Abstract
Being able to correctly model semantic relatedness between texts, and consequently the concepts represented by these texts, has become an important part of many intelligent information retrieval and knowledge processing systems. The need for such systems is especially evident within the biomedical domain, where the sheer amount of scientific publishing contributes to an information overflow. In this paper we present a novel method to approximate semantic relatedness in domain-focused settings. The approach is an extension to a well-known ESA (Explicit Semantic Analysis) method. Our extension successfully leverages the semantics of a domain-specific document corpus. We present the evaluation of the proposed method on a set of reference datasets, that are a de facto reference standard for the task of approximating biomedical semantic relatedness. The proposed method is evaluated in comparison with other state-of-the-art methods, as well as the baselines established with the original ESA method. The results of the experiments suggest that the proposed method combines the semantics of a general and domain-specific corpora to provide significant improvements over the original method.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Approximating semantic relatedness is an important part of various text and knowledge processing tasks. Semantic relatedness is a metric that can be assigned to a pair of labels in order to represent the strength of the relationship of the concepts described by those labels. The automated calculation of the metric is the building block for numerous semantically enhanced data processing techniques such as: word sense disambiguation (Agirre and Rigau 1996) (used for matching word contexts to the best word senses), text summarization (Barzilay and Elhadad 1997) (used for evaluating cohesion of the lexical chains) and information retrieval (Rada et al. 1989) (incorporated in the query-document ranking method). Similar applications of relatedness and similarity (which is a narrower concept) metrics within the scope of Life Sciences include entity–entity relationship extraction (Guo et al. 2006; Mathur and Dinakarpandian 2012), semantic search (Sahay and Ram 2011) and redundancy detection in clinical records (Zhang et al. 2011). An overview of applying semantic similarity to the problem of comparing gene products is discussed by Pesquita et al. (2009). In Pesaranghader et al. (2014) the authors evaluate the application of a relatedness measure as an approximation of semantic similarity in the biomedical domain.
The problem of most state-of-the-art methods used in the biomedical domain for calculating semantic relatedness is their dependence on highly detailed, structured knowledge resources, which are usually tailored to a specific problem or perspective. This dependency makes these methods poorly adaptable to many usage scenarios that do not fit into this predefined mold. On the other hand, the domain knowledge in the biomedical domain is becoming more and more accessible, but mostly in its unstructured form — as texts in large document collections, which makes its use more challenging for automated processing. Overcoming these challenges and harnessing the non-explicit knowledge of the biomedical literature is a step towards adaptable and robust semantic relatedness approximation methods, and consequently, better intelligent systems for accessing and processing the information ‘hidden’ in these corpora.
The ultimate goal of a relatedness measure is to assign a numerical approximation of the relatedness strength to a given pair of input texts (or simply: inputs). In this paper we present a distributional semantic relatedness measure, DomESA (Domain-focused Explicit Semantic Analysis), in which input texts are represented by vectors of concepts, i.e. a distribution of relevance of the given input over a concept space of an encyclopedia-style resource (i.e. Wikipedia). The relatedness approximation is calculated by comparing these distribution vectors. The measure presented here is an extension of a popular state-of-the-art relatedness measure – explicit semantic relatedness (ESA) (Gabrilovich and Markovitch 2007). The distinguishing feature of our approach is that the representation vectors are derived from two sets of vectors: one, which represents texts of a domain-oriented corpus (i.e. Medline), and a second one, which models the semantic relevance between the documents of the domain-oriented corpus and the concepts of the encyclopedic resource. In this way, the final representation vectors, established over the space of concepts of a general domain resource, are obtained through a distributional representation of the input derived from the domain-oriented corpus. This makes the method less dependent on the presence of specific vocabulary within encyclopedic definitions (which are the general domain resource), thus bridging the semantic gap between the intended use (represented by biomedicine-related reference datasets) and the general-domain resource.
DomESA is our key contribution presented in this paper, we also include its detailed evaluation alongside state-of-the-art methods on the reference datasets, as well as an extended discussion of the results obtained in the experimental evaluation.
