Abstract
The chiral separation of terbutaline (TB) using β-cyclodextrins (β-CD) and its derivatives has aroused intensive interest. Herein, the enantioseparation mechanisms of β-CD and heptakis(2,3-di-O-acetyl)-β-CD (HAD-β-CD) towards TB are investigated by a molecular dynamics/quantum mechanics/continuum solvent model (MD/QM/CSM) approach based on the experimental data. One of the two experimentally predicted binding modes of HAD-β-CD/TB is confirmed by our approach and ascribed to the R-complex. R-TB is compactly included by HAD-β- CD: the aromatic ring is within the CD cavity, while the alkyl moiety is also included and almost parallel to the secondary rim of the CD. As indicated by the computed binding affinity, this tight binding mode of R-complex enhances the host–guest hydrophobic interaction and renders the R-complex significantly more stable than the S-complex. In contrast, the binding modes of R- and S-TB with β-CD are similar due to the poor flexibility of the host, which jeopardizes its ability to differentiate R- and S-TB.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
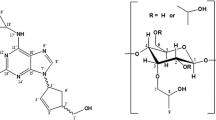
β-cyclodextrin (β-CD) and its derivatives, possessing multiple chiral centres [1], can encapsulate enantiomers to form complexes with varying stabilities, thus achieving chiral separation. [2,3,4,5,6,7] Terbutaline (TB), a chiral drug molecule and a β2-adrenoceptor agonist, can effectively relieve bronchial asthma and chroni obstructive pulmonary disease. In particular, the R-TB functions as the potent enantiomer in generating therapeutic effect, while the S-TB shows side effects [8,9,10]. Naturally, using β-CD and its derivatives to separate the two enantiomers of TB is of great significance.
Several methods have been utilized to study the separation of β-CD and its derivatives with TB enantiomers, such as liquid chromatography, capillary electrophoresis, nuclear magnetic resonance spectroscopy and so on [11,12,13,14,15,16]. On the other hand, theoretical computations and molecular simulations can also act as effective tools in experimental research [17,18,19,20,21,22,23,24]. Therefore, the combination of experiments with theoretical calculations has become appealing in the study of chiral separation. In 2007, Zhang et al. [25] applied β-CD as a chiral selector to separate the enantiomers of TB and utilized molecular docking to obtain their binding modes. In 2013, Venkatesh et al. [26] studied the complexation behaviour of α-CD, 2-hydroxypropylated-α-CD (HP-α-CD), β-CD, and HP-β-CD with TB and orciprenaline using absorption spectroscopy, fluorescence spectroscopy and molecular simulation methods. It was found that 1:1 and 1:2 CD/drug inclusion complexes are formed at low and high CD concentrations, respectively. Furthermore, HP-α-CD and HP-β-CD exhibit stronger inclusion capacity than do the natural CDs. To explain the difference in their binding strength, PM3 method was used to compute the difference in binding energies. In 2014, Guo and coworkers [27] reported the enantioseparations mechanism of ten chiral drugs by β-CD derivatives. Several computational methods, including MMFF, PM3, DFT and ONIOM2 methods, were employed and it was proposed that for the effective chiral separation, the difference in host–guest interaction energy between a pair of enantiomers with a selector should exceed 6 kJ/mol. This provides a beneficial reference for predicting enantiomeric separations. In 2016, Liu et al. [28] used glutamate acid-β-CD as the chiral selector to separate various chiral molecules such as TB by capillary electrophoresis, and further investigated the major influencing factors for the separation efficiency. Based on their PM3 calculations, the difference in the host–guest interaction energies is mainly responsible for the successful separation of chiral molecules.
In 2020, Gogolashvili [29] and coworkers combined CE with NMR spectroscopy to investigate the binding mechanisms of TB enantiomers toward heptakis(2,3-di-O-acetyl)-β-CD (HDA-β-CD) and β-CD (Fig SI1). Experimental results indicated the separation ability of the former is stronger than that of the latter. The binding modes of HDA-β-CD and β-CD with TB were deduced based on 2D ROESY NMR data (Fig SI2). In the two proposed binding modes (Fig SI2 a, b), the NCMe3 protons of TB are close to the H-3 and OAC protons of HDA-β-CD. However, it is not specified whether these binding poses belong to the S-type or R-type complex. Thus, the orientation of TB in the cavity of HDA-β-CD remains inconclusive. Moreover, no energetic data, such as the host–guest interaction energy and solvent effect, has been reported yet.
