Abstract
Mitochondria have to import the vast majority of their proteins, which are synthesized as precursors on cytosolic ribosomes. The translocase of the outer membrane (TOM complex) forms the general entry gate for the precursor proteins, which are subsequently sorted by protein machineries into the mitochondrial subcompartments: the outer and inner membrane, the intermembrane space and the mitochondrial matrix. The transport across and into the inner membrane is driven by the membrane potential, which is generated by the respiratory chain. Recent studies revealed that the lipid composition of mitochondrial membranes is important for the biogenesis of mitochondrial proteins. Cardiolipin and phosphatidylethanolamine exhibit unexpectedly specific functions for the activity of distinct protein translocases. Both phospholipids are required for full activity of respiratory chain complexes and thus to maintain the membrane potential for protein import. In addition, cardiolipin is required to maintain structural integrity of mitochondrial protein translocases. Finally, the low sterol content in the mitochondrial outer membrane may contribute to the targeting of some outer membrane proteins with a single α-helical membrane anchor. Altogether, mitochondrial lipids modulate protein import on various levels involving precursor targeting, membrane potential generation, stability and activity of protein translocases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mitochondria fulfill crucial functions in the eukaryotic cell like ATP production via oxidative phosphorylation, synthesis of lipids, amino acids, heme and iron-sulfur clusters and act as a signaling platform for apoptosis. Mitochondrial functions rely on the presence of about 1000 (yeast) or 1500 (human) different proteins (Reinders et al. 2006; Pagliarini et al. 2008). The vast majority of these proteins is nucleus-encoded, translated on cytosolic ribosomes as precursors and imported into the organelle in a posttranslational manner. Specific targeting signals direct the precursors to the mitochondrial surface. During their journey through the cytosol the precursor proteins are kept in an unfolded state by cytosolic chaperones (Neupert and Herrmann 2007; Chacinska et al. 2009). The translocase of the outer membrane (TOM complex) forms the general entry gate for most of the mitochondrial precursor proteins. The TOM complex consists of the pore-forming Tom40, the receptor proteins Tom20, Tom22 and Tom70 and three small Tom proteins, which are required for stability and assembly of the TOM machinery. After passage through the TOM channel specific protein machineries sort the precursors into the mitochondrial subcompartments: outer and inner membrane, intermembrane space and matrix (Fig. 1) (Koehler 2004; Dolezal et al. 2006; Neupert and Herrmann 2007; Chacinska et al. 2009; Endo and Yamano 2009). The import pathways and mechanisms were studied in baker’s yeast Saccharomyces cerevisiae, but are largely conserved in human mitochondria. Recent studies indicate that the mitochondrial protein machineries are integrated into a sophisticated protein network (Becker et al. 2012). The protein translocases not only directly cooperate with each other to channel precursor proteins, but are also linked to distinct protein complexes like the respiratory chain, the mitochondrial contact site and cristae organizing system (MICOS) and the endoplasmic reticulum-mitochondria encounter structure (ERMES) (van der Laan et al. 2006; Meisinger et al. 2007; von der Malsburg et al. 2011; Harner et al. 2011a). In addition, protein import via the TOM complex is regulated by phosphorylation and thereby closely connected to the metabolic state of the cell (Schmidt et al. 2011; Gerbeth et al. 2013). These observations indicate that various processes can modulate mitochondrial protein import on different levels. Studies of the last years demonstrated that protein biogenesis is not only influenced by protein interactions and modifications, but also by the lipid composition of mitochondrial membranes. The role of mitochondrial lipids for protein biogenesis was underestimated for long time. We are just beginning to understand how the lipid composition affects the membrane-bound protein translocases. Recent studies revealed unexpectedly specific roles of certain lipids for the activity and stability of mitochondrial protein complexes.
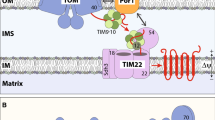
Five pathways sort mitochondrial precursor proteins into the mitochondrial subcompartments. Precursors of multi-spanning outer membrane proteins are recognized by Tom70 and transferred to the MIM machinery, which mediates their integration and assembly into the membrane. The TOM complex forms the general entry gate for most of the other mitochondrial precursors. After passage through the Tom40 channel four sorting pathways branch off. β-barrel precursors are directly transferred to the SAM complex, which mediates their integration into the outer membrane (OM). During the transport small TIM chaperones of the intermembrane space (IMS) shield hydrophobic segments of the β-barrel precursors. Precursors with a cleavable presequence are transferred to the TIM23 complex, which mediates either lateral release into the inner membrane (IM) or transport into the mitochondrial matrix. Precursor translocation via the TIM23 complex into the matrix additionally requires the ATP-consuming action of the PAM module. The presequence is removed upon import by the mitochondrial processing peptidase (MPP). The small TIM chaperones guide precursors of multi-spanning carrier proteins to the TIM22 complex, which facilitates their insertion into the inner membrane. The membrane potential across the inner membrane (Δψ) drives TIM23 and TIM22 dependent import. Mia40 sorts cysteine-rich precursors into the intermembrane space and mediates their oxidative folding. Erv1 shuttles the electrons from Mia40 to cytochrome c of the respiratory chain
Mitochondrial protein import
Protein sorting into the mitochondrial outer membrane
Proteins are integrated into the outer mitochondrial membrane by α-helical or β-barrel domains. These two types of transmembrane proteins follow different import pathways. Precursors of β-barrel proteins are translocated through the TOM channel and transferred with the help of the small TIM chaperones to the sorting and assembly machinery (SAM complex; Fig. 1) (Hoppins and Nargang 2004; Wiedemann et al. 2004). TOM and SAM form a supercomplex to promote efficient biogenesis of β-barrel precursors (Qiu et al. 2013). The SAM complex mediates β-barrel folding and insertion into the outer membrane (Paschen et al. 2003; Ishikawa et al. 2004; Qiu et al. 2013). It consists of the essential β-barrel protein Sam50 (also known as Tob55), which forms the core of the protein translocase. Sam50 associates with the two peripheral subunits Sam35 (Tob38) and Sam37 (Mas37) on the cytosolic surface (Wiedemann et al. 2003; Paschen et al. 2003; Gentle et al. 2004; Klein et al. 2012). While Sam35 has been implicated in promoting precursor recognition, Sam37 was reported to mediate the release of the β-barrel precursor into the outer membrane (Chan and Lithgow 2008; Kutik et al. 2008a).
