Abstract
Glycoproteins are characterized by the heterogeneous and dynamic nature of their glycan moieties, which hamper crystallographic analysis. NMR spectroscopy provides potential advantages in dealing with such complicated systems, given that the target molecules can be isotopically labeled. Methods of metabolic isotope labeling in recombinant glycoproteins have been developed recently using a variety of eukaryotic production vehicles, including mammalian, yeast, insect, and plant cells, each of which has a distinct N-glycan diversification pathway. Yeast genetic engineering has enabled the overexpression of homogeneous high-mannose-type oligosaccharides with 13C labeling for NMR characterization of their conformational dynamics. The utility of stable isotope-assisted NMR spectroscopy has also been demonstrated using the Fc fragment of immunoglobulin G (IgG) as a model glycoprotein, providing useful information regarding intramolecular carbohydrate–protein interactions. Transverse relaxation optimization of intact IgG with a molecular mass of 150 kDa has been achieved by tailored deuteration of selected amino acid residues using a mammalian expression system. This offers a useful probe for the characterization of molecular interaction networks in multimolecular crowded systems typified by serum. Perspectives regarding the development of techniques for tailoring glycoform designs and isotope labeling of recombinant glycoproteins are also discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glycosylation is a major post-translational modification of proteins. More than half of the proteins in nature are believed to undergo carbohydrate modifications (Apweiler et al. 1999; Varki et al. 2017) that confer structural stability and functional diversity. Despite the significant roles of carbohydrate moieties, structural studies of glycoproteins have significantly lagged behind those of other biomacromolecules, as exemplified by the small percentage (approximately 4%) of glycoprotein structures in the total entries deposited in the Protein Data Bank (Kato et al. 2018). This is largely because the glycans of glycoproteins exhibit heterogeneity in terms of covalent structures and possess considerable motional freedom, both of which hinder crystallization of protein and interpretation of electron densities.
In this context, NMR spectroscopy is a promising tool for the structural characterization of glycoproteins because it can deal with heterogeneous and dynamic biomacromolecules in solution. However, it should be noted that glycoproteins can neither be produced by cell-free expression nor by conventional bacterial expression systems such as Escherichia coli. This results in a problematic bottleneck in the stable isotope labeling of glycoproteins as targets of NMR analysis. Therefore, we have been developing stable isotope-assisted NMR methods for studying the conformational dynamics and interactions of glycoproteins using eukaryotic expression systems (Kato and Yamaguchi 2012; Kato et al. 2010, 2018; Yamaguchi and Kato 2010; Yamaguchi et al. 2017).
In the present article, we have outlined current state-of-the-art techniques for the stable isotope labeling of oligosaccharides and glycoproteins and their cutting-edge NMR applications.
N-glycan processing pathways
For the preparation of isotope-labeled glycoproteins, it is essential to consider the biosynthetic and processing pathways of their glycans. In this article, we have focused on protein modification with N-linked oligosaccharides, which is a major type of protein glycosylation. Figure 1 summarizes the N-glycan processing pathways in various organisms. Although E. coli cells do not possess the N-glycosylation mechanism, archaea and some bacteria, including Campylobacter jejuni, share N-glycosylation systems (Fig. 1b), which are distinct from those of eukaryotes in terms of the consensus sequence of glycosylation sites as well as glycan structures (Fig. 1a) (Jarrell et al. 2014; Kowarik et al. 2006). In eukaryotes, the common precursor of N-glycan, which is a triantennary high-mannose-type oligosaccharide containing three glucose and nine mannose residues (G3M9), is transferred from a lipid carrier to the side-chain amide groups of asparagine residues in the endoplasmic reticulum (ER) (Fig. 1a). N-glycan processing occurs in two phases: trimming of high-mannose-type glycans in the ER followed by diversification into various complex-type glycans in the Golgi. The former is common among eukaryotic cells and is associated with the intracellular fate-determination processes of glycoproteins, i.e., folding in the ER, transport from the ER to the Golgi, and degradation in the cytoplasm (Aebi et al. 2010; Kamiya et al. 2012; Kato and Kamiya 2007; Lederkremer 2009; Takeda et al. 2009). Newly synthesized proteins are modified by G3M9 before their release from the ribosome attached to the ER membrane. After its attachment to protein, this glycan undergoes sequential trimming by glucosidases and mannosidases in the ER, giving rise to a series of processing intermediates, which carry determinants of glycoprotein fates recognized by a panel of intracellular lectins operating as molecular chaperones, cargo receptors, and mediators of ER-associated protein degradation (ERAD). For example, the ER chaperone calreticulin specifically recognizes glycoproteins carrying monoglucosylated high-mannose-type intermediates such as GM9 (Kozlov et al. 2010), whereas ERAD lectin OS-9 recognizes glycans after mannose trimming (Hosokawa et al. 2009; Satoh et al. 2010). In contrast, the diversification processes in the Golgi are species-dependent. For example, yeast glycoproteins undergo hyper-mannosylation in the Golgi whereas mammalian glycoproteins are characterized by fucosylation, galactosylation, and sialylation, which are often associated with the functional regulation and fate determination of secretory glycoproteins in extracellular environments (Varki et al. 2017). Therefore, the choice of production vehicle is critical to the preparation of recombinant glycoproteins for NMR studies. Furthermore, genetic engineering of glycan processing pathways is useful for actively controlling glycoprotein glycoforms expressed by eukaryotic cells (Kamiya et al. 2014).

