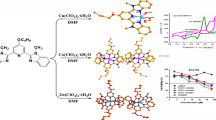
Abstract
Two metal complexes, [Zn(bmbp)Cl2]·H2O (1) and [Co(bmbp)2](ClO4)2·DMF·H2O (2) (bmbp = 4-benzyloxy-2,6-bis(1-methyl-2-benzimidazolyl)pyridine), have been synthesized and characterized. X-ray crystal structure analysis for complex 1 reveals that the Zn(II) atom is penta-coordinated with three nitrogen atoms of the ligand and two chlorine atoms in the coordination sphere, adopting a distorted square pyramidal geometry. For complex 2, the central Co(II) atom is bonded by six nitrogen atoms from two ligands in a distorted octahedral fashion. The two complexes feature 1D chain structures generated by C–H⋯π interactions. The thermal stabilities and spectroscopic properties of the complexes have been studied. Both complexes exhibit inhibition on the growth of the Eca109 cancer cells, but show lower antitumor activity than cisplatin.
Graphic Abstract
Two zinc(II) and cobalt(II) complexes based on 4-benzyloxy-2,6-bis(1-methyl-2-benzimidazolyl)pyridine, which exhibit inhibition on the growth of Eca109 cancer cell, were synthesized and characterized by X-ray single crystal structure analysis.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, benzimidazole derivatives and their metal complexes have attracted a great deal of interest due to their various molecular structures and versatile potential applications. These compounds play an important role in the development of coordination chemistry related to catalysis, biology, medical, magnetics and luminescence [1,2,3,4,5]. Particularly, because of the importance of these compounds as pharmacophores in drugs, much attention has been given to bidentate NN-donor or tridentate NNN-donor chelating ligands containing benzimidazole rings and their metal complexes, which have been shown to display antibacterial [6, 7], antioxidant [8, 9], antiparasitic [10], anti-inflammatory [11] and anticancer activities [12,13,14]. It has been observed that the biological activity of the metal complexes is greater than that of the corresponding free ligands [15], since the metal ions in such complexes not only accelerate the drug action, but also increase the effectiveness of the organic ligands [16]. Among the metals, zinc is one of the most essential trace elements in the human body via acting as structural and functional cofactor of many intracellular proteins. Also cobalt is an important metal in biological systems. Many zinc(II) or cobalt(II) complexes exhibit novel antitumor activities against various human cancer cells, such as MCF-7 breast cancer, HT-29 colon cancer and the Eca109 esophageal cancer cell lines [17, 18]. However, the activities of metal complexes may not just depend on the type of metal ion, but also on the ligand donor atoms, the planarity of ligands and the coordination geometry [19]. A number of benzimidazole metal complexes have been previously reported to be involved in the synthesis, structures and anti-esophageal cancer activities [20,21,22]. Esophageal cancer is the sixth most common cause of cancer deaths worldwide. It is a highly lethal disease and the five-year survival rate rarely exceeds 20% [23]. In 2018, esophageal cancer accounted for 3.2 percent of new diagnoses of cancer and 5.3 percent of deaths from cancer [24]. Although cisplatin is a well-known chemotherapy drug used for the treatment of various malignant cancers, there are undesirable side effects due to its ototoxicity. It is, therefore, important to study platinum-free metal complexes with higher activity and lower toxicity, either by changing the metal or the ligand classes.
To continue our previous work [22], we herein report the synthesis, characterization, and spectroscopic properties studies of zinc(II) and cobalt(II) complexes of 4-benzyloxy-2,6-bis(1-methyl-2-benzimidazolyl)pyridine (bmbp). The crystal structures of the two complexes have been determined by X-ray crystallography. Furthermore, the effects of these complexes on the antitumor viability of esophageal cancer Eca109 cells are investigated and described.