Based on these experimental results [29], we apply a molecular dynamics/quantum mechanics/continuum solvent model algorithm (MD/QM/CSM) approach [30,31,32] to study the separation mechanism of HDA-β-CD and β-CD to TB enantiomers. MD is employed for the conformational sampling and the representative binding conformations (RC) of the host–guest systems are extracted from MD trajectories under the guidance of structural information deduced experimentally. Subsequently the RC is utilized in the QM/CSM computation to obtain the binding affinity. Finally, the chiral separation mechanisms of HDA-β-CD and β-CD for TB are compared from the perspective of binding structure, host–guest interaction and solvent effect.
Theory and methodology
MD simulations
The initial structure of β-CD was obtained from the Protein Data Bank (PDB code: 1DMB), while that of HDA-β-CD was constructed by introducing acetyl groups into the O(2) and O(3) of each glucose unit of β-CD. Moreover, the –NH2–group of TB is hydrogenated, thus carry one positive charge.
Due to the flexibility of the host and guest molecules, the MD trajectories are sensitive to the initial structures and result in different RC. We perform four MD calculations for each system (Fig SI3), where the TB chiral molecules with different orientations are positioned outside but close to the CD cavity. Eventually, the optimal initial structures and RC were determined by taking the experimental results and computation data into comprehensive account.
Amber12 software was used to simulate the host and guest molecules as well as their complexes. The topological structures of β-CD and HDA-β-CD are analyzed using q4md-CD force field with the relevant parameters derived from GLYCAM04 and Amber99SB force field [33]. The generalized Amber force field (gaff) [34] is used for the guest molecules with one positive charge. In addition, the atomic RESP charge [35] is calculated at the HF/6-31G* level using Gaussian09 [36]. Initial structures were dissolved in the TIP3P water periodic box [37] of size 40 × 40 × 40 Å. chloride ion is then introduced into the aqueous solution to keep the system neutral. The electrostatic interaction is treated using the particle mesh Ewald method with the nonbonded cutoff set to 10.0 Å [38]. We used the SHAKE algorithm [39] to constrain the length of bonds involving H atoms. First, the conjugate gradient method and steepest descent method were used to minimize the total energy of the system. Then the system was heated to 300 K during 400 ps. In the production run, the system was under the isobaric isothermal (NPT) ensemble with a pressure of 1 atm and a temperature of 300 K for 6 ns. Andersen barostat and Langevin dynamics were used to control the pressure and temperature, respectively. The integration step was set to 2 fs and snapshots were output every 2 ps. Thus, 3000 conformations were collected from the MD trajectory for each system.
After MD simulation, we use cpptraj module [40] of Amber12 software to cluster the trajectory according to the conformational similarity. Then the local minima on the potential energy surfaces of the two largest clusters were obtained, which were subjected to MM energy minimization using the explicit solvent model. This yielded two relatively stable binding conformations for each CD/TB system. Guided by the binding mode deduced from 2D ROSY NMR (Fig SI2 a and b) [29], one of them was selected as the RC. Water molecules in the RC were removed for further binding free energy calculations at the PM3/PCM level. In the following, we use RCMM and RCPM3 to denote the RC before and after PM3 geometry optimization.
QM/CSM calculations
The free energy model takes into account not only the host–guest interaction but also the solvent effect. The total binding free energies of the host–guest complex in aqueous solution are divided into two parts [31],
Here, \(\Delta {\text{G}}_{{{\text{bind}}}}^{{{\text{solv}}}}\) denotes the change of solvation free energy due to the formation of the inclusion complex, which is computed as.
