Abstract
We propose a novel two-stage method for the classification of hyperspectral images. Pixel-wise classifiers, such as the classical support vector machine (SVM), consider spectral information only. As spatial information is not utilized, the classification results are not optimal and the classified image may appear noisy. Many existing methods, such as morphological profiles, superpixel segmentation, and composite kernels, exploit the spatial information. In this paper, we propose a two-stage approach inspired by image denoising and segmentation to incorporate the spatial information. In the first stage, SVMs are used to estimate the class probability for each pixel. In the second stage, a convex variant of the Mumford–Shah model is applied to each probability map to denoise and segment the image into different classes. Our proposed method effectively utilizes both spectral and spatial information of the data sets and is fast as only convex minimization is needed in addition to the SVMs. Experimental results on three widely utilized real hyperspectral data sets indicate that our method is very competitive in accuracy, timing, and the number of parameters when compared with current state-of-the-art methods, especially when the inter-class spectra are similar or the percentage of training pixels is reasonably high.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Remotely sensed hyperspectral images (HSI) are images taken from drones, airplanes or satellites that record a wide range of electromagnetic spectrum, typically more than 100 spectral bands from visible to near-infrared wavelengths. Since different materials reflect different spectral signatures, one can identify the materials at each pixel of the image by examining its spectral signatures. HSI is used in many applications, including agriculture [1, 2], disaster relief [3, 4], food safety [5, 6], military [7, 8] and mineralogy [9].
One of the most important problems in hyperspectral data exploitation is HSI classification. It has been an active research topic in past decades [10, 11]. The pixels in the hyperspectral image are often labeled manually by experts based on careful review of the spectral signatures and investigation of the scene. Given these ground-truth labels of some pixels (also called “training pixels”), the objective of HSI classification is to assign labels to part or all of the remaining pixels (the “testing pixels”) based on their spectral signatures and their locations.
Numerous methods have been developed for HSI classification. Among these, machine learning is a well-studied approach. It includes multinomial logistic regression [12,13,14], artificial neural networks [15,16,17,18,19], and support vector machines (SVMs) [20,21,22]. Since our method is partly based on SVMs, we will discuss it in more detail here. Early SVM classification methods [23, 24] perform pixel-wise classification that utilizes spectral information but not spatial dependencies. Numerous spectral–spatial SVM classification methods have been introduced since then. They show better performance when compared to the pixel-wise SVM classifiers. Here we discuss some of them.
SVMs with composite kernels [25] use composite kernels that are weighted summations of spectral kernels and spatial kernels. The spatial information is extracted by taking the average of the spectra in a fixed window around each pixel. To further utilize the spatial information, the method in [26] first applies superpixel segmentation to break the hyperspectral image into small regions with flexible shapes and sizes. Then it extracts the spatial information based on the segmentation and finally performs the classification using SVMs with multiple kernels. In [27], a pixel-wise SVM classification is first used to produce classification maps and then a partitional clustering is applied to obtain a segmentation of the hyperspectral image. Then a majority vote scheme is used in each cluster and finally a filter is applied to denoise the result. The method in [28] first produces pixel-wise classification maps using SVMs and then applies edge-preserving filtering to the classification maps. In addition to these methods, techniques based on Markov random fields [29], segmentation [26, 27, 30, 31] and morphological profiles [31, 32] have also been incorporated into SVMs to exploit the spatial information.
Besides machine learning approaches, another powerful approach is sparse representation [33]. It is based on the observation that spectral signatures within the same class usually lie in a low-dimensional subspace; therefore test data can be represented by a few atoms in a training dictionary. A joint sparse representation method is introduced in [34] to make use of the spatial homogeneity of neighboring pixels. In particular, each testing pixel and its neighboring pixels inside a fixed window are jointly sparsely represented. In [35], a kernel-based sparse algorithm is proposed which incorporates the kernel functions into the joint sparse representation method. It uses a fixed size local region to extract the spatial information. Approaches with more flexible local regions were proposed in [36] and [37]. They incorporate a multiscale scheme and superpixel segmentation into the joint sparse representation method, respectively. Multiple-feature-based adaptive sparse representation was proposed in [38]. It first extracts various spectral and spatial features and then the adaptive sparse representations of the features are computed. The method in [39] first estimates the pixel-wise class probabilities using SVMs and then applies sparse representation to obtain superpixel-wise class probabilities in which spatial information is utilized and the final result is obtained by combining both probabilities.
A pixel-wise classifier such as SVM considers only spectral information. It generates results with decent accuracy but would appear “noisy” as spatial information is not used, see [23] and also Fig. 1. Segmentation techniques have been used to incorporate the spatial information, see [26, 27, 30, 31]. Indeed, image segmentation is a well-studied subject in image processing and numerous effective segmentation methods for noisy images have been introduced [40,41,42,43,44,45]. Among them, a variational method called the Mumford–Shah model [40, 41] is one of the most important and successful image segmentation techniques. In this paper, we propose a simple but effective two-stage classification method inspired by our previous methods for image segmentation [43,44,45] which are based on the Mumford–Shah model. In the first stage, we apply a pixel-wise SVM method that exploits the spectral information to estimate a pixel-wise probability map for each class. In the second stage, we apply a convex variant of the Mumford–Shah model to denoise the maps and exploit the spatial information so as to segment the image into different classes accurately. Traditional methods like that in [27] apply a pixel-wise classification to obtain an initialization. Then they use a segmentation algorithm followed by a denoising algorithm to do the classification. In comparison, in our proposed method, since our convex Mumford–Shah model performs denoising and segmentation simultaneously, we just need one step here. Besides, because of the superior segmentation accuracy of our convex Mumford–Shah model, our method has much better classification results.
Our method utilizes spectral information in the first stage and spatial information in the second stage. Experiments show that our method generates very accurate results when compared to the state-of-the-art methods on real HSI data sets, especially when the inter-class spectra are similar. This is because our method can effectively exploit the spatial information even when the other methods cannot distinguish between the spectra. Moreover, our method has a much smaller number of parameters and shorter computation time than the state-of-the-art methods.