Experimental
Materials and Instrumentation
Anhydrous zinc(II) chloride, cobalt(II) perchlorate hexahydrate, anhydrous potassium carbonate and benzyl bromide were purchased from Sigma-Aldrich and used without further purification. N,N-dimethyllformamide was dried using the standard methods. 2,6-bis(1-methyl-2-benzimidazolyl)pyridine-4-ol was prepared by procedures described in the literature [25]. 1H NMR and 13C NMR spectra were recorded with a Bruker DMX 400 MHz instrument at ambient temperature using TMS (SiMe4) as an internal standard. FT-IR spectra were recorded on a Bruker Nicolet 6700 FT-IR spectrometer using KBr pellets. Mass spectra were obtained on a Bruker amaZon SL spectrometer using electrospray ionization (ESI). Elemental analysis was carried out using a Perkin-Elmer 2400 elemental analyzer. The UV–Vis spectra were monitored by a Shimadzu UV-2550 ultraviolet–visible spectrometer. The fluorescence spectra were measured on a Perkin-Elmer LS-55 luminescence spectrometer. Thermogravimetric analysis (TGA) was performed under N2 atmosphere at a heating rate of 10 ℃ min−1 on a Perkin Elmer STA6000 thermal analyzer.
Synthesis of 4-Benzyloxy-2,6-bis(1-methyl-2-benzimidazolyl)pyridine (bmbp)
A solution of 2,6-bis(1-methyl-2-benzimidazolyl)pyridine-4-ol (1.06 g, 3 mmol) in dry DMF(20 mL) was stirred with 5 equivalents of anhydrous K2CO3 (2.07 g, 15 mmol) at 80℃ for 0.5 h under nitrogen. Then, benzyl bromide (0.41 mL, 3.5 mmol.) was added dropwise. After stirring for 5 h, the reaction mixture was allowed to cool to room temperature. Subsequently, water was added and the mixture was stirred until all K2CO3 was dissolved. The white product was then filtered and recrystallized from CH2Cl2/EtOH(1:1). Yield: 0.75 g (56%). Mp: 258.9 ~ 259.3 ℃. Anal calc for C28H23N5O: C, 75.49; H, 5.20; N, 15.72%. Found: C, 75.56; H, 5.28; N, 15.77%. 1H NMR (400 MHz, CDCl3, ppm) δ: 7.97 (s, 2H), 7.82–7.78 (m, 2H), 7.39 (t, J = 7.3 Hz, 2H), 7.37–7.32 (m, 4H), 7.31–7.27 (m, 5H), 5.27 (s, 2H), 4.16 (s, 6H). 13C NMR (101 MHz, CDCl3, ppm) δ: 166.20, 151.21, 150.29, 142.47, 137.16, 135.47, 128.72, 128.35, 127.43, 123.58, 122.83, 120.17, 112.00, 109.93, 70.25, 32.53. IR data (KBr, cm−1): 3437 w, 3037 w, 2925 w, 1587 s, 1475 m, 1448 m, 1416 m, 1381 w, 1327 w, 1179 w, 1032 m, 937 w, 872 w, 740 m, 695 w. ESI–MS: m/z = 468.2 ([M + 23]).
Synthesis of [Zn(bmbp)Cl2]·H2O (1)
A solution of ZnCl2 (0.221 g, 1.0 mmol) in ethanol (10 mL) was added to a hot stirred solution of bmbp (0.445 g, 1.0 mmol) in DMF (20 mL). The reaction mixture was stirred for 30 min. The resulting solution was allowed to stand in air for 3 weeks. White block crystals were obtained, Yield: 0.468 g (78%). Anal calc for C28H25N5Cl2O2Zn: C, 56.07; H, 4.20; N, 11.68%. Found: C, 56.16; H, 4.27; N, 11.74%. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 8.03 (s, 2H), 7.77–7.75 (m, 2H), 7.57–7.49 (m, 2H), 7.47–7.45 (m, 4H), 7.41–7.35 (m, 5H), 5.51 (s, 2H), 4.29 (s, 6H). IR data (KBr, cm−1): 3455 m, 3059 w, 2930 w, 1603 s, 1566 m, 1490 s, 1477 s, 1454 m, 1393 m, 1323 m, 1179 m, 1041 s, 922 w, 860 w, 742 s, 697 w.
Synthesis of [Co(bmbp)2](ClO4)2·DMF·H2O (2)
To a solution of bmbp (0.445 g, 1.0 mmol) in DMF (20 mL), Co(ClO4)2·6H2O (0.183 g, 0.5 mmol) was added and stirred for 30 min at room temperature. The resulting solution was allowed to stand in air for one week. Brown block crystals were obtained, Yield: 0.378 g (62%). Anal calc for C59H55Cl2CoN11O12: C, 57.15; H, 4.47; N, 12.43%. Found: C, 57.22; H, 4.55; N, 12.51%. 1H NMR (400 MHz, DMSO-d6, ppm) δ: 8.10 (s, 2H), 7.94–7.92 (m, 2H), 7.53–7.50 (m, 2H), 7.46–7.45 (m, 4H), 7.44–7.38 (m, 5H), 5.41 (s, 2H), 4.27 (s, 6H). IR data (KBr, cm−1): 3440 w, 3061 w, 2931 w, 1603 s, 1565 w, 1489 s, 1480 m, 1454 w, 1394 w, 1320 w, 1183 w, 1093 s, 1026 m, 923 w, 857 w, 754 m, 696 w.