Reproduced with permission from Kamiya et al. (2014)
Scheme of a the N-glycan processing pathway in the ER common to mammals, insects, yeast, and plants and the divergent pathways in the Golgi complex along with b N-glycosylation in C. jejuni.
Conformational dynamics of oligosaccharides
We employed an engineered strain of Saccharomyces cerevisiae in which the genes involved in hyper-mannosylation in the Golgi were knocked out. This resulted in the overexpression of glycoproteins modified homogeneously by the high-mannose-type oligosaccharide M8B, containing eight mannose residues and lacking the non-reducing terminal mannose residue at the central branch (Fig. 2) (Chiba and Akeboshi 2009). Additional deletion of the gene encoding ER mannosidase gave rise only to glycoproteins with the M9 glycoform. This facilitated the preparation of 13C-labeled oligosaccharides with specific glycoforms from glycoprotein mixtures, which were harvested from genetically engineered yeast cells grown in minimum medium containing [13C]glucose as the sole carbon source (Kamiya et al. 2011, 2013). An isotope-labeled GM9 oligosaccharide can be prepared by treating a yeast-derived glycoprotein mixture exhibiting the M9 glycoform with UDP-[13C]glucose as donor substrate and UGGT as catalyst (Zhu et al. 2015). This ER enzyme catalyzes the re-glucosylation of high-mannose-type oligosaccharides expressed by a yet unfolded protein, thereby giving it an opportunity to revisit the ER chaperones. The 13C-labeled oligosaccharides cleaved from proteins by a hydrazine treatment can be chemically conjugated with a lanthanide-chelating tag at the reducing terminus for observing conformation-dependent paramagnetic effects, providing useful information for the characterization of their conformational dynamics in solution (Kato and Yamaguchi 2015). Based on molecular dynamics simulations validated by paramagnetism-assisted NMR data, dynamic conformational ensembles of the panel of high-mannose-type oligosaccharides have been determined in solution (Suzuki et al. 2017; Yamaguchi et al. 2014). In conjunction with crystallographic data on lectin-bound oligosaccharide conformations, we have proposed mechanisms by which the glycoprotein-fate determinants are recognized by ER chaperones and cargo receptors (Kato et al. 2017).
Schematic of protocols for producing 13C-labeled high-mannose-type oligosaccharides M8B, M9, and GM9 based on metabolic labeling using genetically engineered yeast cells in conjunction with chemoenzymatic synthesis. The genetically engineered yeast cells with deletions of α1–3 mannosyl transferase, α1–6 mannosyl transferase, and mannosyl-phospate transferase produce glycoproteins exclusively expressing the M8B oligosaccharides. The additional deletion of ER α1-2- mannosidase enables the overexpression of glycoproteins possessing only the M9 oligosaccharides. 13C enrichment (98% or higher) of these oligosaccharides can be achieved by cultivating the yeast cells in a medium containing 13C-labeled glucose (Kamiya et al. 2011, 2013). The typical yield of 13C-labeled M9 cleaved from the glycoprotein mixture harvested from the engineered yeast is 380 nmol/l of cell culture. Isotopically labeled GM9 can be prepared by in vitro enzymatic glucosylation catalyzed by UGGT using UDP-[13C6]glucose and M9-expressing glycoproteins thus prepared as donor and acceptor substrates, respectively
Metabolic isotope labeling of glycoproteins
We used monoclonal antibodies as model glycoproteins for developing stable isotope-assisted NMR strategies (Kato et al. 2010). The Fc portion of immunoglobulin G (IgG) possess a conserved N-glycosylation site at Asn297 in each heavy chain, which displays a bi-antennary complex-type oligosaccharide with microheterogeneities resulting from the presence and absence of non-reducing terminal galactose and sialic acid residues as well as the core fucose residue. X-ray crystallographic studies indicate that a pair of N-glycans is packed within the horseshoe-shape of the Fc quaternary structure (Deisenhofer 1981; Matsumiya et al. 2007; Yamaguchi et al. 2007). The Fc glycoforms critically affect IgG effector functions, presumably because the Fc glycans are involved in the optimization of the interaction between IgG and effector molecules (Dekkers et al. 2017; Li et al. 2017). For example, removal of the core fucose residue from the Fc N-glycans causes a dramatic enhancement of antibody-dependent cell-mediated cytotoxicity (ADCC), which is an immune mechanism mediated by the interaction of IgG–Fc with the Fcγ receptor expressed on natural killer cells. (Jefferis 2016; Niwa et al. 2004; Shields et al. 2002; Shinkawa et al. 2003). Bacterially-expressed IgG lacks glycans and therefore cannot maintain structural integrity, which is necessary for interactions with Fcγ receptors (Simmons et al. 2002). Thus, protein glycosylation is now considered to be one of the most important factors in developing biopharmaceuticals typified by therapeutic antibodies.