Other outer membrane proteins contain a single (single-spanning) or several (multi-spanning) transmembrane α-helices. Precursors of multi-spanning outer membrane proteins are recognized by the receptor protein Tom70 of the TOM complex, but are not translocated through the TOM channel (Otera et al. 2007; Becker et al. 2011; Papic et al. 2011; Wenz et al. 2014). Instead, the mitochondrial import machinery (MIM complex; Fig. 1) mediates membrane insertion and assembly of these precursors (Becker et al. 2011; Papic et al. 2011; Dimmer et al. 2012). The MIM complex consists of several copies of Mim1 and the recently identified Mim2 (Becker et al. 2008; Hulett et al. 2008; Popov-Čeleketić et al. 2008; Dimmer et al. 2012). In contrast, no common import pathway for the precursors of single-spanning α-helical outer membrane proteins was found. A role of TOM, SAM and MIM complexes in the import of different precursors was reported (Ott et al. 2007; Stojanovski et al. 2007; Becker et al. 2008; Hulett et al. 2008; Popov-Čeleketić et al. 2008; Thornton et al. 2010; Harner et al. 2011b). Particularly interesting is the biogenesis of the outer membrane protein of 45 kDa (Om45), which is embedded by an N-terminal α-helix and exposes a large soluble domain into the intermembrane space (Lauffer et al. 2012; Song et al. 2014; Wenz et al. 2014). The biogenesis of the Om45 precursor involves three protein translocases: TOM and TIM23 complexes drive the translocation of the soluble domain, while the MIM machinery assembles the precursor into the outer membrane (Song et al. 2014; Wenz et al. 2014). Finally, the insertion of a few α-helical outer membrane proteins was reported to occur in a protein-independent manner (Meineke et al. 2008; Kemper et al. 2008).
Protein sorting into inner mitochondrial compartments
The majority of mitochondrial precursors contain an N-terminal cleavable presequence, which is removed upon import by the mitochondrial processing peptidase (MPP). The receptor proteins Tom20 and Tom22 recognize this type of precursors at the mitochondrial surface (Yamano et al. 2008). After passage through the TOM channel the precursors are directly transferred to the presequence translocase (TIM23 complex; Fig. 1) (Chacinska et al. 2003). The highly dynamic TIM23 complex mediates the sorting of the precursors either into the mitochondrial matrix or into the inner membrane. The intermembrane space domains of Tim23, Tim50 and Tim21 are involved in the interaction with the TOM complex and in precursor recognition (Chacinska et al. 2005; Tamura et al. 2009a; Mokranjac et al. 2009; Marom et al. 2011; Shiota et al. 2011; Schulz et al. 2011; Lytovchenko et al. 2013). Tim23 forms a voltage-sensitive channel, which can be closed by Tim50 to prevent proton leakage (Meinecke et al. 2006; Alder et al. 2008). Tim17 plays a role in motor recruitment and the pore architecture of the TIM23 channel (Chacinska et al. 2005; Martinez-Caballero et al. 2007). The membrane potential drives transport of the positively charged presequence through the TIM23 channel. This step involves activation of the TIM23 channel through membrane-potential mediated conformational changes (Malhotra et al. 2013). Tim21 associates with respiratory chain complexes and thereby couples protein import directly to generation of the membrane potential (van der Laan et al. 2006). The recently identified subunit Mgr2 stabilizes the binding of Tim21 to the TIM23 complex (Gebert et al. 2012). Inner membrane precursors additionally contain an internal hydrophobic sorting sequence, which stops translocation and induces lateral insertion of the preprotein into the membrane. Presequence-containing precursors that have to be fully transported into the mitochondrial matrix require an additional driving force, which is provided by the presequence translocase-associated motor (PAM; Fig. 1) (Neupert and Herrmann 2007; Chacinska et al. 2009). The core component of this import motor is the mitochondrial Hsp70 (mtHsp70) that binds to the TIM23 complex via Tim44. The chaperone facilitates precursor translocation into the matrix by multiple cycles of ATP-dependent precursor binding and release. Thereby, mtHsp70 is alternating between the ADP-bound state with high affinity and the ATP-bound state with low affinity for incoming precursor proteins. This mtHsp70 cycle is controlled by the interaction with several co-factors within the PAM module. The J-domain of Pam18 (Tim14) enhances the ATPase activity of mtHsp70, whereas the nucleotide exchange factor Mge1 stimulates the ADP/ATP exchange of the chaperone. Pam18 forms a stable heterodimer with Pam16 (Tim16), a J-like protein that exhibits an inhibitory role on Pam18 function (Li et al. 2004; Mokranjac et al. 2006). The non-essential Pam17 promotes the association of the Pam16/Pam18 module with the presequence translocase (van der Laan et al. 2005).
Precursors of multi-spanning inner membrane proteins lacking a cleavable presequence are escorted by cytosolic chaperones to the mitochondrial surface, where they are recognized by the Tom70 receptor. After passage through the TOM channel the small TIM chaperones Tim9/Tim10 guide the precursors through the aqueous intermembrane space to the carrier translocase (TIM22 complex; Fig. 1). Tim9 and Tim10 form a hexameric complex, which assembles with Tim12 at the carrier translocase (Lionaki et al. 2008; Gebert et al. 2008). The soluble domain of Tim54 of the TIM22 complex might provide a docking site for the small TIM chaperones (Kerscher et al. 1997). Tim22 is the central component of the carrier translocase and forms a voltage-activated twin-pore in the inner membrane (Rehling et al. 2003; Peixoto et al. 2007). The Tim22 channel facilitates membrane insertion of the precursor proteins in a membrane potential-dependent manner. The subunits Tim18 and Sdh3 play a role in the stability and assembly of the carrier translocase (Wagner et al. 2008; Gebert et al. 2011; Okamoto et al. 2014). Interestingly, Sdh3 is also present in complex II (succinate dehydrogenase) of the respiratory chain, providing an evolutionary link between inner membrane protein sorting and respiration (Gebert et al. 2011).
Precursor proteins with a cysteine-rich motif are transferred after passage of the TOM channel to Mia40, the central component of the mitochondrial intermembrane space import and assembly machinery (MIA; Fig. 1). Mia40 acts as an oxidoreductase to drive oxidative folding by the introduction of disulfide-bridges within its substrates (Banci et al. 2009; Kawano et al. 2009). Mia40 itself is re-oxidized by the sulfhydryl oxidase Erv1 (essential for respiration and viability), which shuttles the electrons towards cytochrome c of the respiratory chain (Mesecke et al. 2005; Bihlmaier et al. 2007; Tienson et al. 2009; Böttinger et al. 2012a).
