available, we can additionally fit \(\varvec{F} = (\varvec{F}_1 \, \dots \, \varvec{F}_N)^\text {T}\) (Sect. 2.2.3).)
In practice, we perform GPR where we assume our observations are corrupted with some independent, normally distributed noise \(\epsilon \sim \mathcal {N}(0, \sigma ^2\varvec{I})\) where \(\varvec{I}\) is the identity matrix, which corresponds to the model
We can equivalently view this from the multivariate Gaussian perspective as using the covariance matrix
because the noise is also normally distributed. The noise \(\epsilon\) can be used to model uncertainty in a SPE calculation (generated by standard quantum chemistry computations), e.g., the energy convergence threshold used or the intrinsic error of the electronic-structure method applied.
The system of equations to solve when fitting energy information is
where \(\mathbf {X}= (\varvec{x}_1 \, \dots \, \varvec{x}_N)\) and \(\mathbf {E}= (E_1 \, \dots E_N)^\text {T}\) are observations, and \(\varvec{\alpha } \in \mathbb {R}^N\) indicates the weights to solve for. These equations can be solved with familiar algorithms such as Cholesky decomposition (cubic complexity, Scipy [66] implementation). When GP surrogates are used in applications such as geometry optimization, we can use incremental Cholesky decomposition (quadratic complexity, custom implementation following Lawson et al. [67]). Note that GPR with noise has the added benefit for increasing numerical stability of linear system solvers.
Gradient-based optimization To evaluate a GP surrogate that is fit to a training set \((\mathbf {X}, \mathbf {E})\), we compute the function
where \(f^*\) is the posterior mean (to distinguish it from the prior f) and \(\varvec{\alpha }\) are weights that are determined during the training process. The posterior mean gives the GPs prediction of the energy.
Gradient-based optimization of the posterior mean corresponds to using the gradient
to perform optimization (\(\nabla _1 k = \frac{\partial k(\varvec{x}, \varvec{y})}{\partial \varvec{x}}\)). Consequently, gradient-based optimization of a GP surrogate requires the first-order derivatives of the mean (\(\nabla \mu\)) and kernel functions (\(\nabla _1 k\)). When mean and kernel functions are sufficiently complicated, a GP user will need to implement these derivatives in order to perform gradient-based optimization of the posterior mean. This highlights the benefit of having a method that enables us to automatically obtain derivatives of mean and kernel functions.
We return now to the problem of ensuring that using hierarchical kernels does not prevent us from using it in applications such as geometry optimization. As a reminder, the issue is that if we use a descriptor d to define a GP kernel, then we will need to somehow recover physical coordinates such as xyz coordinates during geometry optimization. One approach pursued in the literature (e.g., see Meyer et al. [68]) is to use an inverse mapping \(d^{-1}\) to map back into physical coordinates. While this may exist for “simple" descriptors, it is unlikely to exist or be easily computable for more complex descriptors.
Let us define a kernel \(k_d\) with an arbitrary descriptor d as
to highlight that we start in \(\text {xyz}\) coordinates. Then the gradient of this kernel with respect to \(\text {xyz}\) coordinates can be computed by the chain-rule. Because the posterior mean can be written as a linear combination of gradients of the kernel (e.g., see (14)), an optimization procedure will thus be able to optimize directly in xyz coordinates while also factoring the comparison of molecules through a descriptor, giving us the best of both worlds. Note that this works for arbitrarily complex kernels and descriptors so long as they are differentiable. (We will see with AD that strict differentiability is not required.)
So far, there is no need for second-order derivatives. As we will see in the next section, second-order derivatives will appear when we use GPs that fit force information.
2.2.3 Models with conservative forces
GPs have the following remarkable property: if
then
when \(\nabla _1 k \nabla ^\text {T}_2 = \frac{\partial ^2}{\partial \varvec{x}\partial \varvec{y}}k(\varvec{x}, \varvec{y})\) exists. (\(\nabla _1 k = \frac{\partial k(\varvec{x}, \varvec{y})}{\partial \varvec{x}}\) and \(k \nabla _2^\text {T}= \frac{\partial k(\varvec{x}, \varvec{y})}{\partial \varvec{y}^\text {T}}\).) The consequence for PES modeling is that we can define a specific GP kernel that constrains the class of functions a GP describes to be consistent with its gradients so that the physical relationship between energy and force (i.e., the negative gradient of a PES) is encoded. In particular, the GP
with
and
accomplishes this.
Second-order derivatives appear when we leverage force information during both GPR and posterior optimization. To see the former, observe that the system of equations to solve when fitting both energies and forces is
for the weights \(\alpha _i\) and \(\varvec{\beta }_i\) where \(\sigma _e\) is a noise parameter for the energy and \(\sigma _f\) is a noise parameter for the force. The second-order derivatives appear through the kernel \(\tilde{k}\) used to set up the system of linear equations. To see the latter, observe that the posterior predictions of the energy and gradient of the energy are given by
Example of using mad-GP. This is the entire code required for the user to write in order to experiment with a constant mean function and Matérn kernel (\(k_m\), see (5)) which are used in state-of-the-art results
and
Note that the gradient of the posterior mean for the energy is given by the prediction of the posterior mean for the gradient.
Thus the second-order derivative comes into play either by differentiating the posterior mean \(f^*\) or by performing a posterior gradient prediction using \((\nabla f)^*\).
For completeness, we also give the equations for fitting forces exclusively.
We use \(\text {GP}_{\text {E}}\), \(\text {GP}_{\text {F}}\), and \(\text {GP}_{\text {EF}}\) to refer to the GPs that fit energies exclusively (solving (12)), forces only (solving (24)), and energies and forces (solving (21)) respectively.
2.3 A Gaussian process library for representing potential energy surfaces
We introduce mad-GP first via an example (Fig. 1). The code in Fig. 1 corresponds to defining the model
where \(\epsilon _e \sim \mathcal {N}(0, \sigma _e^2)\) and \(\epsilon _f \sim \mathcal {N}(0, \sigma _f^2 \varvec{I})\).
The first portion of the code (lines \(1-7\)) defines a mean function mean implementing a constant 0 mean. The function takes two arguments (1) atoms_x and (2) x. The first argument atoms_x contains a molecular structure’s atomic charges and masses (packaged in the type \(\texttt {Atoms}\)). The second argument \(\texttt {x}\) corresponds to a vector encoding of a molecular structure which may be a function of a molecular structure’s atomic charges, masses, and nuclear positions (packaged in the type \(\texttt {jax.DeviceArray[D]}\) which encodes a \(\mathbb {R}^D\) vector). The reason that \(\texttt {x}\) does not just give the nuclear positions of a molecular structure is because we may want to use a chemical descriptor that is a function of nuclear positions (among other information). Thus \(\texttt {atoms\_x}\) and \(\texttt {x}\) together encode all the information available from a molecular structure. The constant mean function, by definition, ignores all arguments and returns a constant 0.
The second portion of the code (lines \(9-24\)) defines a kernel function kernel that implements the Matérn kernel (\(k_m\), see (5)). The signature of the function is similar to the case for the prior. The arguments \(\texttt {atoms\_x1}\) and \(\texttt {atoms\_x2}\) are not used because the Matérn kernel does not require either atomic charge or atomic mass in a structure.
The last portion of the code (line 26) creates a GP surrogate that fits both energies and forces by supplying (1) the user-defined mean function mean and (2) the user-defined kernel function kernel. The default vector encoding is xyz coordinates, i.e., an identity function on nuclear positions of a molecular structure. This concludes the amount of code that the user is required to write and illustrates the main point: the user does not need to derive or implement derivatives of the mean or kernel functions. Consequently, arbitrarily complex mean and kernel functions can be defined so long as they can be programmed in Python and use mathematical primitives from an AD library.