To produce functionally active antibodies, mammalian cell lines, including hybridoma and Chinese hamster ovary (CHO) cells, have been used as expression vehicles (Kato and Yamaguchi 2012; Kato et al. 2010, 2018; Yamaguchi and Kato 2010; Yamaguchi et al. 2017). These cells can be cultivated in synthetic media in which metabolic precursors such as glucose and amino acids are isotopically labeled, enabling uniform or selective isotope labeling of glycoproteins. While the degree of isotope enrichment in selective labeling critically depends on amino acid types and culture conditions, uniform 13C/15N-labeling of glycoproteins would be possible given that all the relevant metabolic precursors could be substituted with isotope-labeled compounds. A commercially available fully isotope-labeled amino acid mixture, derived from blue-green algal source, is used as metabolic precursor for the cost-effective expression of uniformly 13C/15N-labeled proteins in mammalian cells. However, the mixture needs to be complemented with 13C/15N-labled glutamine, asparagine, cysteine, and tryptophan because these amino acids are decomposed during the acid hydrolysis of algal proteins. Figure 3b displays the 1H–15N HSQC spectrum of the Fc fragment cleaved from mouse antibody of IgG2b subclass, which was prepared using hybridoma cells. Fc glycoforms can be made uniform by in vitro enzymatic trimming using glycosidases. We have reported that step-wise enzymatic trimming of the bi-antennary oligosaccharides of Fc causes progressive spectral changes in its polypeptide backbone, eventually perturbing the Fcγ receptor-binding sites in the hinge region upon cleavage at the innermost GlcNAcβ1-4GlcNAc linkage (Supplementary Fig. 1) (Kato et al. 2010; Yamaguchi et al. 2006). This explains the molecular mechanisms by which the Fcγ receptor-mediated effector functions of IgG are impaired by the removal of the outer Fc carbohydrate branches.
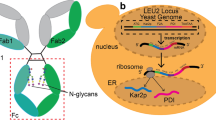
a Schematic diagram of IgG antibody structure. IgG is composed of two identical heavy chains and two identical light chains and can be cleaved by papain into two Fab and one Fc fragment. Asn-297 in each heavy chain provides a conserved glycosylation site. b 1H–15N HSQC and c 1H–13C HSQC spectra of uniformly 13C/15N-labeled mouse IgG2b-Fc. d Part of 2D HSQC-NOESY spectrum of mouse IgG1-Fc (metabolically labeled with [13C6] glucose) exhibiting intramolecular NOE connectivity between Tyr296 and the core fucose residue.
Many attempts have been made to improve expression yields of recombinant glycoproteins by mammalian cells (Kunert and Reinhart 2016; Li et al. 2010; Walsh 2014). For example, the production of human IgG by CHO cells could be increased up to approximately 5 g per liter of cell culture by optimization of medium, bioreactor, and expression vector design (Kunert and Reinhart 2016; Omasa et al. 2010; Stolfa et al. 2017). On the other hand, non-mammalian expression systems that are advantageous in terms of cost, time, and yield, compared with mammalian systems, have been developed for the stable isotope labeling of recombinant glycoproteins. Insect cells can serve as production vehicles for mammalian proteins. In particular, Sf9 cells infected with baculovirus are frequently used for recombinant protein expression with isotope labeling (Saxena et al. 2012; Walton et al. 2006). Furthermore, baculovirus-infected living silkworms are used for producing recombinant proteins for structural studies. We previously reported a protocol to produce an isotope-labeled glycoprotein in the hemolymph of live silkworm larvae reared on an artificial diet using human IgG as a model molecule (Yagi et al. 2015b). In this method, the artificial diet included a protein mixture derived from a yeast strain Candida utilis grown in a culture medium containing 15N-labeled ammonium sulfate as nitrogen source. From a single silkworm larva, 0.1 mg of IgG was harvested with a 15N-enrichment ratio of approximately 80%. Transgenic plants provide an alternative approach to the expression of recombinant glycoproteins, especially those of biopharmaceutical interest. An isotope-labeled IgG glycoprotein could be prepared using transgenic tobacco (Nicotiana benthamiana) (Yagi et al. 2015a); four-week-old seedlings were placed in a drip hydroponic system and cultivated for 49 days in a Murashige and Skoog medium containing 15N-labeled potassium nitrate (K15NO3) and ammonium nitrate (15NH415NO3) as the major nitrogen sources. The yield of IgG from 1 g of the fresh leaves was 0.1 mg with a 15N-enrichment ratio in the range of 52–58%. HSQC spectral comparison of these heterologously produced IgG-Fc glycoproteins with that derived from CHO cells (Yagi et al. 2015c) confirmed their overall structural integrity, but simultaneously indicated significant differences in the microenvironments surrounding N-glycans. The N-glycan diversification processes in the Golgi of these organisms are distinct from those in mammals. Namely, insect cells express pauci-mannose N-glycans, while the plant N-glycans are characterized by β1,2-xylosylation and α1,3-fucosylation. It is conceivable that the observed spectral differences resulted from this glycosylation variation (Supplementary Fig. 2).
