Abstract
The reaction of potassium tetracyanocupprate(I) with triethyltin bromide in presence of phenanthroline (Phen) and quinoxaline (Qox) in acetonitrile via ultrasonic radiation yielded two new nano-supramolecular coordination polymers (SCP) of the type: [Cu2(CN)4(Et3Sn)2(Phen)2] SCP1 and [Cu2(CN)3(Et3Sn)(Qox)] SCP2. The obtained polymers were characterized using elemental and thermal analyses, as well as FT-IR, UV–vis, fluorescence, 1H NMR, and MS spectroscopes. Spectral and analytical features led to the conclusion that both Phen and Qox behave as bidentate ligands and the proposed formulae of SCP1 and SCP2 have the bimetallic nature, in which the geometry around Cu(I) atoms in SCP1 is tetrahedral and planar trigonal. While in SCP2, the Cu(I) has a trigonal plane geometry. Thermal analysis studies indicated that both SCP1 and SCP2 are thermally stable up to 140 °C. Additional confirmation for the structures of SCP1 and SCP2 was obtained from density functional theory (DFT) and molecular mechanics (MM+) calculations. Transmission electron microscopy (TEM) images of SCP1 and SCP2 show regular spherical-like nano-sized particles in the range of 31.24–62.13 nm and 5.16–23.90 nm, respectively. The inhibitory anti-oxidant activities of both SCP1 and SCP2 were investigated using erythrocyte hemolysis and ABTS methods and their cytotoxicity activities towards different tumor cells were also studied by MTT assay. The results showed that both the two polymers have high inhibitory anti-oxidant activities and SCP2 exhibits a significant decrease in surviving fraction of these cancer cell lines compared to SCP1.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Structural variability is one of the most important properties of supramolecular organometallic compounds. There are number of factors contribute to structural variability, for example: number and type of ligands and their combinations as well as the coordination geometry, the number of metal atoms and the presence of ionic charge [1]. Organometallic polymers have unique properties and applications as they depend on their organic segments and the types of metal compounds embedded in their back bones or side chains [2].
Organotin(IV) compounds are characterized by the presence of at least one covalent C–Sn(IV) bond and classified, depending on the number of alkyl (R) or aryl (Ar) moieties into mono-, di-, tri- and tetraorganotin(IV). Organotin(IV) compounds display strong biological activities [3,4,5,6,7,8,9], in which these activates are determined by the number and nature of the organic groups bound to the central stannous atom. Membrane proteins are among the most fascinating structures in biology [10]. The moiety RnSn(IV)(4 − n)+ (n = 2 or 3) may be bound to the membrane proteins, glycoproteins, or cellular proteins; e.g. Et2Sn(IV)2+ to ATPase and hexokinase [11] and the moieties Bu2Sn(IV)2+ and Bu3Sn(IV)+ may be bound to ATPase and acetyl cholinesterase of human erythrocyte membrane [12, 13]. While the moiety Bu2Sn(IV)2+ may be bound to the skeletal muscle membranes. Here, the [Cun(CN)m]x− building block are used for the construction of multi-dimensional supramolecular coordination polymers (SCP) by incorporating the cation R3Sn+ as an essential element for connecting the framework.
Ultrasonic irradiation is considered to be a facile and environmentally friendly energy source, which can provide rather unusual reaction conditions rather the traditional energy sources [14]. These sever conditions can drive the chemical reactions and thus catalyze the formation of nanostructured materials. This strategy supports the formation of uncharged nano-sized structure, in which the channels are remain blocked by the R groups.
Organotin(IV) compounds have been receiving increasing attention for investigating their antitumor activity and showed that tri-organotin(IV) compounds have high potentials as antineoplastic and anti-tuberculosis agents than mono- and di-organotin (IV) analogs due to their ability to bind proteins [15, 16]. Organotin(IV) derivatives have been found to possess anticancer activity on different tumor cell lines and the structures of these organotin(IV) complexes are well characterized in the solid state [17, 18]. Invention of improved drugs become more and more urgent in cancer treatment, due to the fact that most of cancer cells can subsequently survive and gain resistance after initially effective chemotherapy. Discovery of the anti-proliferative and antitumor activity of cisplatin and its derivatives initiated the investigation for the possible therapeutic applications of other metal-based drugs, often organometallic compounds [19,20,21,22,23,24,25], and more particularly organotin(IV) compounds [26,27,28].
Since a limited number of structural motifs [Cun(CN)m(R3Sn).L] where L = bipodal organic ligand [29,30,31,32] is known, the present research work describes the design, synthesize and spectral properties of new fascinating [CuCN]-based nano-supramolecular coordination polymers (SCP) namely, [Cu2(CN)4(Et3Sn)2(Phen)2] SCP1 and [Cu2(CN)3(Et3Sn)(Qox)] SCP2. Phenanthroline (Phen) and quinoxaline (Qox) are chosen as auxiliary N-donor ligands based on the fact that they are rigid, which behave as double-connected ligands and the cation Et3Sn+ acts as a connecting unit. On the other hand, the organometallic (R3M)+ (R = Me or Ph, M = Sn or Pb) unit can act as a connecting moiety by axial anchoring to two donor atoms, exhibiting trigonal bipyramidal configuration, creating structure of high dimensionality [33,34,35]. The inhibitory anti-oxidant activities of both SCP1 and SCP2 were investigated and also the cytotoxicity activities towards different tumor cells (HePG2, MCF-7, PC3, and HCT-116) were also studied.
2 Experimental
2.1 Materials and Instrumentations
Potassium tetracyanocupprate(I) {K3[Cu(CN)4]} was prepared following the literature procedure [9]. Triethyltin bromide and the ligands phenanthroline (Phen) and quinoxaline (Qox) ligands were purchased from Aldrich and Sigma and were used without purification. Techniques and equipment’s, utilized in this study were the same as those used in our previous work [9].