The paper is organized as follows. In the next section we discuss the related body of work. In Section 3 we introduce the basic concepts and the overview of the processing flow of the original ESA method. Section 4 presents DomESA. We also discuss the design differences between DomESA and other related methods. In the following section we outline our experimental setup. Later in the same section we present the results of the experimental evaluation and discuss them. Finally, in the last section the conclusions are presented, and we also outline the possible research lines originating from the work presented in this paper.
2 Related work
There have been numerous efforts made towards a successful replication of human judgment in the assessment of similarity and relatedness between pairs of words or concepts. A relatively recent and up-to-date survey is presented by Zhang et al. (2012). The method we present in this paper can be seen as Wikipedia-based, but in reality it treats Wikipedia as a document corpus, so we would rather classify it as a distributional or corpus-based method.
The idea of leveraging the distributional hypothesis, that words with similar contexts will have similar meanings (‘you shall know a word by the company it keeps’ (Firth, 1957)), has been explored thoroughly in the field of distributional semantics research. Techniques vary from basic word co-occurrence matrices, where a vector associated with a given word is created by counting the words that appear in its immediate context (determined by a window of a certain size), to latent semantic indexing (Dumais 2004). More recently, dense word representations derived through machine learning techniques, e.g. word2vec methods (Mikolov et al. 2013), have gained popularity. In Virginia and Nguyen (2015) the authors propose modeling the semantics of the corpus documents with a Tolerance Rough Set Model (TRSM), an approach, in which the co-occurence data is used to approximate the semantically extended representations of the documents. The label of ‘distributional semantics’ covers an extremely wide scope of methods and models that share the feature conveyed in the label itself — semantics being modeled as a distribution over a large set of features derived from a large body of linguistic evidence (i.e. a large corpus of documents).
Our method extends ESA (Gabrilovich and Markovitch 2007) with an additional computational step. Numerous extensions of ESA have already been proposed, many of which combine the original approach with the Wikipedia-specific features, through the concept-to-concept feature/similarity matrices, e.g. Scholl et al. (2010), Polajnar et al. (2013), and Haralambous and Klyuev (2013). Some of those extensions, e.g. NESA (Asooja et al. 2015) (Non - Orthogonal ESA), also provide variants that are generic enough to be used with any document collection. The main difference is that in our approach we postulate using a combination of corpora to reduce the semantic gap (and consequently increase the performance), while these extensions are intended to increase the method’s performance through a more sophisticated use of the properties of a single knowledge source.
There are Wikipedia-based methods for approximating semantic relatedness other than ESA, such as WikiRelate (Strube and Ponzetto 2006). It is worth noting however, that in the case of algorithms, which by default rely on Wikipedia-specific features (such as link structure), incorporating semantics of an unstructured domain-focused corpus is a much more difficult task.
In this paper, we present a direct evaluation of the proposed extension, i.e. the method is evaluated against reference datasets, which capture scores assigned manually to the pairs of textual inputs by human annotators. We rely on a collection of reference datasets that has become a standard in the evaluation of biomedical semantic relatedness (Pedersen et al. 2007; Pakhomov et al. 2010, 2011).
There is also a significant number of papers that weight in on the application of established methods within biomedical settings. In Muneeb et al. (2015) the authors explore the performance of the aforementioned word2vec and GloVe (Pennington et al. 2014) methods with models trained on a large biomedical corpus. In a recent paper the authors, Sajadi et al. (2015), present an original hybrid node ranking based method, based on Wikipedia structure, applied in biomedical settings. Furthermore, they provide an extensive evaluation of their method in comparison with other state-of-the-art methods, which constitutes an important reference perspective.
In our previous paper (Rybiński and Aldana-Montes 2016), apart from presenting tESA, an extension of the ESA method that works well with the corpus of scientific documents, we analyzed the direct adaptability of the ESA approach to the problem of biomedical relatedness approximation.