Metabolic isotope labeling of glycoprotein carbohydrate moieties can be performed conventionally by cultivating mammalian cells using 13C-labeled glucose as a metabolic precursor (Yamaguchi et al. 1998). Figure 3d shows the HSQC-NOESY spectrum of mouse IgG2b-Fc thus prepared, exhibiting intramolecular NOE connectivities between the side chain of Tyr296 and the core fucose residue of its N-glycan. Knocking out the gene encoding the fucosyltransferase responsible for the core fucosylation in antibody-producing cells yields non-fucosylated IgG. Stable-isotope-assisted NMR data on the non-fucosyated glycoform of human IgG1-Fc thus prepared has revealed that Tyr296 exhibits conformational multiplicity in the absence of the core fucose residue (Matsumiya et al. 2007). X-ray crystallographic data have indicated that this proximal tyrosine residue is involved in interaction with the ADCC-promoting Fcγ receptor (Ferrara et al. 2011; Isoda et al. 2015; Mizushima et al. 2011). These data suggest that the fucosylation masks Tyr296, thereby compromising receptor binding, thus explaining the mechanisms of improved ADCC by removal of the core fucose residue (Supplementary Fig. 3).
Isotope-assisted NMR observation of larger glycoproteins
Glycoproteins generally exhibit slower molecular tumbling because of their bulky glycan moieties and often because of their multidomain and/or oligomeric structures. Moreover, a great majority of membrane proteins are classified as glycoproteins. Therefore, NMR approaches to glycoproteins often involve strategies to cope with rapid relaxation problems resulting from slow molecular tumbling. One promising approach is amino acid-selective labeling using eukaryotic expression systems as exemplified by a methodology developed by Arata and co-workers, which is based on carbonyl 13C labeling of selected amino acid residues. This enables NMR observation of 150 kDa IgG glycoproteins in one-dimensional 13C spectral measurements (Arata et al. 1994; Kato et al. 1989a). Another promising approach is deuteration, which reduces magnetic dipole–dipole interactions as the source of transverse relaxation (Crespi et al. 1968; Kalbitzer et al. 1985; Kato et al. 1989b; LeMaster and Richards 1988; Markley et al. 1968; Sattler and Fesik 1996). Although uniform deuteration of glycoprotein remains challenging because eukaryotic cells are not adapted to D2O media, amino acid-selective deuteration has been successfully applied to suppress line broadening of the remaining proton signals. Metabolic incorporation rates have been estimated for 2H as well as 15N in the model recombinant protein produced by Sf9 cells (Opitz et al. 2015). Such systematic investigation provides useful information and is required for other eukaryotic expression systems.
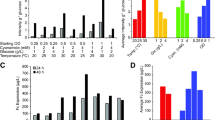
Here, we examined the applicability of regio- and stereo-specifically isotope-labeled amino acids (so-called “SAIL amino acids”) developed by Kainosho and co-workers (Kainosho and Güntert 2009; Miyanoiri et al. 2016), in conjunction with selective amino acid incorporation via a metabolic pathway of mammalian cells, for heteronuclear NMR characterization of larger glycoproteins. We used mouse hybridoma cells for preparation of IgG2b glycoprotein that was metabolically labeled using [δ2-13C; Hα, Hβ, Hγ, Hδ1–2H7]leucine and its nondeuterated counterpart (Taiyo Nippon Sanso Co.) (Supplementary information). Figure 4a, b compare the methyl-transverse relaxation optimized spectroscopy (methyl-TROSY) spectrum of mouse IgG2b glycoprotein thus prepared. Significant improvements in spectral quality were achieved by mere selective deuteration at the leucine residues, enabling the observation of δ2 methyl peaks for all the 42 leucine residues in this antibody (Supplementary Fig. 4). Additional deuteration using [2H7]glucose as a metabolic precursor resulted in little or no further improvement, presumably because the degree of deuteration through metabolic labeling in regular water was not high (estimated as approximately 10%) (Fig. 4c and Supplementary Fig. 4). The isotope-labeled IgG2b was cleaved at the hinge region by papain, giving rise to Fab and Fc fragments (Supplementary information), which were also subjected to spectral measurements. Superposition of the spectra of the Fab and Fc fragments, which contain 28 and 14 non-equivalent leucine residues, respectively, reproduced the spectrum of intact IgG2b in terms of peak positions, allowing us to classify these peaks as Fab and Fc portions (Fig. 4d). Transverse relaxation optimization by tailored deuteration is thus useful for NMR analysis of larger glycoproteins.
Methyl-TROSY spectra of mouse IgG labeled with a [δ2-13C]leucine, b [δ2-13C; Hα, Hβ, Hγ, Hδ1–2H7]leucine, and c [δ2-13C; Hα, Hβ, Hγ, Hδ1–2H7]leucine plus [2H7]glucose. d Superposition of methyl-TROSY spectra of the Fab (orange) and Fc (blue) fragments derived from mouse IgG2b labeled with [δ2-13C; Hα, Hβ, Hγ, Hδ1–2H7]leucine plus [2H7]glucose. Concentration of all the samples was set to 10 mg/ml in 5 mM sodium phosphate buffer containing 50 mM NaCl in 99.8% D2O. The isotopically labeled materials including the SAIL amino acids were from Taiyo Nippon Sanso Co. All the spectra were acquired at 37 °C at pH 7.4, using an AVANCE 800 spectrometer (Bruker BioSpin). In a, the peak indicated by an asterisk appeared during spectral measurements
Application of stable-isotope-assisted NMR techniques for characterizing multimolecular crowded systems
Most soluble glycoproteins are secreted into extracellular environments and function in multimolecular crowded systems typified by IgG, which is a major class of serum protein. NMR offers useful tools for characterizing biomolecular structures and interactions in those systems as exemplified by in-cell NMR approaches (Freedberg and Selenko 2014). Recently, we reported the stable isotope-assisted NMR detection of semi-specific antibody interactions in serum. In this study, uniformly 15N-labeled mouse IgG2b–Fc in human serum was subjected to 1H–15N HSQC spectral measurement, in which many Fc peaks exhibited magnitudes of attenuation varying in intensity, suggesting interactions of IgG2b–Fc with serum component(s) (Yanaka et al. 2017). This spectral change was because of interactions with polyclonal IgG antibodies in serum through Fab portions. These results indicate that there exists a subset of antibodies reactive with mouse IgG2b-Fc, as a potential antigen, in human serum without preimmunization.