Crystal Structure Determination
Single crystals X-ray diffraction data of complexes 1 and 2 were collected on a Bruker SMART APEX CCD-based diffractometer with a graphite-monochromatic Mo-Kα radiation (λ = 0.71073 Å), using φ-ω scans at 293 K. Intensity data were corrected for Lp factors and empirical absorption. The structures of 1 and 2 were solved by direct methods with SHELXS-97 [26] and refined by full-matrix least-squares techniques on F2 with anisotropic thermal parameters, using SHELXL-97 [27]. All non-hydrogen atoms were located with successive difference Fourier maps. The hydrogen atoms bonded to carbons were positioned geometrically and allowed to ride on the parent atoms to which they are attached. In complex 1, the water molecule O1W was disordered over two positions with equal occupancies, but the resulting electron density was uninterpretable. So their contribution was removed with the SQUEEZE routine in PLATON [28]. For complex 2, the water molecule and C50 to C56 were disordered over two positions with occupancies of 0.50, respectively. The experimental crystallographic details are summarized in Table 1. Selected bond lengths and angles for complexes 1 and 2 are listed in Table 2.
Cell Culture
Esophageal cancer cell lines Eca109 were obtained from American Type Culture Collection (ATCC, Manassas, VA) and cultured in DMEM medium supplemented with 10% fetal bovine serum, 100 U mL−1 penicillin and 50 U mL−1 streptomycin. Cells were incubated at 37 ℃ in a humidified incubator with 5% CO2 atmosphere.
MTT Assay
An MTT assay was carried out as described previously [22]. Cells were seeded into 96-well culture plates at a density of 5 × 104 cells/well. The ligand, complexes and cisplatin (positive control) were first solubilized in DMSO and then in medium, and added to final concentrations (0, 3.125, 6.25, 12.5, 25, 50, 100 μM). After treatment with these compounds for 72 h, 20 μL per well of 5 mg·mL−1 MTT in phosphate buffered saline (PBS, pH 7.4) was added and incubated for another 5 h. The medium was aspirated and replaced with 200 μL per well of DMSO to dissolve the formazan salt formed. The color intensity of the formazan solution, which reflects the cell growth condition, was measured at 570 nm using a microplate spectrophotometer (VERSA max). All the experiments were carried out in triplicate at each concentration level and repeated thrice. The IC50 values were calculated by nonlinear regression analysis using Graphpad Prism 5.0 (GraphPad Software Inc., San Diego, CA, USA).
Results and Discussion
Synthesis and Characterization
The ligand bmbp was synthesized by alkylation between 2,6-bis(1-methyl-2-benzimidazolyl)pyridine-4-ol and benzyl bromide in dry K2CO3/DMF. It reacts with ZnCl2 to form a five-coordinated Zn(II) complex (1), and with Co(ClO4)2·6H2O to yield a six-coordinated Co(II) complex (2). In the 1H NMR spectrum of the ligand, the chemical shifts from 7.97 to 7.27 are assigned to the hydrogen atoms of pyridine ring and aromatic ring. The peak at δ = 5.27 is attributed to two hydrogen atoms of –CH2–. The –CH3 proton attached to N atom is seen as a singlet at 4.16 ppm. In the 1H NMR spectrum of complex 1 and 2, the signals of –CH2– shifted to 5.51 ppm (for 1) and 5.41 ppm (for 2), the signals of –CH3 also shifted to 4.29 ppm and 4.27 ppm, respectively.