2.2 General Synthesis of [Cu2(CN)4(Et3Sn)2(Phen)2] SCP1 and [Cu2(CN)3(Et3Sn)(Qox)] SCP2
Ultrasonic synthesis of SCP1 and SCP2 had been carried out in an ultrasonic bath using ultrasonic generator 60 W for 70 min as follows: an aqueous solution (10 mL) containing K3[Cu(CN)4] (0.074 mmol, 22 mg) was added, under a gentle stirring to a solution of Et3SnBr (0.23 mmol, 0.04 mL) in acetonitrile (10 mL), followed by Phen (0.074 mmol, 15 mg) or Qox (0.074 mmol, 10 mg) in 10 mL acetonitrile in an ultrasonic bath. A yellow precipitate started to form after 1 week. Filter, wash with cold water then acetonitrile, and drying in vacuo yielding SCP1 as yellow powder (48 mg, 81.35%). Anal. Calcd. for C40 H46N8Cu2Sn2 (MW = 1002.5 g mol−1): C, 47.88; H, 4.58; N, 11.17; Cu, 12.96. Found: C, 47.86; H, 4.51; N, 11.12; Cu, 12.87 and SCP2 (97 mg (78.22%). Anal. Calcd. for C17 H21N5Cu2Sn (MW = 542 g mol−1): C, 37.63; H, 3.87; N, 12.91; Cu, 23.98. Found: C, 37.58; H, 3.82; N, 12.87; Cu, 23.91.
2.3 Biological Applications
2.3.1 Antioxidant Activity
The assay employs the radical cation derived from 2, 2′-azino-bis (3-ethyl benzthiazoline-6-sulfonic acid) (ABTS) as a stable free radical to assess antioxidant and extracts.
2.3.1.1 ABTS Free Radical Scavenging Activity
The reaction mixture (negative control) consists of 2 mL of ABTS solution (60 µL) and 3 mL of MnO2 solution (25 mg mL−1), all prepared in phosphate buffer (pH 7) [36,37,38]. The mixture is shaken, centrifuged and filtered to remove the excess oxide. The absorbance (Acontrol) of the resulting green-blue solution (ABTS+ radical solution) is recorded at λmax = 734 nm. The absorbance (Atest) was measured upon the addition of 20 µL of 1 mg mL−1 solution of the test sample under investigation in spectroscopic grade MeOH/buffer (1:1 v/v) to the ABTS solution. Ascorbic acid 20 µL (2 mL) solution was used as standard antioxidant (positive control). Blank sample was run using solvent without ABTS. The decrease in absorbance is expressed as % inhibition.
2.3.1.2 Antioxidant Activity Screening Assay for Erythrocyte Hemolysis
The blood was obtained from rats by cardiac puncture and collected in heparinized tubes. Erythrocytes were separated from plasma and the buffy coat was washed three times with 10 volumes of 0.15 M NaCl. During the last wash, the erythrocytes were centrifuged at 3000 rpm min−1 for 7 min to obtain a constantly packed cell preparation. Erythrocyte hemolysis was mediated by peroxyl radicals in this assay system [39]. A 10% suspension of erythrocytes in phosphate buffered saline pH 7.4 (PBS) was added to the same volume of 200 mM AAPH solution in PBS containing samples to be tested at different concentrations. The reaction mixture was shaken gently while being incubated at 37 °C for 2 h. The reaction mixture was then removed, diluted with eight volumes of PBS and was centrifuged at 1500 × g for 10 min. The absorbance of the supernatant was read at 540 nm. Similarly, the reaction mixture was treated with 8 volumes of distilled water to achieve complete hemolysis, and the absorbance of the supernatant obtained after centrifugation was measured at 540 nm. l-ascorbic acid was used as a positive control.
2.4 Cytotoxic Activity
2.4.1 Cell Lines
Four human tumor cell lines namely; hepatocellular carcinoma HePG-2, mammary gland breast cancer MCF-7, human prostate cancer PC3 and colorectal carcinoma HCT-116 were obtained from ATCC via Holding company for biological products and vaccines (VACSERA), Cairo, Egypt.
2.4.2 Chemical Reagents
The reagents RPMI-1640 medium, MTT, DMSO and 5-fluorouracil (Sigma Co., St. Louis, USA), fetal bovine serum (GIBCO, UK). 5-Fluorouracil was used as a standard anticancer drug for comparison.
2.4.3 Cell Proliferation (MTT Assay)
The different cell lines mentioned above were used to determine the inhibitory effects of the tested compound on cell growth using the MTT assay [40, 41]. This colorimetric assay is based on the conversion of the yellow tetrazolium bromide (MTT) to a purple formazan derivative by mitochondrial succinate dehydrogenase in viable cells. The cells were cultured in RPMI-1640 medium with 10% fetal bovine serum. Antibiotics added are 100 units mL−1 penicillin and 100 µg mL−1 streptomycin at 37 °C in a 5% CO2 incubator. The cells were seeded in a 96-well plate at a density of 1.0 × 104 cells/well at 37 °C for 48 h under 5% CO2. After incubation, the cells were treated with different concentrations of the tested drugs and were incubated for 24 h. After 24 h of drug treatment, 20 µL of MTT solution at 5 mg mL−1 was added and was incubated for 4 h. Dimethyl sulfoxide (DMSO) in volume of 100 µL was added into each well to dissolve the purple formazan formed. The colorimetric assay was measured and recorded at absorbance of 570 nm using a plate reader (EXL 800, USA). The relative cell viability in percentage was calculated as (A570 of treated samples/A570 of untreated sample) × 100. The mean IC50 was the concentration of agent that reduces cell growth by 50% under the experimental conditions and is the average from at least three independent determinations. The 50% inhibition concentration (IC50) was determined by curve fitting.