Where, \(\Delta {\text{G}}_{{{\text{comp}}}}^{{{\text{solv}}}}\), \(\Delta {\text{G}}_{{{\text{host}}}}^{{{\text{solv}}}}\), \(\Delta {\text{G}}_{{{\text{guest}}}}^{{{\text{solv}}}}\) denotes the solvation free energies of the complex, CD, and TB, respectively. The solvation free energy was calculated by the polarized continuum model (PCM) [41] as implemented in Gaussian 09 software [36], which is comprised of the electrostatic polarization energy (Gelec) and non-polar interaction energy (Gnp),
We use RCMM of the complex, host, and guest for quantifying the solvent effect via Eq. (2).
The binding free energy in vacuum is computed via,
These thermodynamics quantities are computed from geometry optimization and frequency calculation at the PM3 level.
Results and discussion
MD simulation and extraction of RC
The RCMM of the two enantiomers of TB before inclusion exhibit significant distinction (Fig. 1). R-TB is coiled, while S-TB is extended. The C1-C2*-C3-N4 dihedral angles in R-TB and S-TB are 73.19° and − 176.02°, respectively. This conformational difference can lead to different host–guest binding modes as will be shown later.
Guided by the structures deduced from NMR data (Fig SI2), the optimal RCs of the four systems are obtained from the best MD trajectories (Fig SI3). The dynamic behaviour of the host–guest separation, as measured by the distance between their centroids along the MD trajectories, is compared in Fig. 2. It is noteworthy that the coiled R-TB exhibits less fluctuation than does the extended S-TB either in β-CD or HAD-β-CD cavity. In particular, the largest fluctuation in host–guest separation is seen in β-CD/S-TB complex, indicating worse shape match, which leads to weak host–guest interaction as will be shown later. The RCs for the four CD/TB complexes are extracted from 3200 to 5500 ps, during which MD trajectories become relatively more stable. For all MD trajectories, it is not observed that TB enters the cavity of HDA-β-CD via its side chain. Thus the experimentally deduced binding structure29 in Fig SI2 (b) can be excluded.
The inclusion process of R-TB and HDA-β-CD is illustrated in Fig. 3. At 600 ps, the benzene ring of R-TB approaches wide secondary rim of the HDA-β-CD cavity and is well included at t = 1680 ps. t = At 3500 ps, a conformational change of R-TB is induced: its alkyl group is further bent towards the secondary rim of HAD-β-CD and the C1-C2*-C3-N4 dihedral angle decreases. A binding conformation that resembles the experiment deduced one appears and turns out to be the RC. Notice that the alkyl side chain of R-TB is compactly included and almost parallel to the secondary rim of HDA-β-CD. Afterwards, the RC remains relatively stable until 5500 ps. The inclusion processes of the other three systems are shown by the snapshots along the best MD trajectories in Fig SI4.
Subjected to the cluster analysis, the conformations on the well equilibrated MD trajectories from 3000 to 6000 ps are categorized into different groups based on their structural similarity (Fig SI5). The most stable conformations from the most populated cluster are chosen as the RCs for further structural analyses and binding affinity computations.
Analysis of RC
The RCMM of the four β-CDs/TB complexes in vacuum are optimized at the PM3 level. As expected, this causes the RMSD structural deformation ranging from 1.07 to 1.46 Å (Fig SI6), which is similar to those reported previously. [31, 32]
The structural features of the four RCs are further elucidated in Fig. 4. In all of them, the benzene ring of TB is included in the CD cavity, while the alkyl side chain is exposed to the solvent to different extent. HDA-β-CD shows higher deformation than does β-CD owing to its enhanced flexibility arising from the acetyl substitutions. For both HDA-β-CD/TB chiral complexes, NCMe3 protons of R-TB are close to the H-3 and OAc protons of HDA-β-CD (Fig. 4c and d), which is consistent with the 2D ROESY NMR measurements (Fig SI2) [29], as further elucidated in Fig. 4. The alkyl moiety of R-TB is almost included within the secondary rim of HDA-β-CD indicating the high degree of host–guest shape compensation. The positive charge on –NH2–group from the side chain is well exposed to the aqueous solution, which can lead to a favorable solvation energy as will be shown later. By contrast, S-TB in HAD-β-CD cavity adopts a different orientation. Its side chain is tilted at the secondary rim of HAD-β-CD and its –NH2–group is not sufficiently exposed to solvent.