2.3.1 Support for kernels for atomistic systems
When we introduced kernels for atomistic systems (Sect. 2.2.1), we saw that there were many design choices involved in constructing a GP kernel including (1) choice of descriptor and (2) how to compare descriptors. For example, as a quick test of a simple kernel that is (1) invariant to translation and rotation and (2) takes into account atomic charges, we may choose the kernel
where \(k_m\) is a Matérn kernel, \(\texttt {flatten}: \mathbb {R}^{(M \times L)} \rightarrow \mathbb {R}^{ML}\) flattens a matrix into a vector, \(\texttt {coulomb}\) is a Coulomb descriptor [13], and \(m_{\varvec{x}}\) and \(m_{\varvec{y}}\) are molecular structures. We could decide later that a Matérn kernel on flattened Coulomb matrices is not ideal and opt to compare Coulomb matrices directly as in
for some kernel \(k': \mathbb {R}^{(N_{\text {A}}\times N_{\text {A}})} \times \mathbb {R}^{(N_{\text {A}}\times N_{\text {A}})} \rightarrow \mathbb {R}^K\) defined on matrices directly (e.g., using a matrix norm). To assist the user in managing the multitude of kernels one could construct, mad-GP provides a small combinator library (Fig. 2). A combinator library identifies (1) primitive building blocks and (2) ways to put those building blocks together.
Global and local descriptors The base building blocks that mad-GP’s combinator library provides are local and global descriptors, similar in spirit to libraries such as DScribe [69]. (The difference with libraries like DScribe is that our combinator library supports AD, and consequently, can derive derivatives of descriptors automatically from their implementation.) A global descriptor \(d_g: \texttt {Atoms} \times \mathbb {R}^{3N_{\text {A}}} \rightarrow \mathbb {R}^D\) convert atomic charges, masses, and nuclear positions into vector encodings of the molecule. Cartesian coordinates xyz are a typical example. Another example includes Coulomb matrices (\(\texttt {coulomb}: \texttt {Atoms} \times \mathbb {R}^{3N_{\text {A}}} \rightarrow \mathbb {R}^{(N_{\text {A}}\times N_{\text {A}})}\)). A local descriptor \(d_\ell : \texttt {Atoms} \times \mathbb {R}^{3N_{\text {A}}} \rightarrow \mathcal {D}_1 \times \dots \times \mathcal {D}_{N_{\text {A}}}\) produces atom-centered encodings of the molecules \(\mathcal {D}^{N_{\text {A}}}\). Examples of local descriptors include SOAP descriptors (\(\texttt {soap}: \texttt {Atoms} \times \mathbb {R}^{3N_{\text {A}}} \rightarrow \mathcal {D}_1 \times \dots \times \mathcal {D}_{N_{\text {A}}}\)) which are a differentiable description of a molecular structure that preserves rotational, translational, and permutation invariance.
We have prototype implementations of Coulomb matrix descriptors [13] and SOAP descriptors [14] in mad-GP to demonstrate its flexibility and expressiveness. Note that, in some cases, some of the building blocks of a kernel are not differentiable. For example, when we sort the entries of a Coulomb matrix to obtain permutation invariance (Sect. 3.2) or when we use a topological algorithm such as Munkres algorithm [70] to match the structure of two molecules, these descriptors are not differentiable. They can still be handled with mad-GP because AD will assign a sub-gradient to the points that the descriptor are not differentiable at.
Descriptor-to-vector combinators Once we have described a molecular structure in terms of descriptors, mad-GP provides combinators \(\oplus\) that convert descriptors into vectors. Examples of combinations include \(\texttt {flatten}: \mathbb {R}^{(M \times L)} \rightarrow \mathbb {R}^{ML}\) which flattens a matrix into a vector (as in the Coulomb descriptor example). The combinator \(\texttt {average}: \mathcal {D}^{N_{\text {A}}} \rightarrow \mathcal {D}\) takes a local descriptor and averages the representation over all atom centers as in
For example, we can average each local environment produced by a SOAP descriptor to produce a global descriptor (e.g., see outer average in DScribe’s SOAP implementation [69]).
Vector kernels The kernel functions k are the ordinary symmetric and positive-definite functions used to parameterize GPs and include dot products \(\texttt {dot}\) and Matérn kernels \(\texttt {matern}\).
Structure kernels Finally, the structure kernels are kernels that are used to compare local descriptors. One simple structure kernel \(\texttt {pairwise}: (\mathcal {D}\times \mathcal {D}\rightarrow \mathbb {R}) \times (\mathcal {D}_1 \times \dots \times \mathcal {D}_{N_{\text {A}}}) \times (\mathcal {D}_1 \times \dots \times \mathcal {D}_{N_{\text {A}}}) \rightarrow \mathbb {R}\) performs a pairwise comparison between every pair of atoms in a local environments using a kernel \(k: \mathcal {D}\times \mathcal {D}\rightarrow \mathbb {R}\) that compares the descriptor at two atom centers. There are also more advanced structure matching kernels such as those based on topological algorithms [70] or optimal transport [65] that improve upon pairwise matching by only comparing “suitably-aligned" portions of each molecular structure’s local descriptor. We leave the implementation of more advanced structure kernels for future work.
Using these combinators, the Matérn kernel can be written as\(\begin{aligned} \texttt {kernel}(\text {atoms}_{\varvec{x}}, \text {atoms}_{\varvec{y}}, \varvec{x}, \varvec{y}) = k_{\text {Mat}\acute{\mathrm{e}}\text {rn}}(\texttt {xyz}(\text {atoms}_{\varvec{x}}, \varvec{x}), \texttt {xyz}(\text {atoms}_{\varvec{y}}, \varvec{y})) \end{aligned}\)to explicitly highlight that we are using xyz descriptors of a molecule.
2.3.2 Support for gradient-based surrogate optimization
mad-GP supports gradient-based optimization of a trained GP surrogate by exposing (1) a prediction of the posterior energy and (2) the gradient of the posterior energy. As a reminder, this functionality is useful for geometry optimization. How the gradient is obtained depends on whether we are using \(\text {GP}_{\text {E}}\), \(\text {GP}_{\text {F}}\), or \(\text {GP}_{\text {EF}}\).
Energy For \(\text {GP}_{\text {E}}\), the posterior mean gives the prediction of the energies as
and the gradient of the posterior mean
is computed with (reverse-mode) AD. (Sect. 2.4 for the distinction between reverse-mode and forward-mode.)
Force For \(\text {GP}_{\text {F}}\), observe that the posterior prediction of the gradient is
The second-order derivatives of the kernel are obtained with (reverse-mode then forward-mode) AD. A prediction of the energy is recovered up to a constant via the line integral along the curve \(\gamma _{\varvec{x}_0}^{\varvec{x}}\) starting at \(\varvec{x}_0\) and ending at \(\varvec{x}\) as
where C is some constant. (The base point \(\varvec{x}_0\) is arbitrary and unspecified.) That is, we “compute" the integral using the converse of the gradient theorem for conservative forces: any conservative force field can be written as the gradient of a scalar field. Thus we compute (35) directly by computing \(k\nabla _2^\text {T}\) using (reverse-mode) AD.
Energy and force For \(\text {GP}_{\text {EF}}\), we simply evaluate the posterior mean at a geometry to obtain both the prediction of the energy and (negative) forces. Recall that the gradient of the posterior mean for the energy is given by the prediction of the posterior mean for the gradient.