Crystal Structure of Complex 1
Single-crystal X-ray diffraction analysis reveals that complex 1 crystallizes in the triclinic space group P-1. The local coordination environment of Zn(II) ion is shown in Fig. 1. The central Zn(II) ion is five-coordinated and surrounded by three nitrogen and two chlorine atoms, adopting a slightly distorted square pyramidal geometry with trigonality index τ = 0.080 (τ = (β − α)/60, where β and α are the two largest bond angles around the metal center in five coordinated environment) [29]. The chloride ion Cl(1) and three nitrogen atoms of the ligand occupy the tetragonal basal plane while the other chloride ion Cl(2) is situated in the apical position. As shown in Table 2, the bond lengths of Zn(1)–N(1), Zn(1)–N(3) and Zn(1)–N(4) are, respectively, 2.158 (3), 2.169 (3) and 2.171 (3) Å and similar to previously reported Zn–N distances [18]. The two benzimidazole rings and the pyridine ring are approximately in the same plane (mean deviation = 1.67°, 5.06°), and the dihedral angle between the benzyl ring and the pyridine ring is 63.47°. As shown in Fig. 2, there are two point-to-face C–H⋯π weak interactions between methyl protons and benzimidazole benzene rings, with a C⋯Cg distance of 3.604 Å for C(22)–H(22C)⋯Cg1 [Cg1 is the centroid for C(23) → C(28) ring], and 3.655 Å for C(7)–H(7A)⋯Cg2 [Cg2 is the centroid for C(1) → C(6) ring]. The adjacent molecules are linked through C–H⋯π interactions to form a 1D chain.
Crystal Structure of Complex 2
Single-crystal X-ray diffraction analysis reveals that complex 2 crystallizes in the triclinic space group P-1 and consists of one complex [Co(bmbp)2]2+ cation, two ClO4− anions, one DMF solvent molecule and one crystal water molecule. As shown in Fig. 3, the central Co(II) atom is coordinated by six nitrogen atoms from two ligands in a distorted octahedral geometry. Two ligands coordinate one metal atom each in tridentate mode. The nearly planar ligands are at a dihedral angle of 84.47°. The Co–N bond distances are in the range of 2.105 (3) to 2.146 (3) Å, and compared with Co–N reported in the literature [20]. The bond angles listed in Table 2 indicate that complex 2 has a distorted octahedral geometry. The distortion from regular octahedral geometry is evident from the bond angles, N(4)–Co(1)–N(1) [150.54 (13)°], N(6)–Co(1)–N(9) [150.45 (13)°] and N(8)–Co(1)–N(3) [173.94 (12)°], which are contracted substantially from the ideal value of 180°. This is a common phenomenon in almost all six-coordinated zinc(II) and cobalt(II) complexes of tridentate benzimidazole ligands [18, 20]. The crystal packing of complex 2 is shown in Fig. 4. There are two point-to-face C–H⋯π weak interactions between methyl protons and benzimidazole benzene rings, with a C⋯Cg distance of 3.511 Å for C(43)–H(43A) ⋯Cg1 [Cg1 is the centroid for C(44) → C(49) ring], benzyl benzene protons and benzimidazole benzene ring with a C⋯Cg distance of 3.588 Å for C(28)–H(28) ⋯Cg2 [Cg2 is the centroid for C(16) → C(21) ring]. Two adjacent molecules form a 1D chain through C–H⋯π weak interactions.
IR Spectra and Thermal Analysis
The IR spectra of the ligand and metal complexes were compared and assigned on careful comparison. The ligand and metal complexes show weak bands in the region 2931–2853 cm−1 due to –CH3 and –CH2 stretching vibrations. The aromatic v(C=C) stretching vibrations are observed in the same region, 1490–1448 cm−1. The v(C–O) stretching vibrations can be found at 1032 cm−1,1041 cm−1 and 1026 cm−1 for the ligand, complexes 1 and 2, respectively. The absorption band at 1587 cm−1 in the ligand is attributed to the v (C=N) stretching vibrations. It is shifted to 1603 cm−1 and 1606 cm−1 in complexes 1 and 2, respectively, which is an indication that the ligand coordinated with Zn(II) and Co(II) ions through the nitrogen atom of the benzimidazole and pyridine rings. The appearance of a prominent absorption bands observed at 1093 cm−1 in complex 2, but absent in the free ligand, is assigned to v (ClO4) stretching frequency of the perchlorate ion. The strong bands observed in the range of 754–740 cm−1 in the ligand and the metal complexes are attributed to the C–H bending of the benzimidazole rings.