On the other hand, the secondary rim of β-CD is unable to accommodate the alkyl moiety of either R-TB or S-TB. The binding modes of the two chiral complexes are relatively close. The positive charge on the –NH2–group of R-TB side chain is partially buried.
The conformational changes of the two CDs and TB enantiomers caused by the inclusion are examined by measuring the RMSD of all atomic coordinates of their RCMM (Fig. 5). The shape change of HDA-β-CD is much greater than that of β-CD. In the HDA-β-CD cavity, the deformation of R-TB is smaller than that of S-TB, which is consistent with the observation that R-TB matches that of HDA-β-CD better than does S-TB.
Binding affinities in aqueous solution
The various components of the computed binding affinity are compared for the four CD/TB complexes in Table 1. The binding free energy in vacuum \(\Delta {\text{G}}_{{{\text{bind}}}}^{{{\text{vacu}}}}\) and the solvent effect \(\Delta {\text{G}}_{{{\text{bind}}}}^{{{\text{solv}}}}\) measure the strengths of the non-bonding interaction (van der Waals interaction plus the H-bonding interaction) and hydrophobic interaction (solvation energy change due to inclusion) of the host–guest systems, respectively. The former is usually negative, whereas the latter positive. Less positive \(\Delta {\text{G}}_{{{\text{bind}}}}^{{{\text{solv}}}}\) is a sign of the enhanced hydrophobic interaction.
β-CD/S-TB possesses more positive \(\Delta {\text{G}}_{{{\text{bind}}}}^{{{\text{vacu}}}}\) than does β-CD/R-TB. Thus, the host–guest interaction is stronger for β-CD/R-TB, in which the guest deformation is also larger. However, the stronger interaction between β-CD and R-TB significantly weakens the host–guest hydrophobic interaction as indicated by the more positive \(\Delta {\text{G}}_{{{\text{bind}}}}^{{{\text{solv}}}}\) (Table 2). Consequently, the total binding affinity in aqueous solution for β-CD/R-TB is still close to that of β-CD/S-TB, differing by only 2.47 kcal/mol.
On the other hand, HDA-β-CD/R-TB possesses less negative \(\Delta {\text{G}}_{{{\text{bind}}}}^{{{\text{vacu}}}}\) than does HAD-β-CD/S-TB. This weaker host–guest non-bonding interaction for HAD-β-CD/R-TB is more than compensated by the strongly enhanced host–guest hydrophobic interaction, i.e. the much less positive \(\Delta {\text{G}}_{{{\text{bind}}}}^{{{\text{solv}}}}\) (Table 2). Thus, a much less positive total binding affinity, \(\Delta {\text{G}}_{{{\text{bind}}}}^{{{\text{aqu}}}}\), is eventually obtained for the R-complex than for the S-complex and the difference reaches 7.64 kcal/mol. This explains why HDA-β-CD can separate TB enantiomers better than does β-CD. Thus, the difference in the binding affinities between the two chiral inclusion complexes can be used to judge the chiral separation capacity of the host–guest system [42]. Note that the alkyl side chain of R-TB in is almost parallel to the secondary rim of the CD and the positive charge of –NH2–group is well exposed to the solvent (Fig. 4c). This may be the key for the enhanced hydrophobic interaction between HAD-β-CD and R-TB.
For both HDA-β-CD/TB and β-CD/TB complexes, the relative stability of their R-type and S-type TB complexes depends on the host–guest non-bonding interaction as well as the hydrophobic interaction. Interestingly, these two factors seem to work in opposition. Comparing the two chiral complexes of a particular type, we find if one is strengthened, the other is weakened. Therefore, an ideal binding mode that can coalesce the host–guest non-bonding and hydrophobic interactions is critical to achieve the efficient complexation in aqueous solution. The balance between these two factors plays a vital role in the separation ability for β-CD and its derivative.