3 Results and Discussion
3.1 Characterization of [Cu2(CN)4(Et3Sn) 2(Phen)2]SCP1 and [Cu2(CN)3(Et3Sn)(Qox)] SCP2
The nano-sized particles of SCP1 and SCP2 have been synthesized by sono-chemical method. The analytical results demonstrate that SCP1 and SCP2 have the formulae [Cu2(CN)4(Et3Sn)2(Phen)2] and [Cu2(CN)3(Et3Sn)(Qox)] (Phen = phenanthroline and Qox = quinoxaline), respectively (Scheme 1 ).The obtained structures have been also analyzed by different spectroscopic techniques, which are compatible to the computational mechanical calculations. Both SCP1 and SCP2 are not dissolved in water and most organic solvents while they are dissolved in DMSO and DMF at 60 °C. The magnetic moment values of SCP1 and SCP2 confirmed their diamagnetism, indicating the presence of Cu(I). The molar conductance values of 11.6 and 10.2 Ohm−1 cm2 mol−1 for SCP1 and SCP2, respectively indicate their non-electrolytic nature.
3.2 FT-IR Spectra
The IR spectra of SCP1 and SCP2 indicate the presence of Phen, Qox, Et3Sn, and CuCN fragments (Fig. 1). The presence of the bipodal ligands Phen and Qox in the structures of both SCP1 and SCP2 is realized by υ(CH) vibration bands at 3068 and 3040 cm−1, δ(CH) at 1422 and 1375 cm−1, and δ(CH) at 841–866 and 724–755 cm−1, respectively (Table 1). These bands are shifted to lower wavenumbers compared to those of free ligands Phen and Qox. This is due to the formation of hydrogen bonds between hydrogen atoms of the ligands and the donor sites of the CuCN fragments. The vibration bands in the range of 1627–1620 cm−1, corresponding to υ(C=N) are shifted to lower frequencies from those of the ligands. This supports the coordination of the ligands Phen or Qox to the copper centers. Thus both Phen and Qox may act as bridges connecting the non-linear chains via the copper atoms. In consequence of bridging tetrahedral Cu[N2(Phen)(CN)2] building blocks by the Et3Sn units via coordinate bonds in SCP1 and bridging trigonal plane Cu[N(Qox)(CN)2] building blocks by the Et3Sn units via coordinate Cu ← CN → Sn bonds in SCP2, one can realize the presence of υ(CN) vibration band at 2110 and 2093 cm−1 for SCP1 and at 2120 and 2102 cm−1 for SCP2. The stretching frequencies of these vibration bands are at higher wavenumbers than the band of the genuine salt of the corresponding [Cu(CN)4]3−anion (2076 cm−1) [42], which contains non-bridged cyanide groups. The presence of two cyanide bands indicates the presence of two types of cyanide ligands, where the Cu2 atom in SCP1 may form planar trigonal configuration rather than the tetrahedral structure of Cu1 (Table 1). On the other hand, the structure of SCP2 contains two different cyanide groups, one is coordinated to one Cu atom and one Sn atom. While the other one is coordinated to two Cu atoms (Fig. 1). The presence of Et3Sn units is supported by the observation of υ(CHEt) stretching vibration bands at 2965, 2942, 2868, and 2825 cm−1, δ(CHEt) at 1415 and 1359 cm−1, as well as γ(CHEt) at 768 and 680 cm−1 [43]. A υ(Sn–C) stretching vibration band was also observed at 525 cm−1, indicating the presence of trigonal plane of Et3Sn units. The presence of υ(Cu–C) vibration band at 450 cm−1 [43] also confirms the presence of the (CuCN) fragments.
3.3 1H NMR Spectra
The 1H NMR spectrum of SCP1 showed four broad peaks at 7.60, 8.21, 8.68, and 8.71 ppm, which correspond to aromatic protons of the coordinated Phen (Fig. 2). These are in accord with the 1H NMR spectrum of free Phen, which exhibits sharp and broad peaks at 7.26, 7.55, 8.0, and 8.81 ppm. The 1H NMR spectrum of the SCP2 (S1) showed one singlet peak at 8.93 ppm, which is assigned to the two chemically and magnetically equivalent protons of the CH=N moiety. While the aromatic protons of the coordinated Qox give rise to two broad multiple peaks in the range of 8.09–8.11 and 7.87–7.78 ppm. These assignments were also in accord with 1H NMR spectrum of the free Qox, which exhibits sharp singlet and two multiple peaks at 8.92, 8.04–8.09 and 7.82–7.84 ppm. This significant change in chemical shifts supports the participation of both Phen and Qox ligands in coordination to the Cu centers and the formation of hydrogen bonding.
The 1H NMR spectrum of SCP1 also showed a triplet peak at 1.22 ppm, corresponding to the methyl protons of the ethyl ligand. A strong peak was observed at 2.51 ppm, corresponding to the methylene of the ethyl ligand with two satellite peaks at 2.43 and 2.70 ppm. On the other hand, the protons of the (Et3Sn) units give rise to the quartet peak at 1.46–1.20 ppm and the triplet peak at 1.12–1.04 ppm corresponding to CH2 and CH3 groups, respectively, in the 1H NMR spectrum of 2. The presence of these satellite peaks can be considered as good indication for the presence of Et3Sn connecting units.