2.3.3 Support for GPs with forces
We implement GPs that fit forces (i.e., \(\text {GP}_{\text {F}}\) and \(\text {GP}_{\text {EF}}\)) by implementing their respective kernels using AD where appropriate. For instance, for \(\text {GP}_{\text {EF}}(\mu , k)\), we implement \(\nabla \mu\), \(\nabla _1 k\), and \(k \nabla _2^\text {T}\) with (reverse-mode) AD, and \(\nabla _1 k \nabla _2^\text {T}\) with (reverse-mode then forward-mode) AD. We will see in Sect. 2.4 that the run-time of using AD to calculate a second-order term such as \(\nabla _1 k \nabla _2^\text {T}\) is proportional to DT where D is the number of dimensions of the molecular representation and T is the run-time of k. To fit these GPs to training data, we apply the mean and kernel functions to the training data to construct mean vectors and kernel matrices, and use a generic linear system solver to solve the system of equations. Note that AD does not need to interact with the linear solver.
2.4 Automatic differentiation
Automatic differentiation (AD) is a technique that algorithmically transforms a computer program evaluating a function into one evaluating the derivative of that function that has the same time-complexity as the program evaluating the original function. This statement is a mouthful—let us unpack this by comparison with (1) finite differences (FD) and (2) symbolic differentiation (SD) in the context of an example. Table 1 summarizes the differences between the methods.
2.4.1 AD by example
Consider the function \(f: \mathbb {R}\rightarrow \mathbb {R}\) defined as
for some \(g, h, i: \mathbb {R}\rightarrow \mathbb {R}\). The (simplest) finite differences method approximates the derivative of the function as

This approach can easily be implemented in code (as above), operates on programs, and approximately computes the gradient via a linear approximation. The time-complexity of the function is proportional to the time-complexity of the original function f—we call it twice.
One form of the symbolic derivative for the function f is
where \(g', h', i'\) are the respective (symbolic) derivatives of g, h, i. Thus the symbolic derivative is not unique—depending on how we factor this expression, the symbolic derivative may take worst-case exponential time compared to the evaluation of the original function. Consequently, we should be careful when translating symbolic derivatives into code to pick the appropriate factorization.
Figure 3 provides a pedagogical example of applying source-to-source and reverse-mode AD to f. (mad-GP uses combinations of reverse-mode and forward-mode AD when appropriate, as well as non source-to-source methods of implementation.) We have presented the code in a way such that one can see how this code could be mechanically generated by a computer. Note that each line of code of the original code is translated into
and produces the corresponding derivative code
where \(\texttt {ad\_op\_1st}\) and \(\texttt {ad\_op\_2nd}\) correspond to the derivatives of the operation \(\texttt {op}\) with respect to the first and second arguments. This code can be obtained with a recursive application of \(\texttt {AD}\).
In this form, it is easy to see that the time-complexity of the derivative is proportional to the original code because each line of code in the original function produces at most 2 additional lines of code of the same time-complexity. Upon closer inspection, we see that AD is a method that, as a default, mechanically selects the factorization of the SD that has the appropriate time-complexity. Thus it also computes the derivative exactly, like SD and unlike FD.
AD naturally generalizes from functions of a single variable to functions of multiple variables. Moreover, AD is composable—in the example, we can computed \(\texttt {AD}(f)\) by composing the constituents of its sub-functions \(\texttt {AD}(g)\), \(\texttt {AD}(h)\), \(\texttt {AD}(i)\). Another implication of the composability of AD is that we can use it to compute higher-order derivatives.
2.4.2 AD for higher-order derivatives
\(\texttt {AD}(\texttt {AD}(f))\) computes the second-order derivative of f because the result of the inner call produces a program evaluating the derivative of f, which can be fed back into another call to \(\texttt {AD}\). Note that the time-complexity guarantees of AD hold for higher-order derivatives, whereas the time-complexity guarantees for SD are amplified for higher-order derivatives.
For computing higher-order derivatives of functions with multivariate output spaces, the distinction between reverse-mode (what was presented) and forward-mode AD is important for time-complexity reasons. In particular, for a function \(f: \mathbb {R}^D \rightarrow \mathbb {R}^K\), computing the derivative with forward-mode AD takes D passes whereas it takes K passes for reverse-mode AD. Thus forward-mode is advantageous when \(K > D\) and reverse-mode is advantageous when \(D > K\). When \(D = K\), forward-mode is advantageous because it takes less memory. Consequently, for taking second-order derivatives of scalar-valued functions, it is best to perform reverse-mode AD followed by forward-mode AD. We refer the interested reader to references (e.g., see the survey [71] and the references within) for more background.
2.4.3 AD in mad-GP
For the use case of PES modeling where we require second-order derivatives when taking forces into account, we should be careful to select an AD library that supports both reverse-mode and forward-mode. Popular AD libraries such as Torch [72] that are specialized for neural networks prioritize reverse-mode AD because training neural networks only requires first-order derivatives of a loss function, i.e., single-value output. Jax [73] is an AD library that supports both forward-mode and reverse-mode AD. Consequently, Jax is the default AD library in mad-GP. Jax additionally supports Just-In-Time (JIT) compilation. This means that Jax will generate optimized derivative code based on the sizes of vectors and matrices it sees during run-time, further reducing the cost of using a high-level interpreted language like Python for performing efficient numerical computation.
A visualization of initial geometries from systems (system 01 to system 25 arranged left-to-right and top-to-bottom) in the Baker-Chan dataset [74]. White, grey, blue, red, cyan, green, navy green, orange and yellow balls represent H, C, N, O, F, Cl, Si, P, and S atoms, respectively (Color figure online)
3 Results
In the previous section, we introduced and built machinery that enables systematic exploration of GPs for modeling PESs. In this section, we apply mad-GP to geometry optimization of small molecules to validate its use (Sect. 3.1). We use mad-GP to test the effectiveness of fitting forces (\(\text {GP}_{\text {E}}\) vs. \(\text {GP}_{\text {F}}\) and \(\text {GP}_{\text {EF}}\), Sect. 3.1.1), the effectiveness of fitting energies for GPs that fit forces (\(\text {GP}_{\text {F}}\) vs. \(\text {GP}_{\text {EF}}\), Sect. 3.1.2), and a preliminary study on the use of non-constant priors and hierarchical kernels (Sect. 3.1.3). For completeness, we also qualitatively describe our experiences using AD for constructing GP surrogates for representing molecular PESs (Sect. 3.2).
3.1 Geometry optimization
We use mad-GP to perform geometry optimization on the Baker-Chan dataset [74] of molecules (Fig. 4), a benchmark dataset consisting of initial-guess structures of small molecules, which is tested in prior work on GP surrogates [11]. Note that in Baker et al.’s work, their goal was to optimize to transition-state structures (i.e., first-order saddle points); however, in this work (as well as in Denzel et al.’s prior work), we optimize to stable local minima. There are many geometry optimization algorithms that use GP surrogates [11, 12, 75]. We use the geometry optimization algorithm designed for GPs based on the one given in ASE [12, 75] (Atomistic Simulation Environment). We choose this algorithm as a baseline so we can focus on varying (1) fitting energies/forces and (2) mean and kernel functions to explore mad-GP’s capabilities. We summarize the algorithm below.
Step 1
Update the current training set of N points–\((\mathbf {X}, \mathbf {E})\), \((\mathbf {X}, \varvec{F})\), or \((\mathbf {X}, (\mathbf {E}, \varvec{F}))\)–with a call to an electronic-structure method to obtain energies \(E_{N+1}\) and/or forces \(\varvec{F}_{N+1}\) at a geometry \(\varvec{x}_{N+1}\). In this work, we use the PM7 semi-empirical method [76] implemented in MOPAC2016 [77], which we refer to as MOPAC from now on. To manage the computational complexity of fitting a GP, we only keep the last 100 data points.