We subjected mouse IgG2b labeled with [δ2-13C; Hα, Hβ, Hγ, Hδ1–2H7]leucine to in-serum NMR measurement. As shown in Fig. 5a, IgG2b in human serum exhibited significant reductions of varying magnitudes in peak intensity (Fig. 5b and Supplementary Fig. 5). Interestingly, intensity attenuation was far more pronounced in the peaks originating from the Fab portion. Similar spectral changes were induced by Fab fragments derived from human serum polyclonal IgG. These results suggest that the epitopes recognized by polyclonal antibodies in human serum were more extensively distributed in Fab than in Fc. This raises the possibility that the preexisting antibodies in serum are reactive with the Fab portion of other antibodies and thereby influence their antigen binding activities, which have often been characterized under isolated conditions. The present observation also reminds us of the immune network theory, which states that numerous polyclonal antibodies in the immune system form a network, in which they interact with one another through their antigen recognition sites (Hoffmann 1975; Jerne 1974). Our stable isotope-assisted NMR approach will offer useful probes for characterizing molecular interaction networks involving secretory glycoproteins in multimolecular crowded systems typified by blood environments, providing useful insights for designing and developing biopharamaceuticals, including therapeutic antibodies.
a and b Methyl-TROSY spectra of mouse IgG obtained in 5 mM sodium phosphate buffer containing 50 mM NaCl and 10% (v/v) D2O are shown in black and the NMR spectra with serum or with 10 mg/ml Fab derived from human serum IgG are shown in red, respectively. The peak originating from the serum component is indicated by an asterisk. All the spectra were acquired at 37 °C at pH 7.4, using an AVANCE 800 spectrometer (Bruker BioSpin)
Perspectives
As discussed, stable isotope-assisted NMR studies of glycoproteins have come to fruition using eukaryotic expression systems and provide useful information regarding the dynamic conformations and interactions of glycoproteins. However, several issues such as the cost of isotope labeling, especially uniform 2H labeling, and the task of designing glycoforms in a tailored fashion need to be addressed. An ambitious and promising method would involve bacterial expression of eukaryotic glycoproteins. This has already been achieved to produce recombinant proteins modified by Man3GlcNAc2 moieties in genetically engineered E. coli equipped with a eukaryotic N-glycosylation pathway and PglB, the oligosaccharide transferase from C. jejuni (Valderrama-Rincon et al. 2012). Although mutational modifications are required at the N-glycosylation site because of the distinct consensus sequence recognized by PglB, this line of work will help to overcome the drawbacks of metabolic isotope labeling using eukaryotic expression systems.
For tailoring glycoforms of recombinant glycoproteins, a useful approach is in vitro remodeling combining enzymatic trimming and elongation of the glycans (Kato et al. 2010). Most generally, the innermost GlcNAc-Asn residues into which various N-glycans can be trimmed provide acceptor sites for synthetic N-glycans in in vitro trasnglycosylation catalyzed by endo-β-N-acetylglucosaminidases (Fan et al. 2012; Wang and Lomino 2012). This chemoenzymatic method has been enhanced by developing chemical techniques to convert synthetic N-glycan to an oxazoline analog as an activated donor substrate along with recombinant enzymes engineered for suppression of product hydrolysis (Huang et al. 2012). Moreover, given that a synthetic N-glycan is available in isotope-labeled forms, it can be chemoenzymatically attached onto an isotope-labeled recombinant protein bearing the GlcNAc-Asn acceptor site, which can be prepared using the bacterial expression system and subsequent enzymatic trimming. These techniques, in conjunction with expressed protein ligation (Liu and Cowburn 2017; Liu et al. 2009), will form a technical basis for glycoproteins with multiple glycosylations using tailored designs of glycoform and isotope labeling. Stable isotope-assisted NMR spectroscopy of glycoproteins based on integration of multilateral approaches will thus be an essential tool in the exploration of glycan-mediated biomolecular networks.