Biosynthesis and function of mitochondrial lipids
The major phospholipids of mitochondrial membranes are phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylserine (PS) and the dimeric phospholipid cardiolipin (CL). Sterols like ergosterol in yeast and sphingolipids are only present in minor amounts (van Meer et al. 2008; Horvath and Daum 2013). The signature phospholipid of mitochondria CL is unequally distributed between both mitochondrial membranes. Whereas the mitochondrial inner membrane shows especially high levels of CL, the outer membrane only contains about one quarter of the total mitochondrial CL pool (Gebert et al. 2009). Formation of mitochondrial membranes depends on the import of PC, PS and PI, which are generated in the endoplasmic reticulum. In yeast, the ERMES complex forms a molecular tether between the mitochondrial outer membrane and the endoplasmic reticulum, which may facilitate exchange of lipids between both organelles (Kornmann et al. 2009). Mitochondria are capable of synthesizing CL and PE autonomously (Fig. 2). For PE synthesis, PS is decarboxylated by the phosphatidylserine decarboxylase 1 (Psd1) of the mitochondrial inner membrane (Voelker 1997; Osman et al. 2011; Horvath et al. 2012). Although other biosynthesis pathways exist, the mitochondrially generated PE represents the major fraction of the total PE pool in the yeast cell (Bürgermeister et al. 2004). The biosynthesis of CL comprises a multistep reaction that has been studied in detail (Gohil and Greenberg 2009; Osman et al. 2011; Claypool and Koehler 2012; Horvath and Daum 2013). The CL-precursor phosphatidic acid (PA) is imported from the ER into the mitochondrial outer membrane and further transported via Ups1 (unprocessed 1) across the intermembrane space to the inner mitochondrial membrane (Connerth et al. 2012). Subsequently, Tam41 (translocator assembly and maintenance) converts PA to cytidine diphosphate-diacylglycerol (CDP-DAG) (Tamura et al. 2013), which is used for the generation of phosphatidylglycerolphosphate (PGP) by Pgs1 (PGP synthase). Gep4 dephosphorylates PGP to phosphatidylglycerol (PG), and the cardiolipin synthase (Crd1) catalyzes the formation of CL from PG and CDP-DAG. Finally, the premature CL undergoes remodeling of its acyl-chains involving two enzymes, the CL-specific deacylase Cld1 and the transacylase taffazin (Taz1) (Fig. 2) (Osman et al. 2011; Horvath and Daum 2013).
Mitochondrial phospholipid biogenesis in yeast. Mitochondria synthesize cardiolipin (CL) and phosphatidylethanolamine (PE) in the inner membrane (IM). Psd1 decarboxylates phosphatidylserine (PS) to produce PE. The synthesis of CL involves several steps and is followed by alterations of the acyl-chain composition in a process termed CL remodeling. For details see text. IMS intermembrane space
CL and PE are both so-called non-bilayer forming phospholipids. They are characterized by a relatively small diameter of their head group and a comparably large diameter of their hydrophobic fatty acid tails. Depending on pH and cation concentration, non-bilayer forming phospholipids have the tendency to form local hexagonal phase structures, which can induce an increase of membrane tension and thus substantially influence the properties and architecture of the lipid bilayer (van den Brink-van der Laan et al. 2004; Osman et al. 2011). Consequently, non-bilayer forming phospholipids have been implicated in a multitude of crucial processes such as membrane fusion, transbilayer transport of lipids as well as folding and transport of proteins (van den Brink-van der Laan et al. 2004; Dowhan and Bogdanov 2009). It has been shown that simultaneous deletion of mitochondrial CL and PE synthases is synthetically lethal for the cell, implicating overlapping functions of the two phospholipids (Gohil et al. 2005). Indeed, yeast mitochondria defective in respiration are more frequently developed when PE and CL levels are reduced (Birner et al. 2003; Zhong et al. 2004) indicating that CL and PE are important for mitochondrial functions. Simultaneous depletion of both CL and PE synthases affects mitochondrial morphology, which is likely connected to defects of the mitochondrial fusion machinery (DeVay et al. 2009; Joshi et al. 2012). Furthermore, a genetic interaction of both CL and PE synthases with prohibitins has been detected (Birner et al. 2003; Osman et al. 2009). Prohibitins have been implicated in regulating inner mitochondrial membrane organization by functioning as protein and lipid scaffolds. A recent study reported a role of prohibitins in the remodeling of CL (Richter-Dennerlein et al. 2014). Finally, CL is required for apoptosis, mitophagy and iron-sulfur cluster biogenesis (Gonzalvez et al. 2008; Chu et al. 2013; Patil et al. 2013). Altogether, phospholipids are important for the maintenance of various mitochondrial functions and thus for cell survival.
Role of lipids in protein import
The sorting of precursor proteins into mitochondria involves the action of several membrane-embedded protein transport machineries (Fig. 1). Numerous studies helped to elucidate the composition and function of these translocases, yet the impact of the lipid composition of the mitochondrial membranes on protein sorting was for long time only poorly understood. Recent studies revealed a remarkable importance of certain lipids for specific protein import routes, in which the best-studied lipids are CL and PE.
Outer membrane protein sorting
The biogenesis of β-barrel precursors is sensitive towards alterations of the phospholipid composition of the mitochondrial outer membrane. Particularly, CL and PE are important for efficient import of β-barrel precursors. In CL- and PE-deficient mitochondria early biogenesis steps are impaired (Gebert et al. 2009; Becker et al. 2013). Experimental dissection of the import pathway in PE-depleted mitochondria revealed that already translocation of the β-barrel precursors across the outer membrane is reduced (Becker et al. 2013). The observed import defect for β-barrel precursors in the phospholipid mutants can be partially explained by a decreased activity of the TOM complex. Supporting this view the binding of an arrested precursor to the TOM complex is impaired in both CL- and PE-deficient mitochondria (Gebert et al. 2009; Becker et al. 2013). Moreover, a reconstituted TOM complex requires the lipid composition of the outer mitochondrial membrane for full activity in liposomes (Stan et al. 2000). The molecular role of CL and PE for the function of the TOM complex differs. In the absence of CL the association of the Tom20 receptor with the core of the TOM complex is weakened (Fig. 3) (Gebert et al. 2009). In contrast, depletion of PE does not affect the stability of the TOM complex (Becker et al. 2013). The impaired TOM function contributes to the reduced precursor binding to the SAM complex in mitochondria with decreased PE and CL content (Gebert et al. 2009; Becker et al. 2013). Since CL stabilizes the association of the peripheral subunit Sam37 to Sam50/Sam35 the lipid composition is important for the integrity of this protein translocase (Fig. 3) (Gebert et al. 2009). However, the function of the SAM complex is not generally compromised in CL- or PE-deficient mitochondria, since the assembly of the α-helically anchored Tom22 is not affected (Gebert et al. 2009; Becker et al. 2013). The assembly of the Tom22 precursor with other TOM subunits involves all three SAM components (Stojanovski et al. 2007). The biogenesis of β-barrel precursors is also affected in mitochondria lacking the transacylase Taz1 involved in CL remodeling (Gebert et al. 2009). Impaired function of yeast Taz1 or of its human homolog taffazin causes an altered acyl composition of CL (Ye et al., 2014). Mutations in the human homolog taffazin lead to Barth syndrome, a severe disease that is characterized by cardiac and skeletal myopathies as well as neutropenia (Barth et al. 1983; Claypool et al. 2006). Interestingly, the assembly of β-barrel precursors is impaired in mitochondria isolated from Barth syndrome patients (Gebert et al. 2009). Thus, the phospholipid composition of mitochondrial membranes affects import of β-barrel precursors. Since biogenesis of the β-barrel protein Tom40 is essential for cell survival, the impaired β-barrel biogenesis likely contributes to the synthetically lethal phenotype of the double deletion of the mitochondrial CL and PE synthases (Gohil et al. 2005).