Step 2
Fit the GP with the new data point \((\varvec{x}_{N+1}, E_{N+1})\), \((\varvec{x}_{N+1}, \varvec{F}_{N+1})\), or \((\varvec{x}_{N+1}, E_{N+1}, \varvec{F}_{N+1})\). If we are fitting energies, perform a “set maximum" step: set the mean function \(\mu\) to be the maximum energy seen during geometry optimization
(Note that this does not apply to \(\text {GP}_{\text {F}}\) which we will comment on in Sect. 3.1.2.)
Step 3
Use L-BFGS to search the posterior gradient for a new local minimum. Our current implementation uses Scipy’s L-BFGS algorithm [66] using gradients supplied by mad-GP.
Step 4
If the energy found at the new local minimum is higher than the last energy as determined by a SPE calculation, restart the search for a new local minimum by going to step 2. Otherwise we return the last point \(\varvec{x}_{N+1}\) found.
Step 5
Iterate until convergence. Because we are comparing against MOPAC, the convergence criterion we use is the same as MOPAC’s GNORM keyword:
where the units are in \(\text {eV}/\text{\AA}\) and \(N_{\text {A}}\) is the number of atoms in the system so that \(3N_{\text {A}}\) is the dimensionality of the xyz encoding of a molecule. (A common choice is to also constrain the maximum force, i.e., both the \(l_\infty\) norm and the \(l_2\) norm. We do not do that here to be consistent with MOPAC.)
For the purposes of reducing the number of hyper-parameters we need to tune, we do not use a maximum step-size or adaptive length-scale tuning [11, 78]. Note that smaller length-scales might be needed closer to convergence. We emphasize that the choices of MOPAC and the optimization algorithm above are for demonstrating the use of mad-GP for geometry optimization. Different electronic-structure calculators and geometry optimization algorithms can easily be used/implemented in mad-GP.
3.1.1 Fitting forces
mad-GP supports GP surrogates that fit energies exclusively (\(\text {GP}_{\text {E}}\)), fit forces exclusively (\(\text {GP}_{\text {F}}\)), and fit both energies and forces (\(\text {GP}_{\text {EF}}\)). We use this functionality to compare \(\text {GP}_{\text {E}}\), \(\text {GP}_{\text {F}}\), and \(\text {GP}_{\text {EF}}\) with a baseline implementation of L-BFGS in MOPAC to test the efficacy of fitting forces. We perform SPE calculations for both GP and L-BFGS optimizations by using the semiempirical PM7 Hamiltonian within the unrestricted Hartree-Fock formalism (not to be confused with the Hartree-Fock method, which uses the full non-empirical Fock operator with explicitly calculated one- and two-electron integrals) via the MOPAC keyword "PM7 UHF 1SCF GRAD". The self-consistent field (SCF) energy convergence criterion was set to \(10^{-8}\) kcal \(\text {mol}^{-1}\).
All GPs use a constant 0 mean and a Matérn kernel with \(\sigma _{\text {M}} = 1.0\) and \(l = 0.4\)Å. We set the energy noise \(\sigma _e = 0.002 {\text { eV}}^{1/2}\) and the force noise σf = 0.005 (eV/Å)\({^{\raise0.5ex\hbox{$\scriptstyle 1$} \kern-0.1em/\kern-0.15em \lower0.25ex\hbox{$\scriptstyle 2$}}}\). We have tested \(2 \times 10^{-10} \le \sigma _e \le 2 \times 10^{-5}\) and \(5 \times 10^{-10} \le \sigma _e \le 5 \times 10^{-5}\), and found that they do not significantly affect performance. We refer the reader to the Supporting Information for more details (Table S1).
Table 2 summarizes the results of benchmarking GP surrogates against each other and L-BFGS in MOPAC. We include \(\Delta E\) (eV), which measure the energy difference between the final geometries obtained by each method with respect to the one obtained by MOPAC, as well as the root-mean-squared distance (RMSD in \(\text{\AA}\)). The RMSD between the mad-GP optimized and L-BFGS baseline structures were calculated after the alignment (translate the center-of-mass and perform the proper rotation) of the two molecules using an implementation of the Quaternion algorithm [79] from the rmsd package [80]. We refer the reader to the Supporting Information for more details about the RMSD calculation (Fig. S1).
Recall again that we are not using an adaptive length-scale. In particular, a Matérn kernel with a length-scale of 0.4Å will be quite large once we near the convergence threshold of \(\Vert \varvec{F}_{N+1} \Vert \le 0.0054 \cdot 3N_{\text {A}}\), especially given that we do not dampen the size of the optimizer steps once we near convergence.
Our results show that GPs that fit forces perform better compared to those that do not on the Baker-Chan dataset. In particular, none of the geometry optimizations using \(\text {GP}_{\text {E}}\) converge. The geometry optimizations that use \(\text {GP}_{\text {F}}\) and \(\text {GP}_{\text {EF}}\) converge for all of the molecules. Both \(\text {GP}_{\text {F}}\) and \(\text {GP}_{\text {EF}}\) are competitive with the baseline L-BFGS optimizer in MOPAC with respect to the number of SPE calculations, consistent with previous work demonstrating the effectiveness of GP surrogates. Occasionally, \(\text {GP}_{\text {F}}\) and \(\text {GP}_{\text {EF}}\) find geometries that are lower-energy than the L-BFGS optimizer in MOPAC. All final geometries are stable local minima as verified by frequency calculations conducted with MOPAC except for system id 3, which has a relatively flat PES and has a small imaginary frequency (56.7i cm\(^{-1}\) in \(\text {GP}_{\text {F}}\), 5.5i cm\(^{-1}\) in \(\text {GP}_{\text {EF}}\), and 56.5i cm\(^{-1}\) in L-BFGS baseline) under current convergence criteria. Note that some optimized structures in the Baker-Chan set are pre/post-reaction complex (e.g., 03, 09, 15, 17) with multiple local minima, leading to higher RMSD values and slightly larger energy differences between GP and L-BFGS. This also indicates that the advantage of GP is finding lower-energy structures compared to the L-BFGS algorithm. Next, we will take a closer look at \(\text {GP}_{\text {F}}\) and \(\text {GP}_{\text {EF}}\).
3.1.2 Fitting forces versus energies and forces
It is popular to fit both energies and forces when using GPs as a surrogate function for a PES for geometry optimization. This is a natural design decision to make because knowing both energies and forces gives more information than just forces. Nevertheless, some work [3, 15] considers fitting forces exclusively. To the best of our knowledge, there has been relatively little work comparing the two approaches, and we aim to fill that gap now.
At first glance, the results in Table 2 support the intuition that it is better to fit both energies and forces. \(\text {GP}_{\text {EF}}\) and \(\text {GP}_{\text {F}}\) require roughly the same number of SPE calculations across the Baker-Chan dataset. Nevertheless, there is an important optimization detail that is different between \(\text {GP}_{\text {EF}}\) and \(\text {GP}_{\text {F}}\) which we term the “set maximum" step.
State-of-the-art results on applying \(\text {GP}_{\text {EF}}\) surrogates use constant mean functions and a variation of the following step: update the mean function to be
which sets the mean function to be the constant function that returns the highest energy seen during geometry optimization and c is some constant. The justification for this design decision is that it makes the optimizer more stable by forcing a local minima (e.g., Denzel et al. [11]). We find this reasoning plausible: setting the prior to a high value in unseen regions should encourage the optimizer to stay around regions that it has seen before because regions far away from observed points will take on the mean value (which is high). The reason that the “set maximum" step is important is because it is not applicable to \(\text {GP}_{\text {F}}\). As a reminder, the bias term only affects the energy component when the mean function is constant (the derivative of a constant is 0), and a force-only GP does not have an energy component.