The thermogravimetric analyses (TGA) of the metal complexes were carried out in a nitrogen atmosphere at a heating rate of 10 ℃ min−1 (Fig. 5). For complex 1, the first weight loss of 3.12% (calcd. 3.00%) under 100 ℃ is attributed to the loss of the co-crystallized water molecule, and then the skeleton structure begins to decompose beyond 240 ℃. For complex 2, the first weight loss of 1.72% (calcd. 1.45%) under 100 ℃ is due to the loss of the co-crystallized water molecule, the second weight loss from about 254 to 342 ℃ is corresponding to the total decomposition of DMF molecules and perchlorate ions (observed 22.32%, calculated 21.93%). After that, the skeleton structure decomposes rapidly.
Absorption Spectra and Luminescent Property
The UV–Vis absorption spectra of the free ligand and metal complexes were measured at a concentration of 1.0 × 10–5 mol L−1 in DMF at room temperature (Fig. 6). The ligand and two complexes have absorption with the maximal peaks at about 313 nm and 316 nm, which can be assigned to intraligand π → π* transitions of the benzimidazole rings. There is one additional shoulder peak observed at 358 nm with a lower intensity for complex 1, but absent for the ligand, which probably arises from metal–ligand charge transfer. This behavior was also observed in our earlier reported zinc dichloride complexes [21].
The luminescent properties of the ligand and metal complexes were investigated in DMF solution at room temperature (Fig. 7). The free ligand shows an emission band at 351 nm upon excitation at 310 nm, whereas complexes 1 and 2 exhibit emission band with a maximum at 396 nm (λex = 310 nm) and 415 nm (λex = 320 nm), respectively. The emission of the ligand can be attributed to π* → π transitions, while the emissions of complex 1 and 2 exhibit an obvious red-shift (ca. 45 nm and 64 nm) compared to that of the ligand, which could be assigned to ligand-to-metal charge transfer (LMCT) [30, 31].
Antitumor Activity
The synthesized ligand, corresponding metal(II) complexes and cisplatin were screened by MTT assay for their in vitro antitumor activities against Eca109 cancer cells. As shown in Fig. 8, the Co(II) complex exhibits inhibition on the growth of Eca109 cancer cells but the ligand has no antitumor activity. The IC50 value of the Co(II) complex is 70.14 μM, which is higher than that of cisplatin (43.99 μM) and lower than that of the Zn(II) complex (IC50 value more than 100 μM). The results confirm that the metal complexes are usually more active than the free ligand itself. The discrepancy of the inhibitory activity for the two complexes are associated with the type of metal ion, planarity of ligand as well as the coordination geometry. The results here provide valuable theoretical basis and new ideas for the development of metal-drugs in treatment of esophageal cancer via synthesis and decoration of benzimidazole metal complexes, though the complexes show lower antitumor activity than cisplatin.
Conclusion
Two metal complexes, [Zn(bmbp)Cl2]·H2O(1) and [Co(bmbp)2](ClO4)2·DMF·H2O(2) based on 4-benzyloxy-2,6-bis(1-methyl-2-benzimidazolyl)pyridine were synthesized and characterized by X-ray single-crystal structure analysis. Complex 1 is five-coordinated with a slightly distorted square pyramidal geometry, while complex 2 has a six-coordinate distorted octahedral configuration. The two complexes display 1D chain structures via C–H⋯π interactions. Both complexes exhibit inhibition on the growth of Eca109 cancer cells, but show lower antitumor activity than cisplatin.