This paper is organized as follows. In Sect. 2, support vector machines and variational methods for denoising and segmentation are reviewed. In Sect. 3, our proposed two-stage classification method is presented. In Sect. 4, experimental results are presented to illustrate the effectiveness of our method. Sect. 5 concludes the paper.
2 Support Vector Machines and Variational Methods
2.1 Review of \(\nu \)-Support Vector Classifiers
Support vector machines (SVMs) have been used successfully in pattern recognition [46], object detection [47, 48], and financial time series forecasting [49, 50] etc. This approach also has superior performance in hyperspectral classification, especially when the dimensionality of the data is high and the number of training data is limited [23, 24]. In this subsection, we review the \(\nu \)-support vector classifier (\(\nu \)-SVC) [22] which will be used in the first stage of our method.
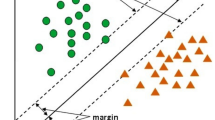
Consider for simplicity a supervised binary classification problem. We are given m training data \(\{\mathbf {x}_{i}\}_{i=1}^m\) in \(\mathbb {R}^{d_1}\), and each data is associated with a binary label \(y_i \in \{-1,+1\}\) for \(i=1,2,\ldots ,m\). In the training phase of SVM, one aims to find a hyperplane to separate the two classes of labels and maximize the distance between the hyperplane and the closest training data, which is called the support vector. In the kernel SVM, the data is mapped to a higher-dimensional feature space by a feature map \(\phi :\mathbb {R}^{d_1}\rightarrow \mathbb {R}^{d_2}\) in order to improve the separability between the two classes.
The \(\nu \)-SVC is an advanced support vector classifier which enables the user to specify the maximum training error before the training phase. Its formulation is given by
where \(\mathbf {w} \in \mathbb {R}^{d_2}\) and \(b\in \mathbb {R}\) are the normal vector and the bias of the hyperplane, respectively, \(\xi _i\)’s are the slack variables which allow training errors, and \(\rho /\Vert \mathbf {w}\Vert _2\) is the distance between the hyperplane and the support vector. The parameter \(\nu \in (0,1]\) is shown to be an upper bound on the fraction of training errors [22].
The optimization problem (1) can be solved through its Lagrangian dual
Its optimal Lagrange multipliers can be calculated using quadratic programming methods [51]. After obtaining them, the parameters of the optimal hyperplane can be represented by the Lagrange multipliers and the training data. The decision function for a test pixel \(\mathbf {x}\) is given by
Mercer’s Theorem [51, p. 423-424] states that a symmetric function K can be represented as an inner product of some feature maps \(\phi \), i.e., \(K(\mathbf {x},\mathbf {y})=\phi (\mathbf {x})\cdot \phi (\mathbf {y})\) for all \(\mathbf {x},\mathbf {y}\), if and only if K is positive semi-definite. In that case, the feature map \(\phi \) need not be known in order to perform the training and classification, but only the kernel function K is required. Examples of K satisfying the condition in Mercer’s Theorem include: \(K(\mathbf {x}_i,\mathbf {x}_j) = \text {exp}(-\Vert \mathbf {x}_i-\mathbf {x}_j\Vert ^2/(2\sigma ^2))\) and \(K(\mathbf {x}_i,\mathbf {x}_j) =(\mathbf {x}_i \cdot \mathbf {x}_j + 1)^p\).
2.2 Review of Variational Methods for Denoising and Segmentation
Let \(\varOmega =\{1,\ldots ,N_1\}\times \{1,\ldots ,N_2\}\) be the index set of pixel locations of an image, \(\mathbf{v}\) be the noisy image and \(\mathbf{u}\) be the restored image. One famous variational method to denoise images with Gaussian noise is the total variation (TV) method [52]. It involves an optimization model with a TV regularization term which corresponds to the function \(\Vert \nabla \cdot \Vert _1\). However, it is known that it reproduces images with staircase effect, i.e., with piecewise constant regions. One approach to improve it is to add a higher-order term, see, e.g., [53,54,55,56,57]. In [56], the authors considered minimizing
Here the first term is the \(\ell _2\) data-fitting term that caters for Gaussian noise. The second term is the TV term while the third term is the extra higher-order term added to introduce smoothness to the restored image \(\mathbf {u}\). By setting the parameters \(\{\alpha _i\}_{i=1}^2\) appropriately, one can control the trade-off between a piece-wise constant and a piece-wise smooth \(\mathbf {u}\).
In [43,44,45], the authors derived the same minimizational function (4) as a convex approximation of the Mumford–Shad model for segmentation. In [43,44,45], (4) is first applied to obtain a smooth denoised image and then thresholding is applied to the restored image to obtain the segmentation. The method is successful for segmenting greyscale and color images corrupted by different noises (Gaussian, Poisson, Gamma), information loss and/or blur. We note that the denoising and segmentation are intimately related. Indeed, Cai and Steidl showed that the famous Chan–Vese segmentation model [58] can be obtained by thresholding the TV denoising model with some properly chosen regularization parameter, see [59] for more details.
The two-stage approach for denoising has also been applied to impulse noise removal, see [60]. In the first stage a standard impulse noise detector, the Adaptive Median Filter [61], is used to detect the locations of possible noisy pixels. Then in the second stage, it restores the noisy pixels while keeping the non-noisy pixels unchanged by minimizing
where \(\varUpsilon \) is the set of non-noisy pixels detected by the Adaptive Median Filter, \(\mathbf {u}|_{\varUpsilon }=(u_i)_{i\in \varUpsilon }\), and \(1 < \alpha \le 2\). We remarked that in [62], Nikolova showed that the 1-norm data-fitting term (used in (5) above) is the correct norm for impulse noise. This two-stage method is the first method that can successfully restore images corrupted with extremely high level of impulse noise (e.g., 90%).
Our proposed method is inspired by the image denoising/segmentation methods in [43,44,45, 56, 60], which apply (4) successfully to denoise/segment images with various noises. In the first stage of our proposed method, we use the spectral classifier \(\nu \)-SVC to generate a pixel-wise probability map for each class. Then in the second stage, we use a combination of (4) and (5) to denoise and segment the result from the first stage.