13C NMR spectra of SCP1 and SCP2 showed strong signals corresponding to the coordinated Phen, Qox ligands, ethyl, and cyanide ligands (Fig. 2). There are three peaks at 131.12, 150.65 and 151.03 ppm, corresponding to the carbon nuclei of the ligands. The cyanide groups exhibit two signals at 143.97 and 144.85 ppm and one signal around 121.83 ppm, indicating the presence of two kinds of the cyanide ligands as also supported by IR spectral data. The multiple peaks at 8.39–13.36 ppm correspond to the ethyl ligands. Thus, 1H- and 13C NMR spectra display the expected proton and carbon signals that support the structures of both SCP1 and SCP2.
3.4 Electronic-Emission Spectra
The electronic absorption spectra of Phen and Qox resemble the spectra of the coordinated polymers SCP1 and SCP2. Substitution of nitrogen for methane in Phen and Qox, just as in naphthalene, produces only relatively small spectroscopic changes (Table 2). The short wavelength band in phenanthrene with a peak at 200 nm, is assigned to the 1Ba ← 1 A transition. The longitudinally polarized 1Bb ← 1A band at 225 nm resembles that observed in the spectrum of phenanthrene at 221 nm [44]. The broad band at 260 nm can be considered as a composite one, since the 1Lb band was submerged under the 1La band like the case of anthracene [44]. This band exhibits blue shift than that observed in the spectra of naphthalene (286 nm) and anthracene (310 nm) [43]. Thus, this broad band can be assigned to 1La ←1A and 1Lb ←1A transitions. The last long wavelength band at 285 nm is weak and corresponds to n–π* transition which disappears on the addition of HCl which blocks the lone pairs of electrons via protonation. The spectrum of SCP1 displays the same bands of phenanthroline at more or less the same positions (Table 2), except the n–π* band which disappears indicating participation of phenanthroline in the coordination sphere of copper. Also, the 1Lb ← 1A band suffers slight red shift (275). Unmodified Phen is a weakly emissive compound but, following several routes, a wide range of highly luminescent phenanthroline compounds with emission bands ranging from the UV to the near infrared region can be obtained [45]. Phenanthroline exhibits close π–π* and n–π* singlet excited states and the emission originates mainly from the former [45]. Excited states n–π* often decay via non-radiative bath ways, and they are usually characterized by vanishingly low emission quantum yields. Phenanthroline itself is characterized by a weak fluorescence quantum yield (Фfl. = 0.0087) and a short singlet lifetime (t < 1 ns) at room temperature [46]. The emission spectrum of Phen displays a well-developed band at 370 nm and a shoulder at 430 (Fig. 3a). The first band corresponds to the π–π* transition. The low-energy, delayed emission band at 430 nm can be assigned to the triplet excimer phosphorescence of Phen. This assignment is substantiated by reference to the excimer phosphorescence of naphthalene, quinoxaline, and phenanthroline [47]. On the other hand, the emission spectrum of SCP1 displays a well-developed band at 375 nm and two broad bands at 425 and 480 nm. The structure of SCP1 contains the CuCN fragment and Phen, both of them luminescent at λmax = 390 nm [48]. The amine bearing CuCN complexes emit in the visible region while their photophysical behavior appears to be close to that of CuCN itself [49]. The emission spectrum of CuCN is assumed to arise from transition between the lowest triplet excited state and the ground state. It is also supposed that a bent triplet state is responsible for the relatively high energy CuCN emission [50]. Photophysical properties of Phen, as well as that of CuCN, can be tuned by formation of SCP. Thus, the emission band of SCP1 at 375 nm corresponds to close lying π–π* transition of Phen. The broad band at 425 nm can be assigned to the triplet excimer phosphorescence of Phen and to So–T transition in the CuCN fragments which suffers a shift to longer wavelength by 35 nm than that of CuCN itself. Thus, the luminescence behavior of Phen shows excellent sensitivity towards copper which makes it attractive as a luminescent sensor. The broad, low energy band at 480 nm can be assigned to the metal to ligand charge transfer, MLCT [51]. However, due to the broadening of this band, the possibility exists of single-metal-centered (MC) transitions of the type 3d10 → 3d9 4s1 and 3d10 → 3d9 4p1 on the Cu(I) center.
The electronic absorption spectra of Qox in absence and presence of HCl display four absorption bands at 210, 230, 265 and 312 nm (Table 2). The band at 210 nm is due to 1Ba ← 1A which is red shifted than that of naphthalene (170 nm) [44]. The band at 230 nm is due to 1Bb ← 1A while those at 265 and 312 nm correspond to 1La ← 1A and 1Lb ← 1A transitions, respectively As it is expected that the π*-levels are appreciably lowered in Qox and that the n–π* transitions are shifted far enough to the red to make them discernible. The electronic absorption spectrum of SCP2 in solid state display two broad composite bands at 240–245 nm and 300–320 nm (Table 2). The first band corresponds to 1Bb ← 1A and MLCT transitions while the second one is attributed to the 1La ← 1A and 1Lb ← 1A transitions, respectively. In this case the 1La band suffers a red shift under the effect of the coordination to the copper atom. The emission spectra of Qox and the SCP2 show emission bands at wavelengths, between 380 and 535 nm (Table 2; Fig. 3b). The emission spectrum of Qox in the solid state displays a well-developed band at 375 nm due to π–π* transition, a shoulder at 430 nm due to the triplet excimer phosphorescence of Qox and a shoulder at 480 nm due to 1(n, π*) → So fluorescence of Qox. [52]. This assignment was substantiated by reference to the excimer phosphorescence of naphthalene, quinoxaline, and phenanthroline [51]. On the other hand, the emission band of SCP2 at 400 nm corresponds to close lying π–π* transition of Qox. The broad band at 450 nm can be assigned to the triplet excimer phosphorescence of Phen and to So–T transition in the CuCN fragments which suffers a shift to longer wavelength by 40 nm than that of CuCN itself [49, 50]. Thus, the luminescence behavior of SCP1 shows excellent sensitivity towards copper which makes it attractive as a luminescent sensor. The broad, low energy band at 530 nm can be assigned to the metal to ligand charge transfer (MLCT). However, due to the broadening of this band, the possibility exists of single-metal-centered (MC) transitions of the type 3d10 → 3d9 4s1 and 3d10 → 3d9 4p1 on the Cu(I) center.