References
Hao JM, Yu BY, Van Hecke K, Cui GH (2015) Cryst Eng Comm 17:2279–2293
Kalinowska-Lis U, Szewczyk EM, Checinska L, Wojciechowski JM, Wolf WM, Ochocki J (2014) Chem Med Chem 9:169–176
Husain A, Rashid M, Shaharyar M, Siddiqui AA, Mishra R (2013) Eur J Med Chem 62:785–798
Chen N, Yang P, He X, Shao M, Li MX (2013) Inorg Chim Acta 405:43–50
Manjunatha MN, Dikundwar AG, Nagasundara KR (2011) Polyhedron 30:1299–1304
Arjmand F, Parveen S, Afzal M, Shahid M (2012) J Photochem Photobiol B 114:15–26
Ou ZB, Lu YH, Lu YM, Chen S, Xiong YH, Zhou XH, Mao ZW, Le XY (2013) J Coord Chem 66:2152–2165
Wu HL, Zhang YH, Zhang JW, Yang ZH, Chen CY, Peng HP, Wang F (2015) Transit Met Chem 40:145–152
Wu HL, Kou F, Jia F, Liu B, Yuan JK, Bai Y (2011) J Photochem Photobiol B 105:190–197
Navarrete-Vázquez G, Cedillo R, Hernández-Campos A, Yépez L, Hernández-Luis F, Valdez J, Morales R, Cortés R, Hernández M, Castillo R (2001) Bioorg Med Chem Lett 11:187–190
Rathore A, Sudhakar R, Ahsan MJ, Ali A, Subbarao N, Jadav SS, Umar S, Shaharyar M (2017) Bioorg Chem 70:107–117
Singla M, Ranjan R, Mahiya K, Mohapatra SC, Ahmad S (2015) New J Chem 39:4316–4327
Rajarajeswari C, Loganathan R, Palaniandavar M, Suresh E, Riyasdeend A, Akbarshae MA (2013) Dalton Trans 42:8347–8363
Hu JY, Guo Y, Zhao JA, Zhang JS (2017) Bioorg Med Chem 25:5733–5742
Ramachandran E, Kalaivani P, Prabhakaran R, Rath NP, Brinda S, Poornima P, Padma VV, Natarajan K (2012) Metallomics 4:218–227
Siddiqi ZA, Kalid M, Kumar S, Shahid M, Novo S (2010) Eur J Med Chem 45:264–269
Bordbar M, Tabatabaee M, Alizadeh-Nouqi M, Mehri-Lighvan Z, Khavasi HR, Yeganeh-Faal A, Fallahian F, Dolati M (2016) J Iran Chem Soc 13:1125–1132
Liu SG, Cao WQ, Yu LL, Zheng WJ, Li LL, Fan CD, Chen TF (2013) Dalton Trans 42:5932–5940
Pyle AM, Barton JK (1990) Prog Inorg Chem Bioinorg Chem 38:413–475
Li GB, Wang SX, Su WY, Pan RK, Liu KD, Liu SG (2014) Russ J Coord Chem 40:764–767
Pan RK, Liu SG, Wang SX, Li GB, Su WY, Huang QW, He YM (2015) Z Anorg Allg Chem 641:627–630
Pan RK, Song JL, Li GB, Lin SQ, Liu SG, Yang GZ (2017) Transit Met Chem 42:253–262
Bashash M, Hislop TG, Shah AM, Le N, Brooks-Wilson A, Bajdik CD (2011) BMC Cancer 11:164
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) CA Cancer J Clin 68:394–424
Pan RK, Li GB, Liu SG, Zhou XP, Yang GZ (2016) Monatsh Chem 147:1189–1196
Sheldrick GM (1997) SHELXS-97, program for the solution of crystal structures. University of Göttingen, Göttingen
Sheldrick GM (1997) SHELXL-97, program for the refinement of crystal structures from diffraction data. University of Göttingen, Göttingen
Spek AL (2009) Acta Cryst D 65:148–155
Addison AW, Rao TN, Reedijk J, Van Rijn J, Verschoor GC (1984) J Chem Soc Dalton Trans 7:1349–1356
Wang D, Liu SX (2007) Polyhedron 26:5469–5476
Zhang LP, Ma JF, Yang J, Liu YY, Wei GH (2009) Cryst Growth Des 9:4660–4673
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (No. 21502085), Special funds for public welfare research and capacity building in Guangdong Province (Nos. 2015A020211038 and 2016A010103042), Research Group of Rare earth resource exploiting and luminescent materials (No. 2017KCXTD022).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10870_2020_824_MOESM2_ESM.doc
Supplementary file2 (DOC 556 kb) CCDC 1401680 and 1449418 contain the supplementary crystallographic data for complexes 1 and 2. These data can be obtained free of charge via https://www.ccdc.cam.ac.uk/conts/retrieving.html, or from the Cambridge Crystallographic Data Centre (CCDC), 12 Union Road, Cam bridge CB2 1EZ, UK.
Rights and permissions
About this article
Cite this article
Pan, RK., Song, JL., Su, WY. et al. Zinc(II) and Cobalt(II) Complexes Derived from 4-Benzyloxy-2,6-bis(1-methyl-2-benzimidazolyl)pyridine: Synthesis, Crystal Structures, Spectroscopic Properties and Antitumour Activities. J Chem Crystallogr 50, 241–248 (2020). https://doi.org/10.1007/s10870-020-00824-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10870-020-00824-7