3 Our Two-Stage Classification Method
SVMs yield decent classification accuracy [23], but their results can be noisy (see Fig. 1) since only spectral information is used. We therefore propose to use an image denoising/segmentation scheme to incorporate the spatial information into the classification. Our method first estimates the pixel-wise probability map for each class using SVMs. Then the spatial positions of the training pixels are used in a variational denoising/segmentation method to effectively segment the image into different classes.
3.1 First Stage: Pixel-Wise Probability Map Estimation
3.1.1 SVM Classifier
HSI classification is a multi-class classification, but the SVM is a binary classifier. To extend SVM to multi-class, we use the One-Against-One (OAO) strategy [63] where \([c(c-1)/2]\) SVMs are built to classify every possible pair of classes. Here c is the number of classes. In this paper, we choose the SVM method \(\nu \)-SVC [22] with OAO strategy for the HSI multiclass classification in our first stage. Moreover, the radial basis function kernel (RBF kernel) [21] is used as the kernel function in our SVM method. The RBF kernel is defined as
We remark that one can use other SVMs, other multiclass strategies such as the One-Against-All strategy in [63], or other kernel functions such as the polynomial kernel [21] instead.
3.1.2 Probability Estimation of SVM Outputs
Given a testing pixel \(\mathbf{x}\) and a SVM classifier with decision function \(f(\mathbf{x})\) in (3), we can label \(\mathbf{x}\) with a class according to the sign of \(f(\mathbf{x})\), see [21]. Under the OAO strategy, there are \([c (c-1)]/2\) such pairwise functions \(f_{h,l}\), \(1\le h,l \le c\), \(h\ne l\). We use them to estimate the probability \({p_h}\) that \(\mathbf {x}\) is in the h-th class. The idea is given in [64, 65]. We first estimate the pairwise class probability \(\mathrm{Prob}(y=h \ | \ y=h \text { or } y=l)\) by computing
where \(\eta \) and \(\tau \) are computed by minimizing a negative log likelihood problem over all the training pixels [64].
Then the probability vector \(\mathbf {p}=[p_1,p_2,\ldots ,p_c]^\intercal \) of the testing pixel \(\mathbf{x}\) is estimated by solving
By [65], its optimal solution can be obtained by solving the simple \((c +1)-(c +1)\) linear system
where
b is the Lagrange multiplier of the equality constraint in (8), \(\mathbf {e}\) is the c-vector of all ones, and \(\mathbf {0}\) is the c-vector of all zeros. In our tests, the probability vectors \(\mathbf {p}(\mathbf{x})\) for all testing pixels \(\mathbf{x}\) are computed by this method using the toolbox of LIBSVM library [66].
We finish Stage 1 by forming the 3D tensor \(\mathcal {V}\) where \(\mathcal {V}_{i,j,k}\) gives the probability that pixel (i, j) is in class k. More specifically, if pixel (i, j) is a testing pixel, then \(\mathcal {V}_{i,j,:}=\mathbf {p}(\mathbf{x}_{i,j})\); if pixel (i, j) is a training pixel belonging to the c-th class, then \(\mathcal {V}_{i,j,c}=1\) and \(\mathcal {V}_{i,j,k}=0\) for all other k’s.
3.2 Second Stage: Denoising/Segmentation of the Pixel-Wise Probability Map
Given the probability tensor \(\mathcal {V}\) obtained in Stage 1, one can obtain an HSI classification by taking the maximum probability for each pixel [28]. However, the result will appear noisy as no spatial information is taken into account. The goal of our second stage is to incorporate the spatial information into \(\mathcal {V}\) by a denoising/segmentation method that keeps the value of the training pixels unchanged during the optimization, as their ground-truth labels are given a priori.
Let \(\mathbf{v} _{k}:=\mathcal {V}_{:,:,k}\), \(k=1,\ldots ,c\), be the probability map of the k-th class obtained from Stage 1. We improve them by minimizing
where \(\beta _1\), \(\beta _2\) are regularization parameters and \({\varUpsilon }\) is the set of training pixels. We choose this minimization functional because it gives superb performance in denoising [56] and segmentation [43,44,45]. The higher-order \(\Vert \nabla \mathbf {u}\Vert _2^2\) term encourages smoothness of the solution and can improve the classification accuracy, see Sect. 4.4. In our tests, we use anisotropic TV [67] and periodic boundary condition for the discrete gradient operator, see [68, p. 258].
Alternating direction method of multipliers (ADMM) [69] is used to solve (10). First, we rewrite (10) as
Here D denotes the discrete operator of \(\nabla \), \(D=\begin{pmatrix}D_x \\ D_y \\ \end{pmatrix} \in \mathbb {R}^{2n\times n}\), where \(D_x\) and \(D_y\) are the first-order difference matrices in the horizontal and vertical directions, respectively, and n is the total number of pixels in the hyperspectral image, \(\iota _\mathbf {w}\) is the indicator function, where \(\iota _\mathbf {w}=0\) if \(\mathbf {w}|_{\varUpsilon }=\mathbf {v}_k|_{\varUpsilon }\) and \(\iota _\mathbf {w}=\infty \) otherwise. Its augmented Lagrangian is given by
where \(\mu >0\) is a positive constant, \(E = \begin{pmatrix}D \\ I \\ \end{pmatrix}\), \(\mathbf{g}=\begin{pmatrix}\mathbf {s} \\ \mathbf {w} \\ \end{pmatrix}\) and \({\varvec{\lambda }}= \begin{pmatrix}{\varvec{\lambda }}_1 \\ {\varvec{\lambda }}_2 \\ \end{pmatrix}\) the Lagrange multipliers.
The formulation (12) allows us to solve \(\mathbf {u}\) and \(\mathbf{g}\) alternately as follows:
The \(\mathbf {u}\)-subproblem (13a) is a least squares problem. Its solution is
Since periodic boundary conditions are used, the solution can be computed efficiently using the two-dimensional fast Fourier transform (FFT) [70] in \(O(n\log n)\) complexity.