3.5 Mass Spectra
SCP1 and SCP2 were also characterized by mass spectrometry (Tables 3, 4). The positive mass spectra of SCP1 exhibits base peaks at m/z = 180 and 178, corresponding to the molecular ions of Phen+ and Et2Sn+, respectively. On the other hand, an ion peak is observed at m/z = 130, corresponding to the molecular ion of Qox+. Fragmentation of Qox+ ion gives rise to peaks at m/z = 76 and 103, which is due to C6H4+ and C7H5N+, respectively. The ion peaks observed in the positive mass spectra of SCP1 and SCP2 at m/z = 289–973 are due to different fragments corresponding to [(Et3Sn)m(Cu)n(CN)x]+ and [(Et3Sn)m(Cu)n(CN)x(L)z]+, L = Phen or Qox, supporting the structure and polymeric nature of SCP1 and SCP2. The mass spectra also exhibit ion peaks at m/z = 802 and 541, which are attributed to the molecular formula of [Cu2(CN)4(Et3Sn)2(Phen)2]+ and [Cu2(CN)3(Et3Sn)Qox]+ ≡ [M]+, respectively. The presence of these ion peaks can be considered as a further support of the structure suggested by elemental analysis data. On the other hand, the negative mass spectra of SCP1 and SCP2 exhibit ion peaks of [Cun(CN)n+1]− and [CN]− units, respectively. The base peaks at m/z = 204 and 230, corresponding to [Cu2(CN)4]− and [Cu2(CN)3]− of the molecular ions represent the building blocks of SCP1 and SCP2, respectively. Thus, the mass spectra of SCP1 and SCP2 confirm the presence of the bipodal ligands Phen and Qox, as well as Et3Sn and CuCN fragments. Also, it supports the polymeric nature of the [Cu2(CN)3] building blocks.
3.6 Thermal Analysis
To evaluate the thermal stability of both SCP1 and SCP2, thermogravimetric analyses (TGA) were performed from 30 to 800 °C under N2. TGA thermograms of SCP1 and SCP2 (Fig. 4) showed that they intact up to 110 and 140 °C, respectively. Both polymers undergo decomposition in four steps (Table 5). The first decomposition step in SCP1 takes place in the range of 120–220 °C, corresponding to the decomposition of the fragments of (Et3Sn)2 + Phen (Found: 591.44 g mol−1 and Calc.: 591.40 g mol−1). This step occurs in two successive processes as indicated by the DrTGA curve. The first one corresponds to the release of the more volatile connecting units (Et3Sn)2 followed by the release of one Phen ligand. The second decomposition step takes place in the range of 240–280 °C, attributing to the loss of three cyanide ligands (Found: 77.84 g mol−1 and Calc.: 78.00 g mol−1). The third decomposition step takes place in the range of 380–460 °C, corresponding to the weight loss of one Phen ligand, one cyanide ligand, and two copper atoms (Found: 333.03 g mol−1 and Calc.: 333.08 g mol−1). Thus, the thermogram of SCP1 indicates the presence of Et3Sn, CN, Phen and copper atoms in its structure. On the other hand, the first decomposition step of SCP2 plays in the range of 140–210 °C, attributing to the decomposition of three ethyl ligands. This step is followed by the loss of Qox ligand and two cyanide molecules in the temperature range of 220–270 °C. The third step corresponds to the loss of one CuCN fragment in the temperature range of 280–370 °C. Over 600 °C, a complete decomposition occurs producing Cu and Sn (Found: 182.50 g mol−1 and Calc.: 182.95 g mol−1) (Table 5).
3.7 Transmission Electron Microscopy (TEM)
The transmission electron microscopy (TEM) images of SCP1 and SCP2 showed that they have regular spherical-like nanostructures with nanoparticle size. As many as possible diameters were measured to estimate the nanoparticles size distribution and the diameter range of the particles which are in the range of 31.24–62.13 nm and 5.16–23.90 nm, respectively (Fig. 5).
3.8 Molecular Modeling
Cluster calculations are performed using DMol3 program [53,54,55] in Materials Studio package [56], which is designed for the realization of large scale density functional theory (DFT) calculations. DFT semi-core pseudopods calculations (dspp) are performed with the double numerical basis sets plus polarization functional (DNP). The DNP basis sets are of comparable quality to 6-31G Gaussian basis sets [57]. Delley et al. showed that the DNP basis sets are more accurate than Gaussian basis sets of the same size [55]. The RPBE functional [58] is so far the best exchange–correlation functional [59] based on the generalized gradient approximation (GGA) which is employed to take account of the exchange and correlation effects of electrons. The geometric optimization is performed without any symmetry restriction.