Conclusion
The chiral recognition mechanisms of β-CD and HDA-β-CD toward enantiomers of TB are investigated by a MD/QM/CSM approach based on the experimental data, i.e. the structural information deduced from 2D ROESY NMR and measured binding affinities. From the computational results, we draw the following conclusions: (1) Before the inclusion, the stable configurations of R-TB and S-TB in aqueous solution exhibit significant difference: the former coiled and the latter extended. Such structural differences can exert great impact on the binding modes of TB with β-CD and −β-CD. (2) For HDA-β-CD/TB complexes, the guest molecule enters the CD cavity via the benzene ring rather than via the side chain in MD simulations, which concludes the orientation of TB in the HDA-β-CD cavity. (3) The NCMe3 group of R-TB is parallel to the secondary rim of HDA-β-CD, forming a tighter binding mode compared to that of HDA-β-CD/S-TB. (4) The difference in binding affinities between the two chiral inclusion complexes can serves a measure of chiral The separation capability of a chiral selector. (5) An ideal binding mode can reconcile the two opposing components of the binding affinity, i.e. the host–guest non-bonding interaction and their hydrophobic interaction.
References
Crini, G.: Review: a history of cyclodextrins. Chem. Rev. 114, 10940–10975 (2014)
Schneiderman, E., Stalcup, A.M.: Cyclodextrins: a versatile tool in separation science. J. Chromatogr. B. 745, 83–102 (2000)
Scriba, G.K.E.: Cyclodextrins in capillary electrophoresis enantioseparations recent developments and applications. J. Sep. Sci. 31, 1991–2201 (2008)
Scriba, G.K.E.: Chiral recognition in separation sciences. Part I: polysaccharide and cyclodextrin selectors. Trends Anal. Chem. 120, 115639 (2019)
Dai, Y., Wang, S., Zhou, J., Liu, Y., Sun, D., Tang, J., Tang, W.: Cationic cyclodextrin as versatile chiral selector for enantiomeric separation in capillary electrophoresis. J. Chromatogr. A. 1246, 98–102 (2012)
Zhou, J., Ai, F., Zhou, B., Tang, J., Ng, S.-C., Tang, W.: Hydroxyethylammonium monosubstituted cyclodextrin as chiral selector for capillary Electrophoresis. Anal. Chim. Acta. 800, 95–102 (2013)
Azhar, N.R., Yahaya, N., Suah, F.B.M.M., Prabu, S., Hui, B.Y., Shahriman, M.S., Zain, N.N.M., Raoov, M.: Enantioseparation of ketoconazole and miconazole by capillary electrophoresis and a study on their inclusion interactions with β-cyclodextrin and derivatives. Chirality 33(1), 37–50 (2021)
Wetterlin, K.: Resolution of terbutaline, a new beta. sympathomimetic amine. J. Med. Chem. 15, 1182–1183 (1972)
Handley, D.A., Mccullough, J.R., Crowther, S.D., Morley, J.: Sympathomimetic enantiomers and asthma. Chirality 10, 262–272 (1998)
Beng, H., Zhang, H., Jayachandra, R., Li, J., Wu, J., Tan, W.: Enantioselective resolution of Rac-terbutaline and evaluation of optically pure R-terbutaline hydrochloride as an efficient antiasthmatic drug. Chirality 30(6), 759–768 (2018)
Walhagen, A., Edholm, L.E., Kennedy, B.M., Xiao, L.C.: Determination of terbutaline enantiomers in biological samples using liquid chromatography with coupled columns. Chirality 1, 20–26 (1989)
Fanali, S.: Use of cyclodextrins in capillary zone electrophoresis resolution of terbutaline and propranolol enantiomers. J. Chromatogr. 545, 431–444 (1991)
Kim, K.H., Seo, S.H., Kim, H.J., Jeun, E.Y., Kang, J.-S., Mar, W., Youm, J.R.: Determination of terbutaline enantiomers in human urine by capillary electrophoresis using hydroxypropyl-β-cyclodextrin as a chiral selector. Arch. Pharm. Res. 26(2), 120–123 (2003)