Cardiolipin (CL) and phosphatidylethanolamine (PE) modulate different steps of mitochondrial protein biogenesis. In the absence of CL (−CL) or upon reduction of PE levels (PE↓. the binding of precursor proteins to the TOM complex is affected. Consequently, the TOM-dependent biogenesis of β-barrel proteins is impaired. In contrast, TOM-independent import of multi-spanning outer membrane (OM) proteins is not disturbed by the altered lipid composition. The activity of the respiratory chain complexes III and IV is decreased, which causes a reduced membrane potential (∆ψ) and impaired protein import across or into the inner membrane (IM). Deletion of CL affects the association of Tom20, Sam37 and Pam16/Pam18 with TOM, SAM and TIM23 complexes. In addition, the stability of the respiratory chain supercomplexes is decreased. Transport defects are indicated by dashed red lines, whereas disturbances of protein complex stability is indicated by jagged red lines
Interestingly, there is a close link between outer membrane protein biogenesis and the ERMES complex, which is important for the maintenance of normal CL and PE levels (Osman et al. 2009; Kornmann et al. 2009). Mitochondria of ERMES mutants are defective in the biogenesis of β-barrel proteins (Meisinger et al. 2007; Wideman et al. 2010). Moreover, one of the core components of the ERMES complex, Mdm10 (mitochondrial distribution and morphology), associates with the SAM complex to mediate the assembly of the TOM complex (Meisinger et al. 2007; Thornton et al. 2010). Thus, Mdm10 provides a molecular link between ERMES and outer membrane protein biogenesis.
In contrast to β-barrel proteins, the biogenesis of outer membrane proteins with α-helical membrane anchors is largely unaffected in mitochondria with defective PL or CL synthesis (Gebert et al. 2009; Becker et al. 2013). However, recent reports indicate an important role of the low ergosterol content in yeast mitochondrial membranes for the targeting of some outer membrane proteins with a C-terminal α-helical membrane anchor (tail-anchored) (Kemper et al. 2008; Krumpe et al. 2012). First, in yeast mutants where the ergosterol content between mitochondria and ER membranes is comparable mitochondrial tail-anchored proteins mislocalize to the ER (Krumpe et al. 2012). Second, an increasing ergosterol content inhibits the insertion of a mitochondrial tail-anchored precursor into liposomes (Kemper et al. 2008). These studies imply a role of the lipid content of the mitochondrial outer membrane in protein targeting.
Inner membrane protein sorting
Protein transport into and across the mitochondrial inner membrane by TIM23 and TIM22 complexes essentially requires the presence of a membrane potential. The membrane potential is generated by the activity of the respiratory chain complexes, which form supercomplexes in the mitochondrial inner membrane (Schägger and Pfeiffer 2000; Cruciat et al. 2000). Structural investigations on cytochrome c reductase (complex III) and cytochrome c oxidase (complex IV) revealed that, among other phospholipids, CL and PE specifically bind to both complexes (Lange et al. 2001; Shinzawa-Itoh et al. 2007). In addition, the activities of complex III and in particular of complex IV depend on the presence of CL and PE (Pfeiffer et al. 2003; Zhong et al. 2004; Wenz et al. 2009; Böttinger et al. 2012b; Tasseva et al. 2013). Yet, both phospholipids might affect the respiratory activity via different mechanisms. CL stabilizes the association of complex III and complex IV within the respiratory chain supercomplexes (Fig. 3) (Zhang et al. 2002; Zhang et al. 2005; Wenz et al. 2009; Bazán et al. 2013). The negatively charged head group of CL was shown to facilitate the interaction of complex III and complex IV (Wenz et al. 2009). In contrast, the role of PE in the stability of respiratory chain complexes is less clear. In mammalian mitochondria, the depletion of PE causes decreased levels of respiratory supercomplexes (Tasseva et al. 2013), whereas in PE-deficient yeast mitochondria higher oligomeric assemblies comprising complexes III and IV can be detected (Böttinger et al. 2012b). Also mitochondria from a taz1 deletion yeast mutant and from Barth syndrome patients show defective assembly and reduced stability of respiratory chain supercomplexes (Brandner et al. 2005; McKenzie et al. 2006). Altogether, the phospholipid composition of the mitochondrial inner membrane is crucial for the activity of the respiratory chain.
The impaired activity of the respiratory chain complexes causes a reduced membrane potential in CL- or PE-deficient mitochondria. Consequently, the membrane potential-dependent import pathways via the TIM23 and TIM22 complexes are impaired (Fig. 3) (Jiang et al. 2000; Tamura et al. 2006; Gallas et al. 2006; Kutik et al. 2008b; Tamura et al. 2009b). Additionally, the presence of CL is important for the stability of the TIM23 complex. In particular, the association of the Pam16/Pam18 module to the TIM23 complex is affected in CL-deficient mitochondria (Fig. 3) (Tamura et al. 2006; Gallas et al. 2006; Kutik et al. 2008b; Tamura et al. 2009b). Oligomerization of the model precursor ADP/ATP carrier (AAC) and its interaction with respiratory chain complexes depend on the presence of CL (Claypool et al. 2008; Kutik et al. 2008a; Tamura et al. 2009b). In contrast, the oligomerization of the AAC precursor and the integrity of the TIM23 complex remain largely unaffected upon reduction of the PE level (Böttinger et al. 2012b). Altogether, both non-bilayer forming phospholipids CL and PE are crucial for the membrane potential-dependent protein import into and across the inner membrane. In addition, deletion of CL affects stability and assembly of inner membrane protein complexes.