For the \(\mathbf{g}\)-subproblem (13b), the optimal \(\mathbf {s}\) and \(\mathbf {w}\) can be computed separately as follows:
and
The solution of (15) can be obtained by soft thresholding [71], i.e.,
where \(\mathbf{r}=D\mathbf {u}^{(t+1)}-{\varvec{\lambda }}_1^{(t)}\). The solution of (16) is simply
The computation of (13c), (17) and (18) has a computational complexity of O(n). Hence, the computational complexity of our ADMM is \(O(n \log n)\) for each iteration, where n is the total number of pixels.
The convergence of our ADMM to the global minimum is guaranteed by [69]. Once it finishes, we obtain the enhanced probability map \(\mathbf {u}\) for class k. We denote it as \(\mathcal {U}_{:,:,k}\). After the map for each class is obtained, we get a 3D tensor \(\mathcal {U}\). The final classification of the (i, j)-th pixel is given by finding the maximum value in \(\mathcal {U}_{i,j,:}\), i.e., \(\underset{k}{\text {argmax }}\, \mathcal {U}_{i,j,k}\). Our proposed method is summarized in Algorithm 1.

We remark that in Stage 1, the operation is along the spectral dimension, i.e., the third index of the tensor, while in Stage 2, the operation is along the spatial dimension, i.e., the first two indices of the tensor. The techniques of Stage 2 are essentially similar to our segmentation methods in [43,44,45], where a smooth denoised image is first computed and then thresholding (here maximizing) is applied to it to segment (here classify) it.
Figure 2 shows the probability maps before the second stage and the enhanced probability maps after the second stage. The figures are in gray scale, i.e., completely white represents probability one and completely black represents probability zero. Note that the second stage does not guarantee the enhanced probability maps to have a sum to one property. In Fig. 2b and d, the enhanced probability maps are normalized to sum to one.
4 Experimental Results
4.1 Experimental Setup
4.1.1 Data Sets
Three commonly tested hyperspectral data sets are used in our experiments. These data sets have pixels labeled so that we can compare the methods quantitatively. The first one is the “Indian Pines” data set acquired by the Airborne Visible/Infrared Imaging Spectrometer (AVIRIS) sensor over the Indian Pines test site in North-western Indiana. It has a spatial resolution of 20 m per pixel and a spectral coverage ranging from 0.2 to 2.4 \(\upmu \)m in 220 spectral bands. However, due to water absorption, 20 of the spectral bands (the 104–108th, 150–163th and 220th bands) are discarded in experiments in previous papers. Therefore our data set is of size \(145 \times 145 \times 200\), and there are 16 classes in the given ground-truth labels.
The second and third images are the “University of Pavia” and “Pavia Center” data sets acquired by the Reflective Optics System Imaging Spectrometer (ROSIS) sensor over Pavia in northern Italy. The sensor has 1.3 m spatial resolution and spectral coverage ranging from 0.43 to 0.86 \(\upmu \)m. The data set sizes are \(610 \times 340 \times 103\) and \(1096 \times 715 \times 102\), respectively, where the third dimension is the spectral dimension. Both sets have nine classes in the ground-truth labels.
4.1.2 Methods Compared and Parameters Used
We have compared our method with five well-known classification methods: \(\nu \)-support vector classifiers (\(\nu \)-SVC) [22, 23] (i.e., the first stage of our method), SVMs with composite kernels (SVM-CK) [25], edge-preserving filtering (EPF) [28], superpixel-based classification via multiple kernels (SC-MK) [26] and multiple-feature-based adaptive sparse representation (MFASR) [38]. All the tests are run on a laptop computer with an Intel Core i5-7200U CPU, 8 GB RAM and the software platform is MATLAB R2016a.
In the experiments, the parameters are chosen as follows. For the \(\nu \)-SVC method, the parameters are obtained by performing a fivefold cross-validation [72]. For the SVM-CK method, the parameters are tuned such that it gives the highest classification accuracy. All parameters of the EPF method, the SC-MK method, and the MFASR method are chosen as stated in [26, 28, 38], respectively, except the window size in the EPF method, the number of superpixels and the parameters of the superpixel segmentation algorithm in the SC-MK method, and the sparsity level of the MFASR are tuned such that the highest classification accuracies are obtained. For our method, the parameters of the \(\nu \)-SVC (1) in the first stage are obtained by performing a fivefold cross-validation and the parameters of the optimization problem (10) in the second stage are tuned such that it gives the highest classification accuracy. The optimal parameters in the second stage \(\beta _1\) and \(\beta _2\) are 0.4 and 3; 0.1 and 3; 0.2 and 4 for Indian Pines, University of Pavia and Pavia Center, respectively. By [69], Algorithm 1 converges for any \(\mu >0\), so \(\mu \) is fixed as 5 for all the tests on the three data sets.
4.1.3 Performance Metrics
To quantitatively evaluate the performance of the methods, we use the following three widely used metrics: (i) overall accuracy (OA): the percentage of correctly classified pixels, (ii) average accuracy (AA): the average percentage of correctly classified pixels over each class, and (iii) kappa coefficient (kappa): the percentage of correctly classified pixels corrected by the number of agreements that would be expected purely by chance [73].
For each method, we perform the classification ten times where each time we randomly choose a different set of training pixels. In the tables below, we give the averages of these metrics over the ten runs. The accuracies are given in percentage, and the highest accuracy of each category is listed in boldface. In order to graphically show the classification results in an objective way, we also count the number of mis-classifications for each testing pixel over the ten runs. The numbers of mis-classifications are shown in the corresponding heatmap figures, with the heatmap colorbar indicating the number of mis-classifications.
4.2 Classification Results
4.2.1 Indian Pines
The Indian Pines data set consists mainly of big homogeneous regions and has very similar inter-class spectra (see Fig. 3 for the spectra of the training pixels of Indian Pines data where there are three similar classes of corns, three similar classes of grasses and three similar classes of soybeans). It is therefore very difficult to classify it if only spectral information is used. In the experiments, we choose exactly the same number of training pixels as in [26, 37] and they amount to about 10% of the pixels from each class. Some classes have small numbers of pixels, and hence 10 pixels are taken as training pixels for each of these classes. The rest of the labeled pixels are used as testing pixels.