Cucinotta, V., Giuffrida, A., Maccarrone, G., Messina, M., Puglisi, A., Torrisi, A., Vecchio, G.: The 6-derivative of β-cyclodextrin with succinic acid:a new chiral selector for CD-EKC. J. Pharm. Biomed. Anal. 37, 1009–1014 (2005)
Tang, K., Zhang, P.: Enantioselective extraction of terbutaline enantiomers with β-cyclodextrin derivatives as hydrophilic selectors. Chem. Pap. 65(3), 273–279 (2001)
Gogolashvili, A., Tatunashvili, E., Chankvetadze, L., Sohajda, T., Szeman, J., Gumustas, M., Ozkan, S.A., Salgado, A., Chankvetadze, B.: Separation of terbutaline enantiomers in capillary electrophoresis with cyclodextrin-type chiral selectors and investigation of structure of selector-selectand complexes. J. Chromatogr. A 1571, 231–239 (2018)
Zhou, Z.-M., Li, X., Chen, X.P., Fang, M., Dong, X.: Separation performance and recognition mechanism of mono(6-deoxy-imino)-beta-cyclodextrins chiral stationary phases in high-performance liquid chromatography. Talanta 82(2), 775–784 (2010)
Shi, J.-H., Ding, Z.-J., Hu, Y.: Experimental and theoretical studies on the enantioseparation and chiral recognition of mandelate and cyclohexylmandelate on permethylated β-cyclodextrin chiral stationary phase. Chromatographia 74, 319–332 (2011)
Li, W., Tan, G., Zhao, L., Chen, X., Zhang, X., Zhu, Z., Chai, Y.: Computer-aided molecular modeling study of enantioseparation of iodiconazole and structurally related triadimenol analogues by capillary electrophoresis: chiral recognition mechanism and mathematical model for predicting chiral separation. Anal. Chim. Acta 718, 138–147 (2012)
Alvira, E.: Influence of valine enantiomer configuration on the molecular dynamics simulation of their separation by β-cyclodextrin. Chem. Phys. Lett. 679, 31–37 (2017)
Seridi, L., Boufelfel, A.: Naringenin encapsulation in β-CD and in heptakis(2,6-di-O-methyl)-β-CD:NMR, NBO and QTAIM analysis. J. Incl. Phenom. Macrocycl. Chem. 90, 287–304 (2018)
Quevedo, M.A., Zoppi, A.: Current trends in molecular modeling methods applied to the study of cyclodextrin complexes. J. Incl. Phenom. Macrocycl. Chem. 90, 1–14 (2018)
Alvira, E.: Molecular simulation of the separation of isoleucine enantiomers by beta-cyclodextrin. Molecules 24, 1021 (2019)
Dodziuk, H., Lukin, O., Nowinski, K.S.: Molecular mechanics calculations of molecular and chiral recognition by cyclodextrins. Is it reliable? The selective complexation of decalins by β-cyclodextrin. J. Mol. Struct. Theochem. 503(3), 221–230 (2000)
Zhang, G., Hong, Z., Chai, Y., Zhu, Z., Song, Y., Liu, C., Ji, S., Yin, X., Wu, Y.: A study on the chiral recognition mechanism of enantioseperation of adrenaline and its analogues using capillary electrophoresis. Chem. Pharm. Bull. 55(2), 324–327 (2007)
Venkatesh, G., Thulasidhasan, J., Rajendiran, N.: A spectroscopic and molecular modeling studies of the inclusion complexes of orciprenaline and terbutaline drugs with native and modified cyclodextrins. J. Incl. Phenom. Macrocycl. Chem. 78, 225–323 (2014)
Guo, X., Wang, Z., Zuo, L., Zhou, Z., Guo, X., Sun, T.: Quantitative prediction of enantioseparation using β-cyclodextrin derivatives as chiral selectors in capillary electrophoresis. Analyst. 139, 6511–6519 (2014)
Liu, Y., Deng, M., Yu, J., Jiang, Z., Guo, X.: Gapillary electrophoretic enantioseparation of basic drugs using a new single-isomer cyclodextrin derivative and theoretical study of the chiral recognition mechanism. J. Sep. Sci. 39, 1766–1775 (2016)