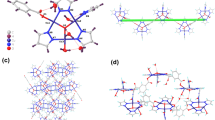
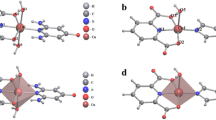
The optimized molecular structure of SCP1 with minimum energy obtained from the calculations is shown in Fig. 6. The Cu(1) and Cu(24) centers acquire N2PhenNCNCCN and N2PhenCCN geometries. The energy minimization results indicated that the coordination environments around the CuI centers are distorted tetrahedral (DT-4) and trigonal plane (TP-3) geometries as also indicated by the bond lengths and angles (Table 6). The distortion is caused due to the different N-atoms coordinated to the Cu-atom that come from the cyanide and Phen ligands. The nitrogen atoms of the Phen ligands cause acute angles N17–Cu1–N18 (83.74°) and N34–Cu24–N35 (90.80°). The bond distances and bond angles are in the normal reported range [58]. The tin atom (Sn6) assumes trigonal bipyramid (TBPY-5) geometry where two ethyl groups (C41, C42) and one cyanide (C39) orient themselves toward the corners of a distorted TP-3. The nitrogen atom of the cyanide group (N4) and the ethyl carbon atom (C44) occupy axial positions which exhibit nearly right angles (Table 6), with respect to the Et3Sn plane, however, the N4–Sn6–C44 angle equals 171.27° which reflects a bent structure of the (CN–Ete3Sn–NC) connecting unit. On the other hand, the tin atom (Sn7) is coordinated to thee ethyl groups forming a distorted TP-3 and two cyanide ligands in axial positions via coordinate bonds forming TBPY-5 geometry. The N22–Sn7–C5 angle is 144.62° exhibiting more bent structure of the (NC–Et3Sn–NC) connecting unit than the case of Sn6 and reflects the fact that the ethyl groups occupy perpendicular positions out of the plane defined by NC–Sn–NC spacer. The structure of 1 may extend to form 3D-SCP network via the (CN–Ete3Sn–NC) connecting units and H-bonds.
The molecular structure along with atom numbering and HOMO–LUMO representation of the SCP2 are shown in Fig. 7. Analysis of the bond lengths and bond angles are listed in Table 7. The Cu atoms acquire slightly distorted trigonal plane geometry where they coordinate to two ordered cyanide groups and one nitrogen atom of Qox ligand. The Sn atom coordinates to three ethyl groups and two cyanide groups creating TBPY-5 configuration (Table 7). The Cu–CN bond angles exhibit slight bent structures which are more pronounced in case of Cu25 and Sn atoms (165.370°, Table 7). Bond lengths and bond angles are in the normal range reported for the proto-type compounds [30,31,32]. The cyanide ligands acquire different bond orders were two cyanide ligands connect one Cu atom (1.121–1021 Å) which is further supported by the presence of two υCN IR-bands at 2120 and 2102 cm−1. The photo-induced charge redistribution over the whole molecular skeleton of the SCP2 is well represented by plot of the Frontier molecular orbitals (HOMO and LUMO) as illustrated in Fig. 7b, c. EHOMO is a quantum chemical descriptor which is often associated with the electron donating ability of the molecule. High value of EHOMO indicates a tendency of the molecule to donate electrons to appropriate acceptor molecule of low empty molecular orbital energy. However, ELUMO indicates the ability of the molecule to accept electrons. The HOMO electronic density distribution for the SCP2 can be represented as localization of charge density on the (Et3Sn) fragment and the bridging cyanide ligand. In the case of LUMO level, the charge is mostly localized that can facilitate the donation from the cyanide ligand to Cu25 atom. Quantum chemical parameters (EHOMO, ELUMO and the energy gap, DE = ELUMO − EHOMO) are found to be − 5.197, − 3.343 and 1.854 eV, respectively. Thus, the cyanide ligands bridge the Cu and Et3Sn forming 1D-chains which further connected by the Qox and H-bonds creating 3D-network as also supported by mass spectra.
3.9 Antioxidant Activities
The SCP1 and SCP2 have been tested for the antioxidant activity using ABTS assay and rate erythrocyte hemolysis. In the case of ABTS assay (Table 8), the data indicated that the tested compounds exhibited potent anti-oxidative activity relative to ascorbic acid (89.4% inhibition) while SCP2 (65.0% inhibition) is significantly more active than SCP1 (65.0% inhibition) and is quite close to ascorbic acid. On the other hand, the results of erythrocyte hemolysis indicate that SCP1 and SCP2 has an excellent anti-hemolytic activity relative to vitamin-C and SCP2 is more active than SCP1 (Table 9).
3.10 Cytotoxic Activities
SCP1 and SCP2 were designed and synthesized to examine the effects on viability and proliferation of hepatocellular carcinoma HePG-2, mammary gland breast cancer MCF-7, human prostate cancer PC3 and colorectal carcinoma HCT-116 human cancer cell lines. Different concentrations of the tested compounds SCP1 and SCP2 are prepared and dissolved in complete RPMI-1640 media with 10% serum. Cell inhibition rates are determined by MTT assay. The results reveal that SCP1 and SCP2 able to inhibit the growth of the different cancer cells under investigation in a dose-dependent manner. A dose–response curves are also constructed in order to determine the effective dose, which may be used for subsequent experiments (Figs. 8, 9). The compound SCP1 shows a sigmoidal dose–response inhibition of cells viability, giving IC50 values of 16.1 ± 1.58 (HePG-2), 33.2 ± 2.95 (MCF-7), 36.5 ± 2.67 (PC3) and 14.0 ± 1.64 (HCT-116) µg mL−1, respectively (Table 10; Fig. 8). It is evident that the tested compound SCP1 has a significant inhibiting effect on HCT-116 and HePG-2 cell lines while it exhibits moderate inhibiting effect on MCF-7 and PC3 cell lines. On the other hand, the SCP2 is found to inhibit cell proliferation and induces very high % of apoptotic cell lines under investigation in vitro model compared with the control group, DOX and 5-Fu (Table 8; Fig. 9). The SCP2 exhibits sigmoidal dose-response inhibition of the investigated cell lines viability as the concentration of SCP2 was increased to 100 µM mL−1 giving very high % of apoptotic cell lines under investigation as also indicated by the low IC50 values of 4.14 ± 0.21 (HePG-2), 3.97 ± 0.17 (MCF-7), 11.12 ± 0.20 (PC3) and 5.08 ± 0.18 (HCT-116) µg mL−1 (Fig. 9). However, total inhibition was not found in vitro model, so that replication of the cancer cells is still possible. So, the tested promising drugs need in vivo investigation to evaluate the maximum tolerated dose and the number of injections which achieve complete inhibition of the tumor masses.