Finally, there is a large group of methods for the approximation of biomedical semantic relatedness, which rely, to different extent, on the use of structured domain specific resources. E.g. in Pedersen et al. (2007) and Liu et al. (2012) the authors propose extracting concept representations from the documents of a large biomedical corpus, in a process, which is guided by a structured knowledge resource. The method relies on ‘scanning’ the corpus for words co-occuring with words associated with specific concepts of the knowledge source. In Sánchez and Batet (2011) the authors showcase the performance of a wide spectrum of ontology-based Information Content (IC) methods, which use SNOMED CT as the knowledge resource. SNOMED CT is the largest medical vocabulary collection, with over 400K systematically organized concepts with their lexical representations and additional information. The IC measures presented by Sánchez and Batet (2011) use the ontological structure of SNOMED (positions of concepts in the ontology, distance between them, number of sub-concepts, etc.) to compute a semantic score between a pair of concepts. In Martinez-Gil (2016) the author proposes a fuzzy logic-based method for combining scores achieved by the ontology based-methods. Our method, although dependent on a specific corpus, does not rely on high-level KB representations of the domain, which makes it more flexible and easier to adapt to non-standard use cases.
3 Preliminaries
3.1 Basic concepts
A system for approximating semantic relatedness can be represented very simply as a black-box, which takes two input texts (words, phrases, paragraphs, etc.) and produces a single output – a relatedness score, which corresponds to the relatedness between the inputs. This basic idea is illustrated in Fig. 1.
Throughout the processing flow we use Tf-Idf (term frequency - inverse document frequency) weighted vectors as a basic tool to represent texts. Texts within the black box are either input texts or the documents of the background corpus (or corpora). We denote the input texts with single letters s and t, while \(\overline {s}\) and \(\overline {t}\) denote their Tf-Idf weighted vectors. An i-th document of a corpus is denoted as \(d^{Med}_{i}\) or \(d^{Wiki}_{i}\) (depending on the corpus it belongs to). Overline is used to denote the corresponding Tf-Idf vectors: \(\overline {d}^{Med}_{i}\) and \(\overline {d}^{Wiki}_{i}\). To compare vectors we use cosine similarity, which is denoted \(cosine(\overline {a}, \overline {b})\) for the example vectors \(\overline {a}\) and \(\overline {b}\):
where a i and b i denote the i-th elements of the n-length vectors.
As suggested, we contemplate the use of two corpora. The general domain corpus is a collection of Wikipedia articles, while the biomedical corpus is a collection of abstracts of biomedical scientific publications – Medline. The original ESA can be set up with either one of the two, while the extended method combines the use of both corpora.
In our experimental evaluation we used the 2015 snapshot of the Medline corpus, which makes our results directly comparable with those obtained by Rybiński and Aldana-Montes (2016). The collection contains more than 14M abstracts. As for the Wikipedia corpus, we used a snapshot from December 2015, which contains over 3.8M English language articles.
We use standard Apache LuceneFootnote 1 mechanisms for pre-processing of texts prior to the Tf-Idf vectors computations. Texts are transformed to lowercase and stopwords (words that occur very commonly, but provide little or no semantic information, e.g. the, of, at and a) are eliminated. Numbers are also eliminated and non-alphanumeric characters (e.g. ‘-’) are normalized. In the case of the titles, we also disregard words that appear in less than 3 different documents of the respective corpora.
3.2 ESA
In ESA set up with an N document corpus (e.g. Wikipedia), the processing for a pair of inputs s and t is as follows:
- 1:
-
Creation of concept vectors \(\overline {c}^{s, Wiki}\), \(\overline {c}^{t, Wiki}\) for each of the inputs.
- 2:
-
Calculation of the relatedness score as \(cosine(\overline {c}^{s, Wiki}, \overline {c}^{t, Wiki})\).
Concept vectors are vectors of N elements, where the i-th element for the vector calculated for an input t, \(\overline {c}_{i}^{t, Wiki}\), is calculated as follows:
As mentioned, the corpora can be used interchangeably, although the original method described by Gabrilovich and Markovitch (2007) was set up with the Wikipedia corpus.
For practical reasons it is enough to consider a certain number of top valued elements within each concept vector (e.g. 10000), with the rest of elements set to 0.