Conclusions
In the last years several studies indicated that the lipid bilayer of biological membranes is much more than a hydrophobic border between cellular compartments. Individual lipids modulate the function of a versatile set of mitochondrial proteins and thereby affect central mitochondrial processes like the import of precursor proteins. Unexpectedly, recent studies revealed that both CL and PE exert specific roles for the functionality of several protein translocases. The TOM complex depends on both phospholipids for optimal function. Furthermore, membrane potential-dependent import across and into the inner membrane is affected in CL- and PE-depleted mitochondria, since both phospholipids are required for full respiratory activity. However, the molecular mechanisms by which CL and PE promote the functionality of distinct protein complexes appear to be different. While CL seems to confer stability to multimeric membrane complexes, depletion of PE does not affect the formation of protein complexes. Thus, the non-bilayer forming character of CL and PE cannot be the only important property modulating protein import machineries. Therefore, it will be important to analyze whether bilayer forming phospholipids such as PC, PS or PI play a role for activity and stability of protein transport machineries and for maintenance of the membrane potential. In addition, structural information on the membrane-embedded protein translocases will likely provide important insights how phospholipids are bound to protein complexes to modulate their activities. Moreover, studies on cellular ergosterol content revealed that the lipid composition affects protein targeting to the mitochondrial outer membrane (Krumpe et al. 2012). Future studies will have to elucidate how the specific lipid composition of membranes is recognized by the incoming precursor to warrant proper targeting. Although we are just at the beginning to understand the role of lipids in mitochondrial protein biogenesis, the first studies already revealed fascinating insights into the specific modulation of protein biogenesis steps by lipids.
References
Alder NN, Jensen RE, Johnson AE (2008) Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell 134:439–450
Banci L, Bertini I, Cefaro C, Ciofi-Baffoni S, Gallo A, Martinelli M, Sideris DP, Katrakili N, Tokatlidis K (2009) MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat Struct Mol Biol 16:198–206
Barth PG, Scholte HR, Berden JA, van der Klei-van Moorsel JM, Luyt-Houwen IE, van ’t Veer-Korthof ET, van der Harten JJ, Sobotka-Plojhar MA (1983) An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J Neurol Sci 62:327–355
Bazán S, Mileykovskaya E, Mallampalli VKPS, Heacock P, Sparagna GC, Dowhan W (2013) Cardiolipin-dependent reconstitution of respiratory supercomplexes from purified Saccharomyces cerevisiae complexes III and IV. J Biol Chem 288:401–411
Becker T, Pfannschmidt S, Guiard B, Stojanovski D, Milenkovic D, Kutik S, Pfanner N, Meisinger C, Wiedemann N (2008) Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J Biol Chem 283:120–127
Becker T, Wenz LS, Krüger V, Lehmann W, Müller JM, Goroncy L, Zufall N, Lithgow T, Guiard B, Chacinska A, Wagner R, Meisinger C, Pfanner N (2011) The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J Cell Biol 194:387–395
Becker T, Böttinger L, Pfanner N (2012) Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem Sci 37:85–91
Becker T, Horvath SE, Böttinger L, Gebert N, Daum G, Pfanner N (2013) Role of phosphatidylethanolamine in the biogenesis of mitochondrial outer membrane proteins. J Biol Chem 288:16451–16459
Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, Herrmann JM (2007) The disulfide relay system of mitochondria is connected to the respiratory chain. J Cell Biol 179:389–395
Birner R, Nebauer R, Schneiter R, Daum G (2003) Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine biosynthetic machinery with the prohibitin complex of Saccharomyces cerevisiae. Mol Biol Cell 14:370–383
Böttinger L, Gornicka A, Czerwik T, Bragoszewski P, Loniewska-Lwowska A, Schulze-Specking A, Truscott KN, Guiard B, Milenkovic D, Chacinska A (2012a) In vivo evidence for cooperation of Mia40 and Erv1 in the oxidation of mitochondrial proteins. Mol Biol Cell 23:3957–3969
Böttinger L, Horvath SE, Kleinschroth T, Hunte C, Daum G, Pfanner N, Becker T (2012b) Phosphatidylethanolamine and cardiolipin differentially affect the stability of mitochondrial respiratory chain supercomplexes. J Mol Biol 423:677–686
Brandner K, Mick DU, Frazier AE, Taylor RD, Meisinger C, Rehling P (2005) Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: implications for Barth syndrome. Mol Biol Cell 16:5202–5214
Bürgermeister M, Birner-Grünberger R, Nebauer R, Daum G (2004) Contribution of different pathways to the supply of phosphatidylethanolamine and phosphatidylcholine to mitochondrial membranes of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1686:161–168
Chacinska A, Rehling P, Guiard B, Frazier AE, Schulze-Specking A, Pfanner N, Voos W, Meisinger C (2003) Mitochondrial translocation contact sites: separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J 22:5370–5381
Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer HE, Truscott KN, Guiard B, Pfanner N, Rehling P (2005) Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120:817–829
Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628–644
Chan NC, Lithgow T (2008) The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol Biol Cell 19:126–136
Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, Wang KZQ, Zhu J, Klein-Seetharaman J, Balasubramanian K, Amoscato AA, Borisenko G, Huang Z, Gusdon AM, Cheikhi A, Steer EK, Wang R, Baty C, Watkins S, Bahar I, Bayir H, Kagan VE (2013) Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol 15:1197–1205
Claypool SM, Koehler CM (2012) The complexity of cardiolipin in health and disease. Trends Biochem Sci 37:32–41
Claypool SM, McCaffery JM, Koehler CM (2006) Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J Cell Biol 174:379–390
Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM (2008) Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J Cell Biol 182:937–950