It is noticed that the SCP2 demonstrated inhibitory activities more than the SCP1. Growth inhibition of 50% (IC50) is calculated as the SCP1 and SCP2 concentrations, which caused a 50% reduction in cell proliferation during the drug incubation. The mean IC50 is the concentration of drug that reduces cell growth by 50% under the experimental conditions and is the average with ± SEM from at least three independent determinations. Thus, the tested SCP1 and SCP2 at a concentration of IC50 value decreased cell viability of the investigated cells compared to the control group but need more investigation in vitro and in vivo. For active Sn complexes, the average Sn–N bond lengths are > 239 pm, whereas inactive complexes had Sn–N bonds < 239 pm [60, 61], implying that pre-dissociation of the ligand may be important in the mode of action of these complexes, while coordinated ligand may favor transport of the active species to the site of action in the cells, where they are released by hydrolysis. The Sn–N bond length of SCP1 equals to 209 pm while SCP2 equals to 269 pm so, SCP2 is more active than SCP1 and it shows higher cytotoxicity than the well-known drug, DOX.
4 Conclusions
Two new hydrogen bonded bimetallic supramolecular coordination polymers, SCP1 and SCP2, have been synthesized and structurally characterized. Spectroscopic and structural features showed that their structures consist of (CN–Ete3Sn–NC) connecting units and [Cun(CN)m](m − n)− building blocks, which depend on the type of the Phen or Qox ligand, creating 3D-network structures. Taken together, the new SCP1 and SCP2 exhibit significant decrease in cell viability in HePG-2, MCF-7, PC3 and HCT-116 human cancer cell lines. Interestingly, the SCP2 was found to inhibit the cell proliferation and induces very high % of apoptotic cell lines under investigation in vitro model compared with the control group, DOX and 5-Fu. The tested compounds, especially SCP2, need more investigations to advance to pre-clinical development. Additional experiments are required to determine the exact mechanism of action of these new coordinated polymers that could be a hope to decrease different cancer invasion and metastasis.
References
D. Braga, F. Grepioni, Chem. Commun. (1996). https://doi.org/10.1039/CC9960000571
A.S. Abd-El-Aziz, P.O. Shipman, B.N. Boden, W.S. McNeil, Prog. Polym. Sci. 35, 714 (2010) and references therein
S. Silver, FEMS Microbiol. Rev. 27, 341 (2003)
M.J.M. Vaerewijck, G. Huys, J.C. Palomino, J. Swings, F. Portaels, FEMS Microbiol. Rev. 29, 911 (2005)
E.S. Raper, Coord. Chem. Rev. 153, 199 (1996)
S. Zartilas, S.K. Hadjikakou, N. Hadjiliadis, N. Kourkoumelis, L. Kyros, M. Kubicki, M. Baril, I.S. Butler, S. Karkabounas, J. Balzarini, Inorg. Chim. Acta 362, 1003 (2009)
M. Cavicchioli, A.C. Massabni, T.A. Heinrich, C.M. Costa-Neto, E.P. Abrảo, B.A.L. Fonseca, E.E. Castellano, P.P. Corbi, W.R. Lustri, C.Q.F. Leite, J. Inorg. Biochem. 104, 533 (2010)
E.E. Tredget, H.A. Shankowsky, A. Groenveld, R. Burrell, J. Burn Care Rehabil. 19, 531 (1998)
S.E.H. Etaiw, S.N. Abdou, A.A. Faheim, J. Coord. Chem. 68, 491 (2015)
K.R. Vinothkumar, R. Henderson, Q. Rev. Biophys. 43, 65 (2010)
K.R. Siebenlist, F. Taketa, Toxicol. Appl. Pharmacol. 58, 67 (1981)
A.A. Ali, R.K. Upreti, A.M. Kidway, Toxicol. Lett. 38, 13 (1987)
A.A. Ali, R.K. Upreti, A.M. Kidway, Bull. Environ. Contam. Toxicol. 44, 29 (1990)
J.H. Bang, K.S. Suslick, Adv. Mater. 22, 1039 (2010)
B.M. Elliot, W.N. Aldridge, J.W. Bridges, Biochem. J. 177, 461 (1979)
S.E.H. Etaiw, M.M. El-bendary, Spectrochim. Acta A 110, 304 (2013)
M. Gielen, E.R.T. Tiekink, Metallotherapeutic Drug and Metal-Based Diagnostic Agents: 50Sn Tin Compounds and Their Therapeutic Potential (Wiley, New York, 2005), pp. 421–439
N. Ogwuru, L.E. Khoo, G. Eng, Appl. Organomet. Chem. 12, 409 (1998)
G. Sava, G. Jaouen, E.A. Hillard, A. Bergamo, Dalton Trans. 41, 8226 (2012)
J.M. Hearn, I. Romero-Canelón, B. Qamar, Z. Liu, I. Hands-Portman, P.J. Sadler, ACS Chem. Biol. 8, 1335 (2013)