Table 3 summarizes the results of comparing \(\text {GP}_{\text {F}}\) and \(\text {GP}_{\text {EF}}\) by varying the prior choices, including the ability to use the “set maximum" step. The entries with a \(\Delta E\) but no corresponding SPE exceed the max cycle limitation before reaching convergence(\(> 300\) steps). The entries with no \(\Delta E\) and no corresponding SPE failed to produce any meaningful results. As we can see, the “set maximum" step where \(c = 0\) drastically improves the performance of \(\text {GP}_{\text {EF}}\) by reducing the number of SPE calls required. We have also tested the “set maximum" step with \(c = 10\)a.u. (e.g., as suggested by Denzel et al. [11]) and found that we improve the results over a baseline where we do not perform a “set maximum" step, but worse when \(c = 0\). (Note that Denzel et al.’s geometry optimization [11] is not the same as ours and contains an over-shooting step that may explain the difference in performance, among other differences.) Thus, at least for constant 0 priors, we can improve the performance of \(\text {GP}_{\text {EF}}\) at the cost of tuning one additional parameter (i.e., the constant c) and performing a “set maximum" step during optimization.
We have also included an example of a non-constant prior in the form of a simplified pairwise Leonard-Jones (LJ) potential
where \(\user2{x} = (\user2{x}^{1} \ldots \user2{x}^{{{N_\text{A}}}} )^{{\text{T}}}\) are the xyz coordinates of a molecular structure and \(r_{i,j} = \Vert \varvec{x}^{i} - \varvec{x}^j \Vert\) is the distance between atoms i and j (in \(\text{\AA}\)). Thus we compute the LJ potential \(V_{\text {LJ}}(\varvec{x})\) as the summation of the pairwise potential of every unique pair of atoms. We do not use a cutoff radius in evaluating the summation. For our tests, we set the effective diameter of the cross-section \(\sigma _{\text {LJ}}= 1 \text { } \text{\AA}\) and the depth of the potential well \(\epsilon _{\text {LJ}} = 1 \text { } \text {eV}\). We implement this LJ potential as a proof-of-concept to demonstrate how to implement a non-constant prior in mad-GP (Fig. S2 in the Supporting Information for more details). We are not claiming that LJ is a good choice of mean function.
The surrogate \(\text {GP}_{\text {EF}}(\text {LJ}, k)\) performs similarly to \(\text {GP}_{\text {EF}}(0, k)\) and \(\text {GP}_{\text {F}}(\text {LJ}, k)\) performs similarly to \(\text {GP}_{\text {F}}(0, k)\). This is not surprising as the LJ potential is a very simple model that is mostly suitable for describing non-covalent interactions between non-bonded molecules, and unsuitable for bond breaking, bond stretching, valence-angle bending, and internal rotations. In this proof-of-concept test, the LJ mean function behaves like a constant 0 mean function. Implementing and systematically testing more complex and physically inspired prior mean functions is current work-in-progress. We also believe that further exploration of the “set maximum" step and how it affects a choice of mean function is worth pursuing in future work that more thoroughly compares \(\text {GP}_{\text {F}}\) and \(\text {GP}_{\text {EF}}\).
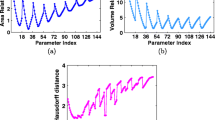
Comparing different choices of mean functions, descriptors, and kernel functions with respect to the average number of steps across the Baker-Chan dataset (lower is better). Each curve, read from left-to-right, passes through each choice and results in an average number of steps (darker is better). The figure was generated with wandb [81]
3.1.3 Gaussian processes with non-constant priors and hierarchical kernels
We use mad-GP to explore several combinations of non-constant priors and hierarchical kernels for \(\text {GP}_{\text {F}}\) and \(\text {GP}_{\text {EF}}\). Figure 5 provides a visualization of the results using wandb [81].
Each curve, read from left-to-right, passes through each choice and results in an average number of steps (color-coded, darker is better) across the Baker-Chan dataset (lower is better). The choices include (1) optimizer: \(\text {GP}_{\text {F}}\) vs. \(\text {GP}_{\text {EF}}\), (2) mean functions, (3) descriptors: COULOMB which gives the Coulomb matrix [13], PRE-COULOMB which gives the Coulomb matrix without atomic charges \(Z_i\), and xyz which gives Cartesian coordinates, and (4) kernel functions: squared-exponential, Matérn kernel with \(p=3\) (three-times differentiable), and Matérn kernel with \(p=2\) (twice differentiable).
For completeness, the Coulomb matrix without atomic charges is defined as the matrix with ij-th entry
where \(\user2{x} = (\user2{x}^{1} \ldots \user2{x}^{{{N_\text{A}}}} )^{{\text{T}}}\) are the xyz coordinates of a molecular structure with \(N_{\text {A}}\) atoms. The Coulomb matrix is defined as the matrix with ij-th entry
where \(\user2{x} = (\user2{x}^{1} \ldots \user2{x}^{{{N_\text{A}}}} )^{{\text{T}}}\) are the xyz coordinates of a molecular structure with \(N_{\text {A}}\) atoms. The squared-exponential kernel is
where \(\sigma _{\text {se}}\) is a weight and l is a length-scale parameter. Finally, the Matérn kernel with \(p=3\) is
where \(\rho = \Vert \varvec{x}- \varvec{y} \Vert\).
For the first choice of optimizer, we see that \(\text {GP}_{\text {F}}\) is better at reducing the average number of steps as compared to \(\text {GP}_{\text {EF}}\). Note that we are not performing the “set maximum" step for \(\text {GP}_{\text {EF}}\) as we hope to compare different priors. These results are consistent with those in Table 3. For prior functions, constant prior functions perform better than LJ potentials. These results are also consistent with those in Table 3.
For the choice of descriptors, xyz coordinates perform better than both the Coulomb descriptor and the pre-Coulomb descriptor without atomic charges. There are at least two points we should highlight about this result. First, AD enables us to perform gradient-based geometry optimization using descriptors of molecules without requiring us to define an inverse transformation from descriptor space back into xyz coordinates. Indeed, the geometry optimization with Coulomb descriptors converges on the Baker-Chan dataset, and thus gives us an example of a chemically-inspired descriptor that can be applied for geometry optimization. Second, while the average number of steps may be higher for a single geometry optimization, note that Coulomb descriptors are invariant to rotation, a property that xyz descriptors do not possess. Consequently, further geometry optimization involving a GP using Coulomb descriptors will be robust to rotations of a molecule while those based on xyz descriptors will not be. Note that, herein, we are referring to the direct usage of the xyz encoding of a structure without pre-fixing the orientation (by defining a standard orientation using principle axis as done in quantum chemistry codes) and the center of mass of the input structure, which certainly eliminate the issue of rotation and translation invariance of the descriptor.
Finally, for the choice of kernel functions, we see that if we use constant 0 mean function, then Matérn kernels perform the best. As a reminder, we are not performing the “set maximum" step here for constant priors so it is better to look at the results for \(\text {GP}_{\text {F}}\) for comparison of mean and kernel functions. For \(\text {GP}_{\text {F}}\), we see that several combinations work well: Coulomb matrices with Matérn \(p=2\), \(p=3\), and squared exponential kernels work. Coulomb descriptors increase the dimensionality of the representation, and so it is interesting to see that kernels that impose smoother constraints still work.
3.2 Using automatic differentiation
One of the design goals of mad-GP is to enable users to more systematically explore different priors and kernel functions for GP PES surrogate models without needing to implement first and second-order derivatives. The key technique for achieving this was AD. We summarize some of our experiences using AD in this section.