Gogolashvili, A., Chankvetadze, L., Takaishvili, N., Salgado, A., Chankvetadze, B.: Separation of terbutaline enantiomers in capillary electrophoresis with neutral cyclodextrin-type chiral selectors and investigation of the structure of selector-selectand complexes using nuclear magnetic resonance spectroscopy. Electrophoresis 41(12), 1023–1030 (2020)
Zhou, Y., Zhou, B., Nie, X., Ye, R., Gong, X., Zhu, W., Xiao, H.: A theoretical study on the microencapsulation of herbicide MCPA with native β-cyclodextrin and its derivatives by a molecular dynamics/quantum mechanics/continuum solvent model approach. Chem. Ind. Eng. Prog. 34(12), 4185–4190 (2015)
Ye, R., Nie, X., Zhou, Y., Wong, C.F., Gong, X., Jiang, W., Tang, W., Wang, Y.A., Heine, T., Zhou, B.: Exploring host-guest complexation mechanisms by a molecular dynamics/quantum mechanics/continuum solvent model approach. Chem. Phys. Lett. 648, 170–177 (2016)
Dang, P., Ye, R., Meng, F., Han, Y., Zhou, Y., Gong, X., Zhou, B.: Microencapsulation thermodynamics of methylated β-cyclodextrins with bile salt: enthalpy, entropy, and solvent effect. J. Incl. Phenom. Macrocycl. Chem. 88, 181–189 (2017)
Ćezard, C., Trivelli, X., Aubry, F., Djedaïni-Pilard, F., Dupradeau, F.Y.: Molecular dynamics studies of native and substituted cyclodextrins in different media: charge derivation and force field performances. Phys. Chem. Chem. Phys. 13(33), 15103–151021 (2011)
Case, D.A., Darden, T.A., Cheatham, T.E., III., Simmerling, C.L., Wang, J., Duke, R.E., Luo, R., Crowley, M., Ross, W.S., Zhang, W., Merz, K.M., Wang, B., Hayik, S., Roitberg, A., Seabra, G., Kolossváry, I., Wong, K.F., Paesani, F., Vanicek, J., Wu, X., Brozell, S.R., Steinbrecher, T., Gohlke, H., Yang, L., Tan, C., Mongan, J., Hornak, V., Cui, G., Mathews, D.H., Seetin, M.G., Sagui, C., Babin, V., Kollman, P.A.: Amber12. University of California, San Francisco (2012)
Besler, B.H., Merz, K.M., Kollman, P.A.: Atomic charges derived from semiempirical methods. J. Comput. Chem 11(4), 431–439 (1990)
Frisch, M.J., Trucks, G.W., Schlegel, H.B., et al.: Gaussian 09, Revision E.01. Gaussian Inc., Wallingford (2013)
Jorgensen, W.L., Chandrasekhar, J., Madura, J.D., Impey, R.W., Klein, M.L.: Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79(2), 926–935 (1983)
Essmann, U., Perera, L., Berkowitz, M.L., Darden, T., Lee, H., Pedersen, L.G.: A smooth particle mesh ewald method. J. Chem. Phys. 103(19), 8577–8593 (1995)
van Gunsteren, W.F., Berendsen, H.J.C.: Algorithms for macro-molecular dynamics and constraint dynamics. Mol. Phys. 34(5), 1311–1327 (1977)
Roe, D.R., Cheatham, T.E., III.: PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9(7), 3084–3095 (2013)
Tomasi, J., Mennucci, B., Cammi, R.: Quantum mechanical continuum solvation models. Chem. Rev. 105(8), 2999–3094 (2005)
Mindrila, G., Mandravel, C., Dobrica, I., Bugheanu, P., Stanculescu, I.R.: Theoretical study of β and γ-cyclodextrins inclusion complexes with nineteen atropisomeric polychlorobiphenyls. J. Incl. Phenom. Macrocycl. Chem. 74, 37–143 (2012)
Acknowledgements
BZ and JL gratefully acknowledge the financial support from the National Natural Science Foundation of China, No. 22075144.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dou, R., Chen, K., Chi, G. et al. Why heptakis(2,3-di-O-acetyl)-β-cyclodextrin can separate terbutaline enantiomers better than β-cyclodextrin: nonbonding and hydrophobic interactions. J Incl Phenom Macrocycl Chem 100, 189–195 (2021). https://doi.org/10.1007/s10847-021-01072-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10847-021-01072-8