4 DomESA - domain-oriented biomedical extension of ESA
4.1 Presentation of the method
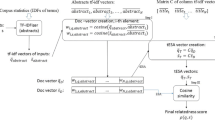
Here, we explain the processing of DomESA, the novel ESA extension. First, we define the matrix K. Each column of K corresponds to a document from the domain-oriented corpus, i.e. Medline. The rows of the matrix correspond to the documents of the general domain corpus, i.e. Wikipedia. Th element at the (i,j) position of the matrix, K i j is defined as a cosine similarity between the i-th document of Wikipedia and j-th document of Medline:
Note, we also introduce a limit on non-zero values per column. Specifically, we only consider top k values per column.Footnote 2 As a result of this cutoff we only deal with a truncated matrix, which is much more convenient resource-wise. In addition, it accounts for less noise being ‘introduced’ into the final representations of input texts. The processing flow of DomESA can be summarized as follows:
- 1:
-
Creation of concept vectors \(\overline {c}^{s, Med}\), \(\overline {c}^{t, Med}\) for each of the inputs.
- 2:
-
Calculation of DomESA vectors \(\overline {m}_{s}\), \(\overline {m}_{t}\); for the input t, \(\overline {m}_{t}\) is calculated as follows:
$$ \overline{m}^{t} = K \overline{c}^{t, Med} $$(4) - 3:
-
Calculation of the relatedness score as \(cosine(\overline {m}^{s}, \overline {m}^{t})\).
This formulation leads us to the formula for the i-th position of the vector \(\overline {m}^{t}\), which corresponds to the i-th document in Wikipedia corpus (i.e. to the i-th concept, represented by this article). The value, denoted by \({\overline {m}^{t}_{i}}\), is given by:
where N denotes the size of the Wikipedia corpus.
A cutoff is also applied to the concept vectors obtained in step 1, similarly as it is done in our implementation of the original ESA method. Both the original ESA and DomESA are represented graphically in Fig. 2.
The intuitive explanation of DomESA’s processing is as follows. In the ESA model the input is represented with a superposition of ’one-hot’ document vectors with each vector multiplied with the document-input relevance score (which is 0 if the input does not have common tokens with the document). DomESA does the same, but each document is represented differently – instead of using the ‘one-hot’ representation, each document from the domain-focused corpus is represented with a ‘k-hot’ vector of the most relevant Wikipedia entries. Consequently, each Medline document, ‘activated’ by the input, ‘activates’ k Wikipedia articles. As a result, DomESA does not require the inputs to appear strictly within the same documents of Medline corpus – it is enough that they eventually ‘activate’ the same Wikipedia articles.
DomESA has been implemented in Java, with the corpus Tf-Idf vector space models implemented with Apache Lucene. The creation of the matrix K had been executed offline with the same technology with traditional Java multi-threading.
4.2 Design differences between DomESA and other corpus-based methods
On a conceptual level the processing in DomESA is similar to the processing in ESA, except that DomESA uses an additional computational step. NESA and XESA, both extensions of ESA, also use document-document similarity matrices in the calculations. Nonetheless, both of these methods use a matrix of similarities computed for a single document corpus, so the domain adaptation is not being contemplated. Furthermore, the similarity matrix of NESA and XESA is used to overcome the supposed orthogonality of Wikipedia articles. DomESA on the other hand, uses a highly non-orthogonal corpus (Medline), so it can be reasonably expected that the orthogonality of Wikipedia should be less of a problem.
DomESA (and other ESA extensions for that matter) provides a flexibility advantage over approaches that rely on context window word counts (Pedersen et al. 2007; Liu et al. 2012), because new representations can be created without having to actually ‘scan’ through all the documents that contain the input terms, so the cost of creating the representation vectors is much lower. It is worth noting though, that on a conceptual level our method is designed to accomplish a goal similar to that of Liu et al. (2012). In Liu et al. (2012), the authors use second order co-occurence vectors to compare their inputs. In other words, to compare two inputs, they actually compare the words that are related to the contexts, in which those inputs appear. In DomESA we employ a similar principle, i.e. we compare inputs by comparing the Wikipedia articles related to the Medline articles that the inputs appear in.