One question we might be interested in is: “what can I do now that I could not do before?" For a standard kernel such as a Matérn kernel, the extra work that is done if AD is not used is (1) deriving or looking up the derivatives, (2) transcribing them to code, (3) and checking that the derivatives are implemented correctly. For completeness, the first-order derivatives are
and
The second-order derivative is
where \(\otimes\) is an outer product.
Although this might be simple enough for a standard kernel, consider implementing the Matérn kernel for arbitrary p. This kernel is defined as
where \(\rho = \Vert \varvec{x}- \varvec{y} \Vert\). We have implemented this kernel for \(1 \le p \le 10\) in mad-GP using 18 lines-of-code. Deriving the first and second-order derivatives with respect to \(\varvec{x}\) and \(\varvec{y}\) and checking that they are correct is harder. Notably, SD tools are less effective on multivariate spaces.
A kernel where we sort the columns of a Coulomb matrix by it’s \(l_2\) norm (lines 2–3 and 8–9). Line 5 gives an example of how to use the molecule metadata atoms_x and atoms_y to obtain the atomic masses for use in a descriptor. The function jax.lax.map is a functional looping construct (line 6). The function \(\texttt {coulomb\_desc}\) (not shown) implements a Coulomb matrix using atomic masses and positions as inputs
Beyond this, consider deriving and implementing first and second-order derivatives for a chemical descriptor that may not be differentiable in the traditional sense. Figure 6 gives an example of a descriptor implemented in mad-GP that is not differentiable because the columns of the Coulomb matrix are sorted by their \(l_2\) norm. (It also gives an example of how to use atomic charges in a GP kernel.) AD, unlike SD, can still handle such descriptors by using the concept of sub-gradients. Thus AD presents us the opportunity to perform geometry optimization using kernels with descriptors that are non-invertible and non-differentiable.
We emphasize again that AD implements an appropriate factorization of the SD in an automatic fashion. Consequently, it implements a particular symbolic derivative and the floating-point precision is identical to implementing that symbolic derivative. The performance cost that one pays when using AD is that the first and second-order derivatives are derived when the code is executed. This is a one-time cost because we can use JIT technology to cache the results of the derivatives. Compared to the cost of SPE calculations and geometry optimization, the one-time cost to compile the first and second-order derivatives is negligible.
4 Discussion
As we demonstrate with our prototype mad-GP, users need only write the mean and kernel functions before being able to explore a variety of GP surrogate models that implement \(\text {GP}_{\text {E}}\), \(\text {GP}_{\text {F}}\), or \(\text {GP}_{\text {EF}}\). In the rest of this section, we will highlight a few interesting discoveries that we were able to uncover due to the flexibility of mad-GP.
AD is crucial for atomistic kernel functions In light of the testing results, one can see the possibility for an abundance of mean functions, kernel functions, their combinations, and their compositions that one can explore using mad-GP. To the best of our knowledge, most prior work explores simple mean functions like constant function and standard kernels such as Matérn kernels. Notably, the usage of AD along with gradient-based optimization of GP surrogates means that we can use arbitrarily complex descriptors so long as we can implement them in mad-GP. We have identified several combinations that work using mad-GP. We leave a more through exploration of this direction as future work.
“Set maximum" step The usage of \(\text {GP}_{\text {EF}}\) only performs well in our tests if the “set maximum" step is applied. In contrast, \(\text {GP}_{\text {F}}\) performs well without this additional step. Consequently, we find that \(\text {GP}_{\text {F}}\) is more robust compared to \(\text {GP}_{\text {EF}}\) because we do not need to modify the geometry optimization algorithm. Crucially, the “set maximum" step affects our ability to explore different mean functions for \(\text {GP}_{\text {EF}}\). We believe that further investigation of this step and its impact on choice of mean function in comparing \(\text {GP}_{\text {EF}}\) and \(\text {GP}_{\text {F}}\) is a good direction of future work. These questions only became apparent to us when we tried to implement a tool that could handle a variety of GP use cases in a generic manner. At a meta-level, we hope that this provides further evidence that better tools can lead to better science.
5 Conclusion
In summary, we introduce mad-GP, a library for constructing GP surrogate models of PESs. A user of mad-GP only needs to write down the functional form of the mean and kernel functions, and the library handles all required derivative implementations. mad-GP accomplishes this with a technique called AD. Our hope is that mad-GP can be used to more systematically explore the large design space of GP surrogate models for PESs.
As an initial case study, we apply mad-GP to perform geometry optimization. We compare GP surrogates that fit forces with those that fit energies and forces. In general, we find that \(\text {GP}_{\text {EF}}\) performs comparably with \(\text {GP}_{\text {F}}\) in terms of the number of SPE calculations required, although \(\text {GP}_{\text {F}}\) is more robust for optimization because it does not require an additional step to be applied during optimization. In our preliminary study on the use of non-constant priors and hierarchical kernels in GP PES surrogates, we also confirm that constant mean functions and Matérn kernels work well as reported in the literature, although our tests also identify several other promising candidates (e.g., Coulomb matrices with three-times differentiable Matérn kernels). Our studies validate that AD is a viable method for performing geometry optimization with GP surrogate models on small molecules.
References
A.P. Bartók, M.C. Payne, R. Kondor, G. Csányi, Gaussian approximation potentials: the accuracy of quantum mechanics, without the electrons. Phys. Rev. Lett. 104, 136403 (2010)
P. Rowe, V.L. Deringer, P. Gasparotto, G. Csányi, A. Michaelides, An accurate and transferable machine learning potential for carbon. J. Chem. Phys. 153, 034702 (2020)
S. Chmiela, A. Tkatchenko, H.E. Sauceda, I. Poltavsky, K.T. Schütt, K.-R. Müller, Machine learning of accurate energy-conserving molecular force fields. Sci. Adv. (2017). https://doi.org/10.1126/sciadv.1603015
S. Chmiela, H.E. Sauceda, K.-R. Müller, A. Tkatchenko, Towards exact molecular dynamics simulations with machine-learned force fields. Nat. Commun. 9, 1–10 (2018)
H. Sugisawa, T. Ida, R.V. Krems, Gaussian process model of 51-dimensional potential energy surface for protonated imidazole dimer. J. Chem. Phys. 153, 114101 (2020)