The number of training and testing pixels and the classification accuracies obtained by different methods are reported in Table 1. We see that our method generates the best results for all three metrics (OA, AA and kappa) and outperforms the comparing methods by a significant margin. They are at least 0.95% higher than the others. Also, the second stage of our method improves the overall accuracy of \(\nu \)-SVC (used in the first stage of our method) by almost 20%.
Figure 4 shows the heatmaps of mis-classifications. The results of the \(\nu \)-SVC, SVM-CK and EPF methods produce large area of mis-classifications. The SC-MK also produces mis-classification at the top-right region and the middle-right region which are soybeans-clean and soybeans-no till, respectively. This shows that SC-MK cannot distinguishing these two similar classes well. The heatmap of MFASR method contains scattered regions of mis-classification. In contrast, our method generates smaller regions of mis-classifications and less errors as it effectively utilizes the spatial information to give an accurate result.
4.2.2 University of Pavia
The University of Pavia data set consists of regions with various shapes, including thin and thick structures and large homogeneous regions. Hence, it can be used to test the ability of the classification methods on handling different shapes. In the experiments, we choose the same number of training pixels (200 for each class) as in [26]. This accounts for approximately 4% of the labeled pixels. The remaining ones are used as testing pixels.
Table 2 reports the classification accuracies obtained by different methods. We see that the performance of SC-MK, MFASR, and our method are very close: approximately 99% in all three metrics (OA, AA and kappa) and they outperform the \(\nu \)-SVC, SVM-CK and EPF methods. However, we note that MFASR requires twice the number of parameters as ours and 12 times longer to run, see Tables 7 and 8. Figure 5 shows the heatmaps of mis-classifications. The \(\nu \)-SVC, SVM-CK and EPF methods produce large regions of mis-classifications. The SC-MK method produces many mis-classifications at the middle and bottom regions where the meadows are. The MFASR method and our method generate smaller regions of mis-classification.
4.2.3 Pavia Center
The Pavia Center data set also consists of regions with various shapes. In the experiments, we use the same number of training pixels as in [31] (150 training pixels per class). This accounts for approximately 1% of the labeled pixels. The rest of the labeled pixels are used as testing pixels. Table 3 reports the number of training/testing pixels and the classification accuracies of different methods. We see that the EPF method gives the highest OA and kappa while our method gives the second highest and their values differ by about 0.1%. However, our method gives the highest AA (99.12%) which outperforms the EPF method by almost 1%. The SC-MK and MFASR methods give slightly worse accuracies than our method. Figure 6 shows the heatmaps of mis-classifications.
4.3 Advantages of Our Two-Stage Method
4.3.1 Percentage of Training Pixels
Since our method improves on the classification accuracy by using spatial information, it is expected to be a better method if the training percentage (percentage of training pixels) is higher. To verify that, Tables 4, 5 and 6 show the overall accuracies obtained by our method on the three data sets with different levels of training percentage. We see that our method outperforms the other methods once the training percentage is reasonably high enough (6% for Example 1, 10% for Example 2, and 3% for Example 3). When it is not high, our method still gives a classification accuracy that is very close to the best method compared.
4.3.2 Model Complexity and Computation Time
Tables 7 and 8 show the computation time required and the number of parameters for all methods. We note that the reported timing does not count the time required to find the optimal set of parameters. The \(\nu \)-SVC, SVM-CK and EPF methods have fast computation time because of the simplicity of their models. They have only a few parameters (2, 3 and 4, respectively). However, from the results in Sect. 4.2, they are worse than the other three methods. The SC-MK method is a good method in terms of accuracy and timing, but it has 9 parameters. The MFASR method has 10 parameters and the longest computation time. In comparison, our method has 5 parameters (2 parameters \(\nu \) and \(\sigma \) for the \(\nu \)-SVC (1) and the RBF kernel (6), respectively, in the first stage, 2 parameters \(\beta _1\) and \(\beta _2\) for the denoising model (10) in the second stage and 1 parameter \(\mu \) for the ADMM algorithm (12)). It has much better (if not the best) classification accuracies with slightly longer computation time than those of \(\nu \)-SVC, SVM-CK and EPF.
4.4 Effect of the Second-Order Term
Here we examine empirically the importance of the term \(\Vert \nabla \mathbf {u}\Vert _2^2\) in (10). Figure 7 shows the heatmaps of mis-classifications on the Indian Pines data by using our method with and without \(\Vert \nabla \mathbf {u}\Vert _2^2\) over ten runs. The training pixels are randomly selected and consist of 2.5% of the labeled pixels. Figure 7a shows the ground-truth labels. Figure 7b–d shows the heatmaps of mis-classifications of the \(\nu \)-SVC classifier (i.e., the first stage of our method), the second stage of our method without the \(\Vert \nabla \mathbf {u}\Vert _2^2\) term, and the second stage of our method with the \(\Vert \nabla \mathbf {u}\Vert _2^2\) term, respectively. Recall the term \(\Vert \nabla \mathbf {u}\Vert _2^2\) control the smoothness of the final probability maps and the final classification result is determined by taking the maximum over this map of each class. By choosing the parameter associated with the term appropriately, we can then control the level of shrinking or expanding the homogeneous regions in the final classification result. From Fig. 7c, when the term is dropped, the mis-classification regions at the top left and bottom left of the first stage result are not only still mis-classified, but the numbers of mis-classification increase. In contrast, when the term is kept, we see from Fig. 7d that the numbers of mis-classification are significantly lowered. Moreover, most of the mis-classified regions of the first stage result are now correctly classified when the parameters are chosen appropriately.
5 Conclusions and Future Work
In this paper, we propose a novel two-stage hyperspectral classification method inspired by image denoising/segmentation. The method is simple yet performs effectively. In the first stage, a support vector machine method is used to estimate the pixel-wise probability map of each class. The result in the first stage has decent accuracy but is noisy. In the second stage, a convex variant of the Mumford–Shah model is applied to denoise and classify the hyperspectral image into different classes. Since both spectral and spatial information are effectively utilized, our method is very competitive when compared with state-of-the-art hyperspectral data classification methods. It also has a simpler framework with fewer numbers of parameters and faster computation times. It performs particularly well when the inter-class spectra are close or when the training percentage is high.