Connerth M, Tatsuta T, Haag M, Klecker T, Westermann B, Langer T (2012) Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science 338:815–818
Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA (2000) The cytochrome bc 1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J Biol Chem 275:18093–18098
DeVay RM, Dominguez-Ramirez L, Lackner LL, Hoppins S, Stahlberg H, Nunnari J (2009) Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J Cell Biol 186:793–803
Dimmer KS, Papic D, Schumann B, Sperl D, Krumpe K, Walther DM, Rapaport D (2012) A crucial role for Mim2 in the biogenesis of mitochondrial outer membrane proteins. J Cell Sci 125:3464–3473
Dolezal P, Likic V, Tachezy J, Lithgow T (2006) Evolution of the molecular machines for protein import into mitochondria. Science 313:314–318
Dowhan W, Bogdanov M (2009) Lipid-dependent membrane protein topogenesis. Annu Rev Biochem 78:515–540
Endo T, Yamano K (2009) Multiple pathways for mitochondrial protein traffic. Biol Chem 390:723–730
Gallas MR, Dienhart MK, Stuart RA, Long RM (2006) Characterization of Mmp37p, a Saccharomyces cerevisiae mitochondrial matrix protein with a role in mitochondrial protein import. Mol Biol Cell 17:4051–4062
Gebert N, Chacinska A, Wagner K, Guiard B, Koehler CM, Rehling P, Pfanner N, Wiedemann N (2008) Assembly of the three small Tim proteins precedes docking to the mitochondrial carrier translocase. EMBO Rep 9:548–554
Gebert N, Joshi AS, Kutik S, Becker T, McKenzie M, Guan XL, Mooga VP, Stroud DA, Kulkarni G, Wenk MR, Rehling P, Meisinger C, Ryan MT, Wiedemann N, Greenberg ML, Pfanner N (2009) Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Curr Biol 19:2133–2139
Gebert N, Gebert M, Oeljeklaus S, von der Malsburg K, Stroud DA, Kulawiak B, Wirth C, Zahedi RP, Dolezal P, Wiese S, Simon O, Schulze-Specking A, Truscott KN, Sickmann A, Rehling P, Guiard B, Hunte C, Warscheid B, van der Laan M, Pfanner N, Wiedemann N (2011) Dual function of Sdh3 in the respiratory chain and TIM22 protein translocase of the mitochondrial inner membrane. Mol Cell 44:811–818
Gebert M, Schrempp SG, Mehnert CS, Heisswolf AK, Oeljeklaus S, Ieva R, Bohnert M, von der Malsburg K, Wiese S, Kleinschroth T, Hunte C, Meyer HE, Haferkamp I, Guiard B, Warscheid B, Pfanner N, van der Laan M (2012) Mgr2 promotes coupling of the mitochondrial presequence translocase to partner complexes. J Cell Biol 197:595–604
Gentle I, Gabriel K, Beech P, Waller R, Lithgow T (2004) The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol 164:19–24
Gerbeth C, Schmidt O, Rao S, Harbauer AB, Mikropoulou D, Opalińska M, Guiard B, Pfanner N, Meisinger C (2013) Glucose-induced regulation of protein import receptor Tom22 by cytosolic and mitochondria-bound kinases. Cell Metabol 18:578–587
Gohil VM, Greenberg ML (2009) Mitochondrial membrane biogenesis: phospholipids and proteins go hand in hand. J Cell Biol 184:469–472
Gohil VM, Thompson MN, Greenberg ML (2005) Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae. J Biol Chem 280:35410–35416
Gonzalvez F, Schug ZT, Houtkooper RH, MacKenzie ED, Brooks DG, Wanders RJA, Petit PX, Vaz FM, Gottlieb E (2008) Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J Cell Biol 183:681–696
Harner M, Körner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, Neupert W (2011a) The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J 30:4356–4370
Harner M, Neupert W, Deponte M (2011b) Lateral release of proteins from the TOM complex into the outer membrane of mitochondria. EMBO J 30:3232–3241
Hoppins SC, Nargang FE (2004) The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J Biol Chem 279:12396–12405
Horvath SE, Daum G (2013) Lipids of mitochondria. Prog Lipid Res 52:590–614
Horvath SE, Böttinger L, Vögtle FN, Wiedemann N, Meisinger C, Becker T, Daum G (2012) Processing and topology of the yeast mitochondrial phosphatidylserine decarboxylase 1. J Biol Chem 287:36744–36755
Hulett JM, Lueder F, Chan NC, Perry AJ, Wolynec P, Likić VA, Gooley PR, Lithgow T (2008) The transmembrane segment of Tom20 is recognized by Mim1 for docking to the mitochondrial TOM complex. J Mol Biol 376:694–704
Ishikawa D, Yamamoto H, Tamura Y, Moritoh K, Endo T (2004) Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J Cell Biol 166:621–627
Jiang F, Ryan MT, Schlame M, Zhao M, Gu Z, Klingenberg M, Pfanner N, Greenberg ML (2000) Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J Biol Chem 275:22387–22394
Joshi AS, Thompson MN, Fei N, Huttemann M, Greenberg ML (2012) Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J Biol Chem 287:17589–17597
Kawano S, Yamano K, Naoé M, Momose T, Terao K, Nishikawa S-I, Watanabe N, Endo T (2009) Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space. Proc Natl Acad Sci U S A 106:14403–14407
Kemper C, Habib SJ, Engl G, Heckmeyer P, Dimmer KS, Rapaport D (2008) Integration of tail-anchored proteins into the mitochondrial outer membrane does not require any known import components. J Cell Sci 121:1990–1998
Kerscher O, Holder J, Srinivasan M, Leung RS, Jensen RE (1997) The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J Cell Biol 139:1663–1675
Klein A, Israel L, Lackey SWK, Nargang FE, Imhof A, Baumeister W, Neupert W, Thomas DR (2012) Characterization of the insertase for β-barrel proteins of the outer mitochondrial membrane. J Cell Biol 199:599–611
Koehler CM (2004) New developments in mitochondrial assembly. Annu Rev Cell Dev Biol 20:309–335
Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P (2009) An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325:477–481
Krumpe K, Frumkin I, Herzig Y, Rimon N, Ozbalci C, Brugger B, Rapaport D, Schuldiner M (2012) Ergosterol content specifies targeting of tail-anchored proteins to mitochondrial outer membranes. Mol Biol Cell 23:3927–3935
Kutik S, Stojanovski D, Becker L, Becker T, Meinecke M, Krüger V, Prinz C, Meisinger C, Guiard B, Wagner R, Pfanner N, Wiedemann N (2008a) Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell 132:1011–1024
Kutik S, Rissler M, Guan XL, Guiard B, Shui G, Gebert N, Heacock PN, Rehling P, Dowhan W, Wenk MR, Pfanner N, Wiedemann N (2008b) The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J Cell Biol 183:1213–1221