A. Denzel, B. Haasdonk, J. Kästner, Gaussian process regression for minimum energy path optimization and transition state search. J. Phys. Chem. A 123, 9600–9611 (2019). (PMID: 31617719)
A. Denzel, J. Kästner, Gaussian process regression for transition state search. J. Chem. Theory Comput. 14, 5777–5786 (2018). (PMID: 30351931)
O.-P. Koistinen, V. Ásgeirsson, A. Vehtari, H. Jónsson, Nudged elastic band calculations accelerated with gaussian process regression based on inverse interatomic distances. J. Chem. Theory Comput. 15, 6738–6751 (2019). (PMID: 31638795)
O.-P. Koistinen, V. Ásgeirsson, A. Vehtari, H. Jónsson, Minimum mode saddle point searches using gaussian process regression with inverse-distance covariance function. J. Chem. Theory Comput. 16, 499–509 (2020). (PMID: 31801018)
O.T. Unke, S. Chmiela, H.E. Sauceda, M. Gastegger, I. Poltavsky, K.T. Schütt, A. Tkatchenko, K.-R. Müller, Machine learning force fields. Chem. Rev. 121, 10142–10186 (2021)
A. Denzel, J. Kästner, Gaussian process regression for geometry optimization. J. Chem. Phys. 148, 1–32 (2018)
E. Garijo del Río, J.J. Mortensen, K.W. Jacobsen, Local Bayesian optimizer for atomic structures. Phys. Rev. B 100, 104103 (2019)
M. Rupp, A. Tkatchenko, K.-R. Müller, O.A. von Lilienfeld, Fast and accurate modeling of molecular atomization energies with machine learning. Phys. Rev. Lett. 108, 058301 (2012)
A.P. Bartók, R. Kondor, G. Csányi, On representing chemical environments. Phys. Rev. B (2013). https://doi.org/10.1103/PhysRevB.87.184115
H.E. Sauceda, S. Chmiela, I. Poltavsky, K. Müller, A. Tkatchenko, Molecular force fields with gradient-domain machine learning: construction and application to dynamics of small molecules with coupled cluster forces. J. Chem. Phys. 150(11)(2019)
Smith Jr, V. H.; Schaefer III, H. F.; Morokuma, K. (2012). Applied Quantum Chemistry: Proceedings of the Nobel Laureate Symposium on Applied Quantum Chemistry in Honor of G. Herzberg, RS Mulliken, K. Fukui, W. Lipscomb, and R. Hoffman, Honolulu, HI, 16–21 December 1984 ; Springer Science & Business Media
K. Fukui, Formulation of the reaction coordinate. J. Phys. Chem. 74, 4161–4163 (1970)
K. Fukui, S. Kato, H. Fujimoto, Constituent analysis of the potential gradient along a reaction coordinate. Method and an application to methane + tritium reaction. J. Am. Chem. Soc. 97, 1–7 (1975)
K. Ishida, K. Morokuma, A. Komornicki, The intrinsic reaction coordinate. An ab initio calculation for HNC\(\rightarrow\)HCN and H-+CH4\(\rightarrow\)CH4+H-. J. Chem. Phys. 66, 2153–2156 (1977)
N.C. Blais, D.G. Truhlar, B.C. Garrett, Improved parametrization of diatomics-in-molecules potential energy surface for Na(3p 2P)+H2 \(\rightarrow\) Na(3s 2S)+H2. J. Chem. Phys. 78, 2956–2961 (1983)
D.G. Truhlar, R. Steckler, M.S. Gordon, Potential energy surfaces for polyatomic reaction dynamics. Chem. Rev. 87, 217–236 (1987)
A.J.C. Varandas, F.B. Brown, C.A. Mead, D.G. Truhlar, N.C. Blais, A double many-body expansion of the two lowest-energy potential surfaces and nonadiabatic coupling for H3. J. Chem. Phys. 86, 6258–6269 (1987)
S.C. Tucker, D.G. Truhlar, A six-body potential energy surface for the SN2 reaction Cl-(g) + CH3Cl(g) and a variational transition-state-theory calculation of the rate constant. J. Am. Chem. Soc. 112, 3338–3347 (1990)
G.C. Lynch, R. Steckler, D.W. Schwenke, A.J.C. Varandas, D.G. Truhlar, B.C. Garrett, Use of scaled external correlation, a double many-body expansion, and variational transition state theory to calibrate a potential energy surface for FH2. J. Chem. Phys. 94, 7136–7149 (1991)
E.E. Dahlke, D.G. Truhlar, Electrostatically embedded many-body expansion for simulations. J. Chem. Theory Comput. 4, 1–6 (2008)
P.G. Mezey, Reactive domains of energy hypersurfaces and the stability of minimum energy reaction paths. Theor. Chim. Acta 54, 95–111 (1980)
P.G. Mezey, Catchment region partitioning of energy hypersurfaces. I. Theor. Chim. Acta 58, 309–330 (1981)
P.G. Mezey, The isoelectronic and isoprotonic energy hypersurface and the topology of the nuclear charge space. Int. J. Quant. Chem. 20, 279–285 (1981)
P.G. Mezey, Manifold theory of multidimensional potential surfaces. Int. J. Quant. Chem. 20, 185–196 (1981)
P.G. Mezey, Critical level topology of energy hypersurfaces. Theor. Chim. Acta 60, 97–110 (1981)
P.G. Mezey, Lower and upper bounds for the number of critical points on energy hypersurfaces. Chem. Phys. Lett. 82, 100–104 (1981)
P.G. Mezey, Quantum chemical reaction networks, reaction graphs and the structure of potential energy hypersurfaces. Theor. Chim. Acta 60, 409–428 (1982)
P.G. Mezey, Topology of energy hypersurfaces. Theor. Chim. Acta 62, 133–161 (1982)
P.G. Mezey, The topology of energy hypersurfaces II. Reaction topology in euclidean spaces. Theor. Chim. Acta 63, 9–33 (1983)
P. Mezey, Potential Energy Hypersurfaces. Studies in Physical and Theoretical Chemistry (Elsevier, New York, 1987)
R. Duchovic, Y. Volobuev, G. Lynch, D. Truhlar, T. Allison, A. Wagner, B. Garrett, J. Corchado, POTLIB 2001: a potential energy surface library for chemical systems. Comput. Phys. Commun. 144, 169–187 (2002)
Ö.F. Alış, H. Rabitz, Efficient implementation of high dimensional model representations. J. Math. Chem. 29, 127–142 (2001)
K. Yagi, C. Oyanagi, T. Taketsugu, K. Hirao, Ab initio potential energy surface for vibrational state calculations of H 2 CO. J. Chem. Phys. 118, 1653–1660 (2003)
K. Yagi, S. Hirata, K. Hirao, Multiresolution potential energy surfaces for vibrational state calculations. Theor. Chem. Accounts 118, 681–691 (2007)
S. Carter, S.J. Culik, J.M. Bowman, Vibrational self-consistent field method for many-mode systems: a new approach and application to the vibrations of CO adsorbed on Cu (100). J. Chem. Phys. 107, 10458–10469 (1997)
J.M. Bowman, S. Carter, X. Huang, MULTIMODE: a code to calculate rovibrational energies of polyatomic molecules. Int. Rev. Phys. Chem. 22, 533–549 (2003)
J.M. Bowman, T. Carrington, H.-D. Meyer, Variational quantum approaches for computing vibrational energies of polyatomic molecules. Mol. Phys. 106, 2145–2182 (2008)
B.J. Braams, J.M. Bowman, Permutationally invariant potential energy surfaces in high dimensionality. Int. Rev. Phys. Chem. 28, 577–606 (2009)
A. Jäckle, H.-D. Meyer, Product representation of potential energy surfaces. II. J. Chem. Phys. 109, 3772–3779 (1998)
F. Otto, Multi-layer Potfit: an accurate potential representation for efficient high-dimensional quantum dynamics. J. Chem. Phys. 140, 014106 (2014)
G. Avila, T. Carrington Jr., Using multi-dimensional Smolyak interpolation to make a sum-of-products potential. J. Chem. Phys. 143, 044106 (2015)
B. Ziegler, G. Rauhut, Efficient generation of sum-of-products representations of high-dimensional potential energy surfaces based on multimode expansions. J. Chem. Phys. 144, 114114 (2016)
D.G. Truhlar, C.J. Horowitz, Functional representation of Liu and Siegbahn’s accurate ab initio potential energy calculations for H+H2. J. Chem. Phys. 68, 2466–2476 (1978)