For future work, we plan to investigate the use of deep learning methods in the first stage [16,17,18,19]. We will also investigate the use of automated parameter selection [74,75,76,77] of the variational method in the second stage. Additionally, we plan on using our methods for classifying fused hyperspectral and LiDAR data [18, 78, 79].
References
Patel, N., Patnaik, C., Dutta, S., Shekh, A., Dave, A.: Study of crop growth parameters using airborne imaging spectrometer data. Int. J. Remote Sens. 22(12), 2401–2411 (2001)
Datt, B., McVicar, T.R., Van Niel, T.G., Jupp, D.L., Pearlman, J.S.: Preprocessing EO-1 hyperion hyperspectral data to support the application of agricultural indexes. IEEE Trans. Geosci. Remote Sens. 41(6), 1246–1259 (2003)
Trierscheid, M., Pellenz, J., Paulus, D., Balthasar, D.: Hyperspectral imaging or victim detection with rescue robots. In: IEEE International Workshop on Safety, Security and Rescue Robotics, 2008. SSRR 2008, pp. 7–12. IEEE (2008)
Eismann, M.T., Stocker, A.D., Nasrabadi, N.M.: Automated hyperspectral cueing for civilian search and rescue. Proc. IEEE 97(6), 1031–1055 (2009)
Lu, R., Chen, Y.-R.: Hyperspectral imaging for safety inspection of food and agricultural products. In: Pathogen Detection and Remediation for Safe Eating, vol. 3544, pp. 121–134. International Society for Optics and Photonics (1999)
Gowen, A., O’Donnell, C., Cullen, P., Downey, G., Frias, J.: Hyperspectral imaging-an emerging process analytical tool for food quality and safety control. Trends Food Sci. Technol. 18(12), 590–598 (2007)
Manolakis, D., Shaw, G.: Detection algorithms for hyperspectral imaging applications. IEEE Signal Process. Mag. 19(1), 29–43 (2002)
Stein, D.W., Beaven, S.G., Hoff, L.E., Winter, E.M., Schaum, A.P., Stocker, A.D.: Anomaly detection from hyperspectral imagery. IEEE Signal Process. Mag. 19(1), 58–69 (2002)
Hörig, B., Kühn, F., Oschütz, F., Lehmann, F.: Hymap hyperspectral remote sensing to detect hydrocarbons. Int. J. Remote Sens. 22(8), 1413–1422 (2001)
Mountrakis, G., Im, J., Ogole, C.: Support vector machines in remote sensing: a review. ISPRS J. Photogramm. Remote Sens. 66(3), 247–259 (2011)
Fauvel, M., Tarabalka, Y., Benediktsson, J.A., Chanussot, J., Tilton, J.C.: Advances in spectral-spatial classification of hyperspectral images. Proc. IEEE 101(3), 652–675 (2013)
Li, J., Bioucas-Dias, J.M., Plaza, A.: Semisupervised hyperspectral image segmentation using multinomial logistic regression with active learning. IEEE Trans. Geosci. Remote Sens. 48(11), 4085–4098 (2010)
Li, J., Bioucas-Dias, J.M., Plaza, A.: Spectral-spatial hyperspectral image segmentation using subspace multinomial logistic regression and markov random fields. IEEE Trans. Geosci. Remote Sens. 50(3), 809–823 (2012)
Li, J., Bioucas-Dias, J.M., Plaza, A.: Semisupervised hyperspectral image classification using soft sparse multinomial logistic regression. IEEE Trans. Geosci. Remote Sens. 10(2), 318–322 (2013)
Benediktsson, J.A., Palmason, J.A., Sveinsson, J.R.: Classification of hyperspectral data from urban areas based on extended morphological profiles. IEEE Trans. Geosci. Remote Sens. 43(3), 480–491 (2005)
Yue, J., Zhao, W., Mao, S., Liu, H.: Spectral-spatial classification of hyperspectral images using deep convolutional neural networks. Remote Sens. Lett. 6(6), 468–477 (2015)
Makantasis, K., Karantzalos, K., Doulamis, A., Doulamis, N.: Deep supervised learning for hyperspectral data classification through convolutional neural networks. In: 2015 IEEE International on Geoscience and Remote Sensing Symposium (IGARSS), pp. 4959–4962. IEEE (2015)
Morchhale, S., Pauca, V.P., Plemmons, R.J., Torgersen, T.C.: Classification of pixel-level fused hyperspectral and LiDAR data using deep convolutional neural networks. In: 2016 8th Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), pp. 1–5 (2016)
Pan, B., Shi, Z., Xu, X.: R-vcanet: a new deep-learning-based hyperspectral image classification method. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 10(5), 1975–1986 (2017)
Boser, B.E., Guyon, I.M., Vapnik, V.N.: A training algorithm for optimal margin classifiers. In: Proceedings of the Fifth Annual Workshop on Computational Learning Theory, pp. 144–152. ACM (1992)
Cortes, C., Vapnik, V.: Support-vector networks. Mach. Learn. 20(3), 273–297 (1995)
Schölkopf, B., Smola, A.J., Williamson, R.C., Bartlett, P.L.: New support vector algorithms. Neural Comput. 12(5), 1207–1245 (2000)
Melgani, F., Bruzzone, L.: Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 42(8), 1778–1790 (2004)
Camps-Valls, G., Bruzzone, L.: Kernel-based methods for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 43(6), 1351–1362 (2005)
Camps-Valls, G., Gomez-Chova, L., Muñoz-Marí, J., Vila-Francés, J., Calpe-Maravilla, J.: Composite kernels for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 3(1), 93–97 (2006)
Fang, L., Li, S., Duan, W., Ren, J., Benediktsson, J.A.: Classification of hyperspectral images by exploiting spectral-spatial information of superpixel via multiple kernels. IEEE Trans. Geosci. Remote Sens. 53(12), 6663–6674 (2015)