References
Aebi M, Bernasconi R, Clerc S, Molinari M (2010) N-glycan structures: recognition and processing in the ER. Trends Biochem Sci 35:74–82
Apweiler R, Hermjakob H, Sharon N (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta 1473:4–8
Arata Y, Kato K, Takahashi H, Shimada I (1994) Nuclear magnetic resonance study of antibodies: a multinuclear approach. Methods Enzymol 239:440–464
Chiba Y, Akeboshi H (2009) Glycan engineering and production of ‘humanized’ glycoprotein in yeast cells. Biol Pharm Bull 32:786–795
Crespi HL, Rosenberg RM, Katz JJ (1968) Proton magnetic resonance of proteins fully deuterated except for 1H-leucine side chains. Science 161:795–796
Deisenhofer J (1981) Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-Å resolution. Biochemistry 20:2361–2370
Dekkers G, Treffers L, Plomp R, Bentlage AEH, de Boer M, Koeleman CAM, Lissenberg-Thunnissen SN, Visser R, Brouwer M, Mok JY, Matlung H, van den Berg TK, van Esch WJE, Kuijpers TW, Wouters D, Rispens T, Wuhrer M, Vidarsson G (2017) Decoding the human immunoglobulin G-Glycan repertoire reveals a spectrum of Fc-receptor- and complement-mediated-effector activities. Front Immunol 8:877
Fan SQ, Huang W, Wang LX (2012) Remarkable transglycosylation activity of glycosynthase mutants of endo-D, an endo-beta-N-acetylglucosaminidase from Streptococcus pneumoniae. J Biol Chem 287:11272–11281
Ferrara C, Grau S, Jäger C, Sondermann P, Brünker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M, Ũmana P, Benz J (2011) Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcγRIII and antibodies lacking core fucose. Proc Natl Acad Sci USA 108:12669–12674
Freedberg DI, Selenko P (2014) Live cell NMR. Ann Rev Biophys 43:171–192
Hoffmann GW (1975) A theory of regulation and self-nonself discrimination in an immune network. Eur J Immunol 5:638–647
Hosokawa N, Kamiya Y, Kamiya D, Kato K, Nagata K (2009) Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J Biol Chem 284:17061–17068
Huang W, Giddens J, Fan SQ, Toonstra C, Wang LX (2012) Chemoenzymatic glycoengineering of intact IgG antibodies for gain of functions. J Am Chem Soc 134:12308–12318
Isoda Y, Yagi H, Satoh T, Shibata-Koyama M, Masuda K, Satoh M, Kato K, Iida S (2015). Importance of the side chain at position 296 of antibody Fc in interactions with FcγRIIIa and other Fcγ receptors. PLoS ONE 10:e0140120
Jarrell KF, Ding Y, Meyer BH, Albers SV, Kaminski L, Eichler J (2014) N-linked glycosylation in Archaea: a structural, functional, and genetic analysis. Microbiol Mol Biol Rev 78:304–341
Jefferis R (2016) Glyco-engineering of human IgG-Fc to modulate biologic activities. Curr Pharm Biotechnol 17:1333–1347
Jerne NK (1974) Towards a network theory of the immune system. Ann Immunol 125C:373–389
Kainosho M, Güntert P (2009) SAIL—stereo-array isotope labeling. Quart Rev Biophys 42:247–300
Kalbitzer HR, Leberman R, Wittinghofer A (1985) 1H-NMR spectroscopy on elongation factor Tu from Escherichia coli. FEBS Lett 180:40–42
Kamiya Y, Yamamoto S, Chiba Y, Jigami Y, Kato K (2011) Overexpression of a homogeneous oligosaccharide with 13C labeling by genetically engineered yeast strain. J Biomol NMR 50:397–401
Kamiya Y, Satoh T, Kato K (2012) Molecular and structural basis for N-glycan-dependent determination of glycoprotein fates in cells. Biochim Biophys Acta 1820:1327–1337
Kamiya Y, Yanagi K, Kitajima T, Yamaguchi T, Chiba Y, Kato K (2013) Application of metabolic 13C labeling in conjunction with high-field nuclear magnetic resonance spectroscopy for comparative conformational analysis of high mannose-type oligosaccharides. Biomolecules 3:108–123
Kamiya Y, Satoh T, Kato K (2014) Recent advances in glycoprotein production for structural biology: toward tailored design of glycoforms. Curr Opin Struct Biol 26:44–53
Kato K, Kamiya Y (2007) Structural views of glycoprotein-fate determination in cells. Glycobiology 17:1031–1044
Kato K, Yamaguchi Y (2012) Glycoproteins and antibodies: solution NMR studies. In: Encyclopedia of magnetic resonance. Wiley, Chichester, pp 1779–1790
Kato K, Yamaguchi T (2015) Paramagnetic NMR probes for characterization of the dynamic conformations and interactions of oligosaccharides. Glycoconjugate J 32:505–513
Kato K, Matsunaga C, Nishimura Y, Waelchli M, Kainosho M, Arata Y (1989a) Application of 13C nuclear magnetic resonance spectroscopy to molecular structural analyses of antibody molecules. J Biochem 105:867–869
Kato K, Nishimura Y, Waelchli M, Arata Y (1989b) Proton nuclear magnetic resonance study of a selectively deuterated mouse monoclonal antibody: use of two-dimensional homonuclear Hartmann-Hahn spectroscopy. J Biochem 106:361–364