T.C. Thompson, G. Izmirlian, S.J. Lemon, D.G. Truhlar, C.A. Mead, Consistent analytic representation of the two lowest potential energy surfaces for Li3, Na3, and K3. J. Chem. Phys. 82, 5597–5603 (1985)
K.A. Nguyen, I. Rossi, D.G. Truhlar, A dual-level shepard interpolation method for generating potential energy surfaces for dynamics calculations. J. Chem. Phys. 103, 5522–5530 (1995)
S. Manzhos, T. Carrington Jr., A random-sampling high dimensional model representation neural network for building potential energy surfaces. J. Chem. Phys. 125, 084109 (2006)
K.T. Schütt, P.-J. Kindermans, H.E. Sauceda, S. Chmiela, A. Tkatchenko, K.R. Müller, SchNet: A continuous-filter convolutional neural network for modeling quantum interactions. Proceedings of the 31st international conference on neural information processing systems. Red Hook, NY, USA, 2017; pp 992–1002
K.T. Schütt, H.E. Sauceda, P.-J. Kindermans, A. Tkatchenko, K.-R. Müller, Schnet-a deep learning architecture for molecules and materials. J. Chem. Phys. 148, 241722 (2018)
K. Schutt, P. Kessel, M. Gastegger, K. Nicoli, A. Tkatchenko, K.-R. Müller, SchNetPack: a deep learning toolbox for atomistic systems. J. Chem. Theory Comput. 15, 448–455 (2018)
J. Behler, M. Parrinello, Generalized neural-network representation of high-dimensional potential-energy surfaces. Phys. Rev. Lett. 98, 146401 (2007)
J. Behler, Representing potential energy surfaces by high-dimensional neural network potentials. J. Phys. 26, 183001 (2014)
J. Behler, Perspective: Machine learning potentials for atomistic simulations. J. Chem. Phys. 145, 170901 (2016)
L. Zhang, J. Han, H. Wang, W. Saidi, R.E. Car, End-to-end symmetry preserving inter-atomic potential energy model for finite and extended systems. Advances in Neural Information Processing Systems. 2018
J.S. Smith, O. Isayev, A.E. Roitberg, ANI-1: an extensible neural network potential with DFT accuracy at force field computational cost. Chem. Sci. 8, 3192–3203 (2017)
O.T. Unke, M. Meuwly, PhysNet: a neural network for predicting energies, forces, dipole moments, and partial charges. J. Chem. Theory Comput. 15, 3678–3693 (2019)
B. Anderson, T.S. Hy, R. Kondor, Cormorant: covariant molecular neural networks. Adv. Neural Inf. Process. Syst. 32, 14537–14546 (2019)
J. Klicpera, J. Groß, S. Günnemann, Directional message passing for molecular graphs. International conference on learning representations. 2019
M.A. Wood, A.P. Thompson, Extending the accuracy of the SNAP interatomic potential form. J. Chem. Phys. 148(2018)
C.K. Williams, C.E. Rasmussen, Gaussian Processes for Machine Learning, vol. 2 (MIT Press, Cambridge, MA, 2006)
S. De, A.P. Bartók, G. Csányi, M. Ceriotti, Comparing molecules and solids across structural and alchemical space. Phys. Chem. Chem. Phys. 18, 13754–13769 (2016)
P. Virtanen, R. Gommers, T.E. Oliphant, M. Haberland, T. Reddy, D. Cournapeau, E. Burovski, P. Peterson, W. Weckesser, J. Bright, S.J. van der Walt, M. Brett, J. Wilson, K.-J. Millman, N. Mayorov, A.R.J. Nelson, E. Jones, R. Kern, E. Larson, C.J. Carey, I. Polat, Y. Feng, E.W. Moore, J. VanderPlas, D. Laxalde, J. Perktold, R. Cimrman, I. Henriksen, E. Quintero, A.C.R. Harris, A.M. Archibald, A.H. Ribeiro, F. Pedregosa, P. van Mulbregt, ciPy 1.0 Contributors, SciPy 1.0: fundamental algorithms for scientific computing in python. Nature Methods 2020, 17, 261–272
C.L. Lawson, R.J. Hanson, Solving least squares problems (SIAM, Philadelphia, 1995)
R. Meyer, A.W. Hauser, Geometry optimization using Gaussian process regression in internal coordinate systems. J. Chem. Phys. 152, 084112 (2020)
L. Himanen, M.O.J. Jäger, E.V. Morooka, F. Federici Canova, Y.S. Ranawat, D.Z. Gao, P. Rinke, A.S. Foster, DScribe: library of descriptors for machine learning in materials science. Comput. Phys. Commun. 247, 106949 (2020)
H.W. Kuhn, The Hungarian method for the assignment problem. Naval Res. Logist. Q 2, 83–97 (1955)
A.G. Baydin, B.A. Pearlmutter, A.A. Radul, J.M. Siskind, Automatic differentiation in machine learning: a survey. J. Mach. Learning Res. 18, 1–43 (2018)
A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury, G. Chanan, T. Killeen, Z. Lin, N. Gimelshein, L. Antiga, A. Desmaison, A. Kopf, E. Yang, Z. DeVito, M. Raison, A. Tejani, S. Chilamkurthy, B. Steiner, L. Fang, J. Bai, S. Chintala, In Advances in Neural Information Processing Systems 32, H. Wallach, H. Larochelle, A. Beygelzimer, F. d’ Alché-Buc, E. Fox, R. Garnett, (Eds.), Curran Associates, Inc., pp. 8024-8035 (2019)
J. Bradbury, R. Frostig, P. Hawkins, M. Johnson, J.C. Leary, D. Maclaurin, G. Necula, A. Paszke, J. VanderPlas, S. Wanderman-Milne, Q. Zhang, JAX: composable transformations of Python+NumPy programs. 2018; http://github.com/google/jax
J. Baker, F. Chan, The location of transition states: a comparison of Cartesian, Z- matrix, and natural internal coordinates. J. Comput. Chem. 17, 888–904 (1996)
A.H. Larsen, J.J. Mortensen, J. Blomqvist, I.E. Castelli, R. Christensen, M. Dułak, J. Friis, M.N. Groves, B. Hammer, C. Hargus, E.D. Hermes, P.C. Jennings, P.B. Jensen, J. Kermode, J.R. Kitchin, E.L. Kolsbjerg, J. Kubal, K. Kaasbjerg, S. Lysgaard, J.B. Maronsson, T. Maxson, T. Olsen, L. Pastewka, A. Peterson, C. Rostgaard, J. Schiøtz, O. Schütt, M. Strange, K.S. Thygesen, T. Vegge, L. Vilhelmsen, M. Walter, Z. Zeng, K.W. Jacobsen, The atomic simulation environment—a Python library for working with atoms. J. Phys. 29, 273002 (2017)
J.J. Stewart, Optimization of parameters for semiempirical methods VI: more modifications to the NDDO approximations and re-optimization of parameters. J. Mol. Modeling 19, 1–32 (2013)
J.J.P. Stewart, MOPAC2016. Stewart Computational Chemistry, Colorado Springs, CO, USA (2016)
I. Fdez Galván, G. Raggi, R. Lindh, Restricted-variance constrained, reaction path, and transition state molecular optimizations using gradient-enhanced kriging. J. Chem. Theory Comput. 17:571–582
M.W. Walker, L. Shao, R.A. Volz, Estimating 3-D location parameters using dual number quaternions. CVGIP: Image Understanding 54, 358–367 (1991)
J.C. Kromann, Calculate Root-mean-square deviation (RMSD) of Two Molecules Using Rotation. 2021; Software available from http://github.com/charnley/rmsd,v1.4
L. Biewald, Experiment Tracking with Weights and Biases. (2020) https://www.wandb.com/, Software available from wandb.com
Acknowledgements
This work was supported by Oracle Labs and by the Schiller Institute Grant for Exploratory Collaborative Scholarship (SIGECS). The authors thank Darius Russell Kish, Weiming Qin and Yang Wang for feedback on usage of mad-GP. Finally, the authors thank the Boston College Linux Cluster for computing resources.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, D., Teng, C., Bao, J.L. et al. mad-GP: automatic differentiation of Gaussian processes for molecules and materials. J Math Chem 60, 969–1000 (2022). https://doi.org/10.1007/s10910-022-01334-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10910-022-01334-x