Tarabalka, Y., Benediktsson, J.A., Chanussot, J.: Spectral-spatial classification of hyperspectral imagery based on partitional clustering techniques. IEEE Trans. Geosci. Remote Sens. 47(8), 2973–2987 (2009)
Kang, X., Li, S., Benediktsson, J.A.: Spectral-spatial hyperspectral image classification with edge-preserving filtering. IEEE Trans. Geosci. Remote Sens. 52(5), 2666–2677 (2014)
Tarabalka, Y., Fauvel, M., Chanussot, J., Benediktsson, J.A.: Svm-and mrf-based method for accurate classification of hyperspectral images. IEEE Geosci. Remote Sens. Lett. 7(4), 736–740 (2010)
Ghamisi, P., Benediktsson, J.A., Ulfarsson, M.O.: Spectral-spatial classification of hyperspectral images based on hidden markov random fields. IEEE Trans. Geosci. Remote Sens. 52(5), 2565–2574 (2014)
Liu, T., Gu, Y., Chanussot, J., Mura, M Dalla: Multimorphological superpixel model for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 55(12), 6950–6963 (2017)
Fauvel, M., Benediktsson, J.A., Chanussot, J., Sveinsson, J.R.: Spectral and spatial classification of hyperspectral data using svms and morphological profiles. IEEE Trans. Geosci. Remote Sens. 46(11), 3804–3814 (2008)
Bruckstein, A.M., Donoho, D.L., Elad, M.: From sparse solutions of systems of equations to sparse modeling of signals and images. SIAM Rev. 51(1), 34–81 (2009)
Chen, Y., Nasrabadi, N.M., Tran, T.D.: Hyperspectral image classification using dictionary-based sparse representation. IEEE Trans. Geosci. Remote Sens. 49(10), 3973–3985 (2011)
Chen, Y., Nasrabadi, N.M., Tran, T.D.: Hyperspectral image classification via kernel sparse representation. IEEE Trans. Geosci. Remote Sens. 51(1), 217–231 (2013)
Fang, L., Li, S., Kang, X., Benediktsson, J.A.: Spectral-spatial hyperspectral image classification via multiscale adaptive sparse representation. IEEE Trans. Geosci. Remote Sens. 52(12), 7738–7749 (2014)
Fang, L., Li, S., Kang, X., Benediktsson, J.A.: Spectral-spatial classification of hyperspectral images with a superpixel-based discriminative sparse model. IEEE Trans. Geosci. Remote Sens. 53(8), 4186–4201 (2015)
Fang, L., Wang, C., Li, S., Benediktsson, J.A.: Hyperspectral image classification via multiple-feature-based adaptive sparse representation. IEEE Trans. Instrum. Meas. 66(7), 1646–1657 (2017)
Li, S., Lu, T., Fang, L., Jia, X., Benediktsson, J.A.: Probabilistic fusion of pixel-level and superpixel-level hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 54(12), 7416–7430 (2016)
Mumford, D., Shah, J.: Boundary detection by minimizing functionals. In: IEEE Conference on Computer Vision and Pattern Recognition, vol. 17, pp. 137–154, San Francisco (1985)
Mumford, D., Shah, J.: Optimal approximations by piecewise smooth functions and associated variational problems. Commun. Pure Appl. Math. 42(5), 577–685 (1989)
Morar, A., Moldoveanu, F., Gröller, E.: Image segmentation based on active contours without edges. In: 2012 IEEE 8th International Conference on Intelligent Computer Communication and Processing, pp. 213–220. IEEE (2012)
Cai, X., Chan, R., Zeng, T.: A two-stage image segmentation method using a convex variant of the mumford-shah model and thresholding. SIAM J. Imaging Sci. 6(1), 368–390 (2013)
Chan, R., Yang, H., Zeng, T.: A two-stage image segmentation method for blurry images with poisson or multiplicative gamma noise. SIAM J. Imaging Sci. 7(1), 98–127 (2014)
Cai, X., Chan, R., Nikolova, M., Zeng, T.: A three-stage approach for segmenting degraded color images: smoothing, lifting and thresholding (SLaT). J. Sci. Comput. 72(3), 1313–1332 (2017)
Pontil, M., Verri, A.: Support vector machines for 3d object recognition. IEEE Trans. Pattern Anal. Mach. Intell. 20(6), 637–646 (1998)
El-Naqa, I., Yang, Y., Wernick, M.N., Galatsanos, N.P., Nishikawa, R.M.: A support vector machine approach for detection of microcalcifications. IEEE Trans. Med. Imaging 21(12), 1552–1563 (2002)
Osuna, E., Freund, R., Girosit, F.: Training support vector machines: an application to face detection. In: 1997 Proceedings of IEEE Computer Society Conference on Computer Vision and Pattern Recognition, pp. 130–136. IEEE (1997)
Tay, F.E., Cao, L.: Application of support vector machines in financial time series forecasting. Omega 29(4), 309–317 (2001)
Kim, K.-J.: Financial time series forecasting using support vector machines. Neurocomputing 55(1), 307–319 (2003)
Vapnik, V.N., Vapnik, V.: Statistical Learning Theory, vol. 1. Wiley, New York (1998)
Rudin, L.I., Osher, S., Fatemi, E.: Nonlinear total variation based noise removal algorithms. Phys. D Nonlinear Phenomena 60(1–4), 259–268 (1992)
Mumford, D.: Elastica and computer vision. In: Algebraic Geometry and Its Applications, pp. 491–506. Springer (1994)
Chan, T., Marquina, A., Mulet, P.: High-order total variation-based image restoration. SIAM J. Sci. Comput. 22(2), 503–516 (2000)
Shen, J., Kang, S.H., Chan, T.F.: Euler’s elastica and curvature-based inpainting. SIAM J. Appl. Math. 63(2), 564–592 (2003)
Hintermüller, M., Stadler, G.: An infeasible primal-dual algorithm for total bounded variation-based inf-convolution-type image restoration. SIAM J. Sci. Comput. 28(1), 1–23 (2006)
Bredies, K., Kunisch, K., Pock, T.: Total generalized variation. SIAM J. Imaging Sci. 3(3), 492–526 (2010)
Chan, T.F., Vese, L.A.: Active contours without edges. IEEE Trans. Image Process. 10(2), 266–277 (2001)