Lange C, Nett JH, Trumpower BL, Hunte C (2001) Specific roles of protein-phospholipid interactions in the yeast cytochrome bc 1 complex structure. EMBO J 20:6591–6600
Lauffer S, Mäbert K, Czupalla C, Pursche T, Hoflack B, Rödel G, Krause-Buchholz U (2012) Saccharomyces cerevisiae porin pore forms complexes with mitochondrial outer membrane proteins Om14p and Om45p. J Biol Chem 287:17447–17458
Li Y, Dudek J, Guiard B, Pfanner N, Rehling P, Voos W (2004) The presequence translocase-associated protein import motor of mitochondria. Pam16 functions in an antagonistic manner to Pam18. J Biol Chem 279:38047–38054
Lionaki E, de Marcos LC, Baud C, Vougioukalaki M, Panayotou G, Tokatlidis K (2008) The essential function of Tim12 in vivo is ensured by the assembly interactions of its C-terminal domain. J Biol Chem 283:15747–15753
Lytovchenko O, Melin J, Schulz C, Kilisch M, Hutu DP, Rehling P (2013) Signal recognition initiates reorganization of the presequence translocase during protein import. EMBO J 32:886–898
Malhotra K, Sathappa M, Landin JS, Johnson AE, Alder NN (2013) Structural changes in the mitochondrial Tim23 channel are coupled to the proton-motive force. Nat Struct Mol Biol 20:965–972
Marom M, Dayan D, Demishtein-Zohary K, Mokranjac D, Neupert W, Azem A (2011) Direct interaction of mitochondrial targeting presequences with purified components of the TIM23 protein complex. J Biol Chem 286:43809–43815
Martinez-Caballero S, Grigoriev SM, Herrmann JM, Campo ML, Kinnally KW (2007) Tim17p regulates the twin pore structure and voltage gating of the mitochondrial protein import complex TIM23. J Biol Chem 282:3584–3593
McKenzie M, Lazarou M, Thorburn DR, Ryan MT (2006) Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J Mol Biol 361:462–469
Meinecke M, Wagner R, Kovermann P, Guiard B, Mick DU, Hutu DP, Voos W, Truscott KN, Chacinska A, Pfanner N, Rehling P (2006) Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 312:1523–1526
Meineke B, Engl G, Kemper C, Vasiljev-Neumeyer A, Paulitschke H, Rapaport D (2008) The outer membrane form of the mitochondrial protein Mcr1 follows a TOM-independent membrane insertion pathway. FEBS Lett 582:855–860
Meisinger C, Pfannschmidt S, Rissler M, Milenkovic D, Becker T, Stojanovski D, Youngman MJ, Jensen RE, Chacinska A, Guiard B, Pfanner N, Wiedemann N (2007) The morphology proteins Mdm12/Mmm1 function in the major β-barrel assembly pathway of mitochondria. EMBO J 26:2229–2239
Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM (2005) A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121:1059–1069
Mokranjac D, Bourenkov G, Hell K, Neupert W, Groll M (2006) Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor. EMBO J 25:4675–4685
Mokranjac D, Sichting M, Popov-Čeleketić D, Mapa K, Gevorkyan-Airapetov L, Zohary K, Hell K, Azem A, Neupert W (2009) Role of Tim50 in the transfer of precursor proteins from the outer to the inner membrane of mitochondria. Mol Biol Cell 20:1400–1407
Neupert W, Herrmann JM (2007) Translocation of proteins into mitochondria. Annu Rev Biochem 76:723–749
Okamoto H, Miyagawa A, Shiota T, Tamura Y, Endo T (2014) Intramolecular disulfide bond of Tim22 protein maintains integrity of the TIM22 complex in the mitochondrial inner membrane. J Biol Chem 289:4827–4838
Osman C, Haag M, Potting C, Rodenfels J, Dip PV, Wieland FT, Brugger B, Westermann B, Langer T (2009) The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J Cell Biol 184:583–596
Osman C, Voelker DR, Langer T (2011) Making heads or tails of phospholipids in mitochondria. J Cell Biol 192:7–16
Otera H, Taira Y, Horie C, Suzuki Y, Suzuki H, Setoguchi K, Kato H, Oka T, Mihara K (2007) A novel insertion pathway of mitochondrial outer membrane proteins with multiple transmembrane segments. J Cell Biol 179:1355–1363
Ott M, Norberg E, Walter KM, Schreiner P, Kemper C, Rapaport D, Zhivotovsky B, Orrenius S (2007) The mitochondrial TOM complex is required for tBid/Bax-induced cytochrome c release. J Biol Chem 282:27633–27639
Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong S-E, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134:112–123
Papic D, Krumpe K, Dukanovic J, Dimmer KS, Rapaport D (2011) Multispan mitochondrial outer membrane protein Ugo1 follows a unique Mim1-dependent import pathway. J Cell Biol 194:397–405
Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W (2003) Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426:862–866
Patil VA, Fox JL, Gohil VM, Winge DR, Greenberg ML (2013) Loss of cardiolipin leads to perturbation of mitochondrial and cellular iron homeostasis. J Biol Chem 288:1696–1705
Peixoto PMV, Graña F, Roy TJ, Dunn CD, Flores M, Jensen RE, Campo ML (2007) Awaking TIM22, a dynamic ligand-gated channel for protein insertion in the mitochondrial inner membrane. J Biol Chem 282:18694–18701
Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schägger H (2003) Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem 278:52873–52880
Popov-Čeleketić J, Waizenegger T, Rapaport D (2008) Mim1 functions in an oligomeric form to facilitate the integration of Tom20 into the mitochondrial outer membrane. J Mol Biol 376:671–680
Qiu J, Wenz L-S, Zerbes RM, Oeljeklaus S, Bohnert M, Stroud DA, Wirth C, Ellenrieder L, Thornton N, Kutik S, Wiese S, Schulze-Specking A, Zufall N, Chacinska A, Guiard B, Hunte C, Warscheid B, van der Laan M, Pfanner N, Wiedemann N, Becker T (2013) Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell 154:596–608
Rehling P, Model K, Brandner K, Kovermann P, Sickmann A, Meyer HE, Kühlbrandt W, Wagner R, Truscott KN, Pfanner N (2003) Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 299:1747–1751
Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A (2006) Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res 5:1543–1554
Richter-Dennerlein R, Korwitz A, Haag M, Tatsuta T, Dargazanli S, Baker M, Decker T, Lamkemeyer T, Rugarli EI, Langer T (2014) DNAJC19, a mitochondrial cochaperone associated with cardiomyopathy, forms a complex with prohibitins to regulate cardiolipin remodeling. Cell Metabol 20:158–171
Schägger H, Pfeiffer K (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19:1777–1783