A. Hottin, D.W. Wright, A. Steenackers, P. Delannoy, F. Dubar, C. Biot, G.J. Davies, J.-B. Behr, Chem. Eur. J. 19, 9526 (2013)
M.M. Meier, C. Rajendran, C. Malisi, N.G. Fox, C. Xu, S. Schlee, D.P. Barondeau, B. Höcker, R. Sterner, F.M. Raushel, J. Am. Chem. Soc. 135, 11670 (2013)
M.A. Jakupec, M. Galanski, V.B. Arion, C.G. Hartinger, B.K. Keppler, Dalton Trans. (2008). https://doi.org/10.1039/B712656P
K. Strohfeldt, M. Tacke, Chem. Soc. Rev. 37, 1174 (2008)
C.G. Hartinger, P.J. Dyson, Chem. Soc. Rev. 38, 391 (2009)
L. Pellerito, L. Nagy, Coord. Chem. Rev. 224, 111 (2002)
V. Dokorou, A. Primikiri, D. Kovala-Demertzi, J. Inorg. Biochem. 105, 195 (2011)
M. Gielen, E.R.T. Tiekink, Tin compounds and their therapeutic potential, in Metallotherapeutic Drugs and Metal-Based Diagnostic Agents: The Use of Metals in Medicine. ed. by M. Gielen, E.R.T. Tiekink (Wiley, West Sussex, 2005), pp. 421–439
A.M.A. Ibrahim, E. Siebel, R.D. Fischer, Inorg. Chem. 37, 3521 (1998)
E. Siebel, A.M.A. Ibrahim, R.D. Fischer, Inorg. Chem. 38, 2530 (1999)
H. Hanika-Heidl, S.E.H. Etaiw, MSh. Ibrahim, R.D. Fischer, A.S. Badr El-din, J. Organomet. Chem. 684, 329 (2003)
S.E.H. Etaiw, S.N. Abdou, J. Inorg. Organomet. Polym. 20, 622 (2010)
M. Zurro, S. Asmus, S. Beckendorf, C.M. Lichtenfeld, O.G. Mancheno, J. Am. Chem. Soc. 136, 13999 (2014)
B.A. Maynard, R.E. Sykora, J.T. Maguec, A.E.V. Gorden, Chem. Commun. 46, 4944 (2010)
M.L. Toma, R. Lescouëzec, F. Lloret, M. Julve, J. Vaissermann, M. Verdaguer, Chem. Commun. 1850 (2003)
E. Lissi, B. Modak, R. Torres, J. Escobar, A. Urza, Free Radic. Res. 30, 471 (1999)
A.B.A. El-Gazzar, A.M.S. Youssef, M.M. Youssef, A.A. Abu-Hashem, F.A. Badria, Eur. J. Med. Chem. 44, 609 (2009)
R. Aeschlach, J. Loliger, C.B. Scott, A. Murcia, J. Butler, B. Halliwel, I.O. Aruoma, Food Chem. Toxicol. 32, 31 (1994)
Y. Morimoto, K. Tanaka, Y. Iwakiri, S. Tokuhiro, S. Fukushima, Y. Takeuchi, Biol. Pharm. Bull. 18, 1417 (1995)
T. Mosmann, J. Immunol. Methods 65, 55 (1983)
F. Denizot, R. Lang, J. Immunol. Methods 22, 271 (1986)
R.A. Penneman, L.H. Jones, J. Chem. Phys. 24, 293 (1956)
S.E.H. Etaiw, S.N. Abdou, Appl Organomet. Chem. 32, 4053 (2018)
H.H. Jalfe, M. Orechin, Theory and Applications of Ultraviolet Spectroscopy, 5th edn. (Wiley, New York, 1970)
N. Armaroli, L.D. Cola, V. Balzani, J.-P. Sauvage, C. Dietrich-Buchecker, J.-M. Kern, J. Chem. Soc. Faraday Trans. 88, 553 (1992)
B.N. Bandyopadhyay, A. Harriman, J. Chem. Soc. Faraday Trans. 1 73, 663 (1977)
G.M. Badger, I.S. Walker, J. Chem. Soc. (1956). https://doi.org/10.1039/JR9560000122
K. Yamamote, T. Takemura, H. Baba, Bull. Chem. Soc. Jpn 51, 729 (1978)
M.J. Lim, C.A. Murray, T.A. Tronic, K.E. deKrafft, A.N. Ley, J.C. deButts, R.D. Pike, H. Lu, H.H. Patterson, Inorg. Chem. 47, 6947 (2008)
C.A. Bayse, T.P. Brewster, R.D. Pike, Inorg. Chem. 48, 174 (2009)
S.E.H. Etaiw, S.A. Amer, M.M. El-Bendary, J. Inorg. Organomet. Polym. 21, 662 (2011)
B. Delley, J. Chem. Phys. 92, 508 (1990)
B. Delley, Int. J. Quantum Chem. 69, 433 (1998)
A. Kessi, B. Delley, Int. J. Quantum Chem. 68, 135 (1998)
X. Wu ana, A.K. Ray, Phys. Rev. B 65, 85403 (2002)
Materials, Studio v 5.0 Copyright 2009 (Accelrys Software, Inc., 2009)
W.J. Hehre, L. Radom, P.V.R. Schleyer, J.A. Pople, Ab Initio Molecular Orbital Theory (Wiley, New York, 1986)
B. Hammer, L.B. Hansen, J.K. Nørskov, Phys. Rev. B 59, 7413 (1999)
A. Matveev, M. Staufer, M. Mayer, N. Rösch, Int. J. Quantum Chem. 75, 863 (1999)
S.E.H. Etaiw, S.N. Abdou, A.S. Badr El-din, J Inorg. Organomet. Polym. 25, 1478 (2015)
C. Pellerito, L. Nagy, L. Pellerito, A. Azorcsik, J. Organomet. Chem. 691, 1733 (2006)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Abdou, S.N. Ultrasonic Assisted Nano-structures of Novel Organotin Supramolecular Coordination Polymers as Potent Antitumor Agents. J Inorg Organomet Polym 31, 3962–3975 (2021). https://doi.org/10.1007/s10904-021-02055-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10904-021-02055-5