Kato K, Yamaguchi Y, Arata Y (2010) Stable-isotope-assisted NMR approaches to glycoproteins using immunoglobulin G as a model system. Prog Nucl Mag Res Spectrosc 56:346–359
Kato K, Yagi H, Yamaguchi T (2017). NMR Characterization of the dynamic conformations of oligosaccharides. In: Webb GA (ed) Modern magnetic resonance, Springer, New York, pp 1–18
Kato K, Yanaka S, Yagi H (2018) Technical basis for nuclear magnetic resonance approach for glycoproteins. In: Naito A, Asakura T, Shimada I, Takegoshi K, Yamamoto Y (eds) Experimental approaches of nmr spectroscopy-methodology and application to life science and materials science. Springer, Tokyo, pp 415–438
Kowarik M, Young NM, Numao S, Schulz BL, Hug I, Callewaert N, Mills DC, Watson DC, Hernandez M, Kelly JF, Wacker M, Aebi M (2006) Definition of the bacterial N-glycosylation site consensus sequence. EMBO J 25:1957–1966
Kozlov G, Pocanschi CL, Rosenauer A, Bastos-Aristizabal S, Gorelik A, Williams DB, Gehring K (2010) Structural basis of carbohydrate recognition by calreticulin. J Biol Chem 285:38612–38620
Kunert R, Reinhart D (2016) Advances in recombinant antibody manufacturing. Appl Microbiol Biotechnol 100:3451–3461
Lederkremer GZ (2009) Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol 19:515–523
LeMaster DM, Richards FM (1988) NMR sequential assignment of Escherichia coli thioredoxin utilizing random fractional deuteriation. Biochemistry 27:142–150
Li F, Vijayasankaran N, Shen AY, Kiss R, Amanullah A (2010) Cell culture processes for monoclonal antibody production. MAbs 2:466–479
Li T, DiLillo DJ, Bournazos S, Giddens JP, Ravetch JV, Wang LX (2017) Modulating IgG effector function by Fc glycan engineering. Proc Natl Acad Sci USA 114:3485–3490
Liu D, Cowburn D (2017) Segmental isotopic labeling of proteins for NMR study using intein technology. Methods Mol Biol 1495:131–145
Liu DS, Xu R, Cowburn D (2009) Segmental isotopic labeling of proteins for nuclear magnetic resonance. Method Enzymol 462:151–175
Markley JL, Putter I, Jardetzky O (1968) High-resolution nuclear magnetic resonance spectra of selectively deuterated staphylococcal nuclease. Science 161:1249–1251
Matsumiya S, Yamaguchi Y, Saito J, Nagano M, Sasakawa H, Otaki S, Satoh M, Shitara K, Kato K (2007) Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1. J Mol Biol 368:767–779
Miyanoiri Y, Ishida Y, Takeda M, Terauchi T, Inouye M, Kainosho M (2016) Highly efficient residue-selective labeling with isotope-labeled Ile, Leu, and Val using a new auxotrophic E. coli strain. J Biomol NMR 65:109–119
Mizushima T, Yagi H, Takemoto E, Shibata-Koyama M, Isoda Y, Iida S, Masuda K, Satoh M, Kato K (2011) Structural basis for improved efficacy of therapeutic antibodies upon defucosylation of their Fc glycans. Genes Cells 16:1071–1080
Niwa R, Shoji-Hosaka E, Sakurada M, Shinkawa T, Uchida K, Nakamura K, Matsushima K, Ueda R, Hanai N, Shitara K (2004) Defucosylated chimeric anti-CC chemokine receptor 4 IgG1 with enhanced antibody-dependent cellular cytotoxicity shows potent therapeutic activity to T-cell leukemia and lymphoma. Cancer Res 64:2127–2133
Omasa T, Onitsuka M, Kim WD (2010) Cell engineering and cultivation of chinese hamster ovary (CHO) cells. Curr Pharm Biotechnol 11:233–240
Opitz C, Isogai S, Grzesiek S (2015) An economic approach to efficient isotope labeling in insect cells using homemade 15N-, 13C- and 2H-labeled yeast extracts. J Biomol NMR 62:373–385
Satoh T, Chen Y, Hu D, Hanashima S, Yamamoto K, Yamaguchi Y (2010) Structural basis for oligosaccharide recognition of misfolded glycoproteins by OS-9 in ER-associated degradation. Mol Cell 40:905–916
Sattler M, Fesik SW (1996) Use of deuterium labeling in NMR: overcoming a sizeable problem. Structure 4:1245–1249
Saxena K, Dutta A, Klein-Seetharaman J, Schwalbe H (2012) Isotope labeling in insect cells. Methods Mol Biol 831:37–54
Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, Weikert SH, Presta LG (2002) Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J Biol Chem 277:26733–26740
Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, Uchida K, Anazawa H, Satoh M, Yamasaki M, Hanai N, Shitara K (2003) The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J Biol Chem 278:3466–3473
Simmons LC, Reilly D, Klimowski L, Raju TS, Meng G, Sims P, Hong K, Shields RL, Damico LA, Rancatore P, Yansura DG (2002) Expression of full-length immunoglobulins in Escherichia coli: rapid and efficient production of aglycosylated antibodies. J Immunol Methods 263:133–147
Stolfa G, Smonskey MT, Boniface R, Hachmann AB, Gulde P, Joshi AD, Pierce AP, Jacobia SJ, Campbell A (2017). CHO-omics review: the impact of current and emerging technologies on chinese hamster ovary based bioproduction. Biotechnol J. https://doi.org/10.1002/biot.201700227.