Schmidt O, Harbauer AB, Rao S, Eyrich B, Zahedi RP, Stojanovski D, Schönfisch B, Guiard B, Sickmann A, Pfanner N, Meisinger C (2011) Regulation of mitochondrial protein import by cytosolic kinases. Cell 144:227–239
Schulz C, Lytovchenko O, Melin J, Chacinska A, Guiard B, Neumann P, Ficner R, Jahn O, Schmidt B, Rehling P (2011) Tim50’s presequence receptor domain is essential for signal driven transport across the TIM23 complex. J Cell Biol 195:643–656
Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, Yamashita E, Tsukihara T, Yoshikawa S (2007) Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J 26:1713–1725
Shiota T, Mabuchi H, Tanaka-Yamano S, Yamano K, Endo T (2011) In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc Natl Acad Sci U S A 108:15179–15183
Song J, Tamura Y, Yoshihisa T, Endo T (2014) A novel import route for an N-anchor mitochondrial outer membrane protein aided by the TIM23 complex. EMBO Rep 15:670–677
Stan T, Ahting U, Dembowski M, Künkele KP, Nussberger S, Neupert W, Rapaport D (2000) Recognition of preproteins by the isolated TOM complex of mitochondria. EMBO J 19:4895–4902
Stojanovski D, Guiard B, Kozjak-Pavlovic V, Pfanner N, Meisinger C (2007) Alternative function for the mitochondrial SAM complex in biogenesis of α-helical TOM proteins. J Cell Biol 179:881–893
Tamura Y, Harada Y, Yamano K, Watanabe K, Ishikawa D, Ohshima C, Nishikawa S-I, Yamamoto H, Endo T (2006) Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. J Cell Biol 174:631–637
Tamura Y, Harada Y, Shiota T, Yamano K, Watanabe K, Yokota M, Yamamoto H, Sesaki H, Endo T (2009a) Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J Cell Biol 184:129–141
Tamura Y, Endo T, Iijima M, Sesaki H (2009b) Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J Cell Biol 185:1029–1045
Tamura Y, Harada Y, Nishikawa S-I, Yamano K, Kamiya M, Shiota T, Kuroda T, Kuge O, Sesaki H, Imai K, Tomii K, Endo T (2013) Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell Metabol 17:709–718
Tasseva G, Bai HD, Davidescu M, Haromy A, Michelakis E, Vance JE (2013) Phosphatidylethanolamine deficiency in mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J Biol Chem 288:4158–4173
Thornton N, Stroud DA, Milenkovic D, Guiard B, Pfanner N, Becker T (2010) Two modular forms of the mitochondrial sorting and assembly machinery are involved in biogenesis of α-helical outer membrane proteins. J Mol Biol 396:540–549
Tienson HL, Dabir DV, Neal SE, Loo R, Hasson SA, Boontheung P, Kim S-K, Loo JA, Koehler CM (2009) Reconstitution of the Mia40-Erv1 oxidative folding pathway for the small TIM proteins. Mol Biol Cell 20:3481–3490
van den Brink-van der Laan E, Killian JA, de Kruijff B (2004) Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim Biophys Acta 1666:275–288
van der Laan M, Chacinska A, Lind M, Perschil I, Sickmann A, Meyer HE, Guiard B, Meisinger C, Pfanner N, Rehling P (2005) Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol Cell Biol 25:7449–7458
van der Laan M, Wiedemann N, Mick DU, Guiard B, Rehling P, Pfanner N (2006) A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr Biol 16:2271–2276
van Meer G, Voelker DR, Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9:112–124
Voelker DR (1997) Phosphatidylserine decarboxylase. Biochim Biophys Acta 1348:236–244
von der Malsburg K, Müller JM, Bohnert M, Oeljeklaus S, Kwiatkowska P, Becker T, Loniewska-Lwowska A, Wiese S, Rao S, Milenkovic D, Hutu DP, Zerbes RM, Schulze-Specking A, Meyer HE, Martinou J-C, Rospert S, Rehling P, Meisinger C, Veenhuis M, Warscheid B, van der Klei IJ, Pfanner N, Chacinska A, van der Laan M (2011) Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell 21:694–707
Wagner K, Gebert N, Guiard B, Brandner K, Truscott KN, Wiedemann N, Pfanner N, Rehling P (2008) The assembly pathway of the mitochondrial carrier translocase involves four preprotein translocases. Mol Cell Biol 28:4251–4260
Wenz T, Hielscher R, Hellwig P, Schägger H, Richers S, Hunte C (2009) Role of phospholipids in respiratory cytochrome bc 1 complex catalysis and supercomplex formation. Biochem Biophys Acta 1787:609–616
Wenz LS, Opalinski L, Schuler MH, Ellenrieder L, Ieva R, Böttinger L, Qiu J, van der Laan M, Wiedemann N, Guiard B, Pfanner N, Becker T (2014) The presequence pathway is involved in protein sorting to the mitochondrial outer membrane. EMBO Rep 15:678–685
Wideman JG, Go NE, Klein A, Redmond E, Lackey SWK, Tao T, Kalbacher H, Rapaport D, Neupert W, Nargang FE (2010) Roles of the Mdm10, Tom7, Mdm12, and Mmm1 proteins in the assembly of mitochondrial outer membrane proteins in Neurospora crassa. Mol Biol Cell 21:1725–1736
Wiedemann N, Kozjak V, Chacinska A, Schönfisch B, Rospert S, Ryan MT, Pfanner N, Meisinger C (2003) Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424:565–571
Wiedemann N, Truscott KN, Pfannschmidt S, Guiard B, Meisinger C, Pfanner N (2004) Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J Biol Chem 279:18188–18194
Yamano K, Yatsukawa Y-I, Esaki M, Hobbs AEA, Jensen RE, Endo T (2008) Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J Biol Chem 283:3799–3807
Zhang M, Mileykovskaya E, Dowhan W (2002) Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem 277:43553–43556
Zhang M, Mileykovskaya E, Dowhan W (2005) Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J Biol Chem 280:29403–29408
Zhong Q, Gohil VM, Ma L, Greenberg ML (2004) Absence of cardiolipin results in temperature sensitivity, respiratory defects, and mitochondrial DNA instability independent of pet56. J Biol Chem 279:32294–32300
Acknowledgments
Work included in this study has also been performed in partial fulfillment of the requirements for the doctoral thesis of L.B. and L.E. at the University of Freiburg. This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 746 and by the Excellence Initiative of the German Federal and State Governments (EXC 294 BIOSS Centre for Biological Signalling Studies).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Böttinger, L., Ellenrieder, L. & Becker, T. How lipids modulate mitochondrial protein import. J Bioenerg Biomembr 48, 125–135 (2016). https://doi.org/10.1007/s10863-015-9599-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10863-015-9599-7