Cai, X., Steidl, G.: Multiclass segmentation by iterated rof thresholding. In: International Workshop on Energy Minimization Methods in Computer Vision and Pattern Recognition, pp. 237–250. Springer (2013)
Chan, R.H., Ho, C.-W., Nikolova, M.: Salt-and-pepper noise removal by median-type noise detectors and detail-preserving regularization. IEEE Trans. Image Process. 14(10), 1479–1485 (2005)
Hwang, H., Haddad, R.A.: Adaptive median filters: new algorithms and results. IEEE Trans. Image Process. 4(4), 499–502 (1995)
Nikolova, M.: A variational approach to remove outliers and impulse noise. J. Math. Imaging Vis. 20(1–2), 99–120 (2004)
Hsu, C.-W., Lin, C.-J.: A comparison of methods for multiclass support vector machines. IEEE Trans. Neural Netw. 13(2), 415–425 (2002)
Lin, H.-T., Lin, C.-J., Weng, R.C.: A note on platt’s probabilistic outputs for support vector machines. Mach. Learn. 68(3), 267–276 (2007)
Wu, T.-F., Lin, C.-J., Weng, R.C.: Probability estimates for multi-class classification by pairwise coupling. J. Mach. Learn. Res. 5(Aug), 975–1005 (2004)
Chang, C.-C., Lin, C.-J.: Libsvm: a library for support vector machines. ACM Trans. Intell. Syst. Technol. (TIST) 2(3), 27 (2011)
Zhao, X.-L., Wang, F., Huang, T.-Z., Ng, M.K., Plemmons, R.J.: Deblurring and sparse unmixing for hyperspectral images. IEEE Trans. Geosci. Remote Sens. 51(7), 4045–4058 (2013)
Gonzales, R.C., Woods, R.E.: Digital Image Processing. Addison-Wesley, Reading, MA (1992)
Boyd, S., Parikh, N., Chu, E., Peleato, B., Eckstein, J., et al.: Distributed optimization and statistical learning via the alternating direction method of multipliers. Found. Trends® Mach. Learn. 3(1), 1–122 (2011)
Chan, R.H.-F., Jin, X.-Q.: An Introduction to Iterative Toeplitz Solvers, vol. 5. SIAM, Philadelphia (2007)
Combettes, P.L., Wajs, V.R.: Signal recovery by proximal forward–backward splitting. Multiscale Model. Simul. 4(4), 1168–1200 (2005)
Kohavi, R., et al.: A study of cross-validation and bootstrap for accuracy estimation and model selection. In: Ijcai, Montreal, vol. 14, pp. 1137–1145 (1995)
Cohen, J.: A coefficient of agreement for nominal scales. Educ. Psychol. Measur. 20(1), 37–46 (1960)
Liao, H., Li, F., Ng, M.K.: Selection of regularization parameter in total variation image restoration. JOSA A 26(11), 2311–2320 (2009)
Dong, Y., Hintermüller, M., Rincon-Camacho, M.M.: Automated regularization parameter selection in multi-scale total variation models for image restoration. J. Math. Imaging Vis. 40(1), 82–104 (2011)
Wen, Y.-W., Chan, R.H.: Parameter selection for total-variation-based image restoration using discrepancy principle. IEEE Trans. Image Process. 21(4), 1770–1781 (2012)
Bredies, K., Dong, Y., Hintermüller, M.: Spatially dependent regularization parameter selection in total generalized variation models for image restoration. Int. J. Comput. Math. 90(1), 109–123 (2013)
Gader, P., Zare, A., Close, R., Aitken, J., Tuell, G.: Muufl gulfport hyperspectral and LiDAR airborne data set. Univ. Florida, Gainesville, FL, Tech. Rep. REP-2013-570 (2013)
Debes, C., Merentitis, A., Heremans, R., Hahn, J., Frangiadakis, N., van Kasteren, T., Liao, W., Bellens, R., Pižurica, A., Gautama, S., et al.: Hyperspectral and LiDAR data fusion: outcome of the 2013 grss data fusion contest. IEEE J. Sel. Topics Appl. Earth Obs. Remote Sens. 7(6), 2405–2418 (2014)
Acknowledgements
The authors would like to thank the Computational Intelligence Group from the Basque University for sharing the hyperspectral data sets in their website (http://www.ehu.eus/ccwintco/index.php/Hyperspectral_Remote_Sensing_Scenes), Prof. Leyuan Fang from College of Electrical and Information Engineering at Hunan University for providing the programs of the SC-MK and MFASR methods in his homepage (http://www.escience.cn/people/LeyuanFang) and Prof. Xudong Kang from College of Electrical and Information Engineering at Hunan University for providing the program of the EPF method in his homepage (http://xudongkang.weebly.com/). Raymond H. Chan’s research is supported by HKRGC Grants No. CUHK14306316, CityU Grant: 9380101, CRF Grant C1007-15G, AoE/M-05/12. Kelvin K. Kan’s research is supported by US Air Force Office of Scientific Research under grant FA9550-15-1-0286. Mila Nikolova’s research is supported by the French Research Agency (ANR) under grant No ANR-14-CE27-001 (MIRIAM) and by the Isaac Newton Institute for Mathematical Sciences for support and hospitality during the programme Variational Methods and Effective Algorithms for Imaging and Vision, EPSRC grant no EP/K032208/1. Robert J. Plemmons’ research is supported by HKRGC Grant No. CUHK14306316 and US Air Force Office of Scientific Research under grant FA9550-15-1-0286.
Author information
Authors and Affiliations
Corresponding author
Additional information
In memory of Mila Nikolova.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chan, R.H., Kan, K.K., Nikolova, M. et al. A two-stage method for spectral–spatial classification of hyperspectral images. J Math Imaging Vis 62, 790–807 (2020). https://doi.org/10.1007/s10851-019-00925-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10851-019-00925-9