Suzuki T, Kajino M, Yanaka S, Zhu T, Yagi H, Satoh T, Yamaguchi T, Kato K (2017) Conformational analysis of a high-mannose-type oligosaccharide displaying glucosyl determinant recognised by molecular chaperones using NMR-validated molecular dynamics simulation. Chembiochem 18:396–401
Takeda Y, Totani K, Matsuo I, Ito Y (2009) Chemical approaches toward understanding glycan-mediated protein quality control. Curr Opin Chem Biol 13:582–591
Valderrama-Rincon JD, Fisher AC, Merritt JH, Fan YY, Reading CA, Chhiba K, Heiss C, Azadi P, Aebi M, DeLisa MP (2012) An engineered eukaryotic protein glycosylation pathway in Escherichia coli. Nat Chem Biol 8:434–436
Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, TKinoshita a., Packer NH, Prestegard JH, Schnaar RL, Seeberger PH (eds) (2017) Essentials of glycobiology, 3rd edn. Cold Spring Harbor Laboratory Press, La Jolla
Walsh G (2014) Biopharmaceutical benchmarks 2014. Nat Biotechnol 32:992–1000
Walton WJ, Kasprzak AJ, Hare JT, Logan TM (2006) An economic approach to isotopic enrichment of glycoproteins expressed from Sf9 insect cells. J Biomol NMR 36:225–233
Wang LX, Lomino JV (2012) Emerging technologies for making glycan-defined glycoproteins. ACS Chem Biol 7:110–122
Yagi H, Fukuzawa N, Tasaka Y, Matsuo K, Zhang Y, Yamaguchi T, Kondo S, Nakazawa S, Hashii N, Kawasaki N, Matsumura T, Kato K (2015a) NMR-based structural validation of therapeutic antibody produced in Nicotiana benthamiana. Plant Cell Rep 34:959–968
Yagi H, Nakamura M, Yokoyama J, Zhang Y, Yamaguchi T, Kondo S, Kobayashi J, Kato T, Park EY, Nakazawa S, Hashii N, Kawasaki N, Kato K (2015b) Stable isotope labeling of glycoprotein expressed in silkworms using immunoglobulin G as a test molecule. J Biomol NMR 62:157–167
Yagi H, Zhang Y, Yagi-Utsumi M, Yamaguchi T, Iida S, Yamaguchi Y, Kato K (2015c) Backbone 1H, 13C, and 15N resonance assignments of the Fc fragment of human immunoglobulin G glycoprotein. Biomol NMR Assign 9:257–260
Yamaguchi Y, Kato K (2010) Dynamics and interactions of glycoconjugates probed by stable-isotope-assisted NMR spectroscopy. Methods Enzymol 478:305–322
Yamaguchi Y, Kato K, Shindo M, Aoki S, Furusho K, Koga K, Takahashi N, Arata Y, Shimada I (1998) Dynamics of the carbohydrate chains attached to the Fc portion of immunoglobulin G as studied by NMR spectroscopy assisted by selective 13C labeling of the glycans. J Biomol NMR 12:385–394
Yamaguchi Y, Nishimura M, Nagano M, Yagi H, Sasakawa H, Uchida K, Shitara K, Kato K (2006) Glycoform-dependent conformational alteration of the Fc region of human immunoglobulin G1 as revealed by NMR spectroscopy. Biochim et Biophysica Acta 1760:693–700
Yamaguchi Y, Takahashi N, Kato K (2007). Molecular interactions: antibody structures. In: Kamerling JP (ed) Comprehensive glycoscience. Elsevier, Oxford, pp 745–763
Yamaguchi T, Sakae Y, Zhang Y, Yamamoto S, Okamoto Y, Kato K (2014) Exploration of conformational spaces of high-mannose-type oligosaccharides by an NMR-validated simulation. Angew Chem 53:10941–10944
Yamaguchi Y, Yagi H, Kato K (2017). Stable isotope labeling of glycoproteins for NMR study. In: Kato K, Peters T (eds) NMR in glycoscience and glycotechnology. RSC Publishing, Cambridge, pp 194–205
Yanaka S, Yamazaki T, Yogo R, Noda M, Uchiyama S, Yagi H, Kato K (2017) NMR detection of semi-specific antibody interactions in serum environments. Molecules 22(10):1619
Zhu T, Yamaguchi T, Satoh T, Kato K (2015) A hybrid strategy for the preparation of 13C-labeled high-mannose-type oligosaccharides with terminal glucosylation for NMR study. Chem Lett 44(12):1744–1746
Acknowledgements
Stable isotope labeling was supported by Taiyo Nippon Sanso Co. We particularly thank Dr. Tsutomu Terauchi for useful discussion. We also thank Dr. Hajime Sato (Bruker BioSpin) for his help in the NMR measurements. We thank Ms. Kiyomi Senda and Ms. Kumiko Hattori (Nagoya City University) for their help in protein purification. This work was partly supported by the Nanotechnology Platform Program (Molecule and Material Synthesis) of MEXT, MEXT/JSPS Grants in Aid for Scientific Research (JP25102008, JP15K07935, JP17H06414 and JP17H05893).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yanaka, S., Yagi, H., Yogo, R. et al. Stable isotope labeling approaches for NMR characterization of glycoproteins using eukaryotic expression systems. J Biomol NMR 71, 193–202 (2018). https://doi.org/10.1007/s10858-018-0169-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-018-0169-2