Word embeddings (i.e. word2vec) have the advantage of using dense representation vectors of a relatively low dimension (typically around 200), which makes these methods computationally appealing. However, the use of machine learning to pre-train the model hinders the flexibility of these methods to a certain degree. For example, switching from unigram to bigram inputs requires either re-training the entire model or using some kind of composition strategy involving unigram vectors (addition, multiplication), while ESA and similar methods can be adapted relatively easily or need no adaptation at all, depending on the actual implementation.
tESA is computationally similar to DomESA, but the conceptual difference is evident and, as a result, the semantics of the distributional representations are different in these two methods. Both methods obtain their output vectors by multiplying the ESA-style concept vectors with a matrix. In the case of DomESA this is a matrix of Medline-Wikipedia document similarity, while tESA uses a matrix of Medline document titles. This means that the tESA vectors are expressed over the title vocabulary of a biomedical corpus, while DomESA vectors are still concept vectors (over Wikipedia concept space), although obtained indirectly.
5 Experiments and results
5.1 Experimental setup
As mentioned, the proposed method is evaluated directly on the reference datasets. The summary of the reference datasets is presented in Table 1. Each of the datasets contains pairs of inputs with an associated human-assigned relatedness score. The relatedness scores included in the datasets were calculated as an average between answers supplied by certain number of human annotators.
The task of the system is to replicate the average scores assigned by the human annotators. The quality of this replication is measured as a pair of correlation scores between the system assigned scores and average human scores - with Spearman’s rank correlation and Pearson’s correlation coefficients. Most studies only use Spearman’s rank correlation, as, given a dataset, the problem can be seen as a ranking creation – ranking pairs of concepts from the least related to the most related. Notwithstanding, we believe that using both coefficients is justifiable, because: (a) there are some studies that use Pearson’s correlation coefficient (e.g. Muneeb et al. 2015; Martinez-Gil 2016); (b) using both correlations does provide a fuller perspective of the data.
We present the scores obtained with DomESA alongside with those obtained by relevant state-of-the-art methods, i.e. word2vec’s CBOW and tESA. The evaluation is completed with two important sets of baseline scores, obtained with the original ESA method. One set is calculated for the ESA method set up with Wikipedia, the second one is calculated with the ESA method set up with the Medline corpus.
Comparing the results obtained with different methods effectively involves comparing the correlations those methods generate w.r.t. the model answers. We evaluate the statistical significance of correlation comparisons. Specifically we construct a 0,95 confidence level confidence intervals (CI) for dependent overlapping correlations (as for a pair of methods, both of them produce their correlation against the same reference dataset). If we consider two correlations result A r A and result B r B , our null hypothesis can be formulated as r A − r B = 0. Therefore, if for a given triple of correlations (result A-to-model, result B-to-model, result A-to-result B), the constructed confidence interval does not include 0, the test allows us to refute, under the assumed confidence level, the null hypothesis of the two correlations being equal. This methodology (Zou 2007) is one of few options applicable for testing the comparisons between dependent overlapping correlations.Footnote 3
The results presented in the following section were obtained for concept vectors with a limit of 10000 values; the matrix K was truncated at 10 values per column. The parameter values were calibrated experimentally, nonetheless the experience gained from the calibration process seems to indicate that neither of the methods is especially sensitive to the values of those parameters (i.e. a relatively small change in parameters will only have little, or no, effect on the method’s performance).
5.2 Results and discussion
In Table 2 we have included pair-by-pair results of DomESA, together with the ‘model’ scores generated by medical coders. This set of results can be seen as an example of how the evaluation has been performed. Specifically, it corresponds to a single set of correlation pairs presented in the overview of the results in Tables 3 and 4.
Tables 3 and 4 present the overview of the results obtained in the experimental evaluation, with the tables corresponding to Spearman’s rank correlation and Pearson’s correlation, respectively. The best respective values within our evaluation are highlighted in bold, whereas the correlation scores, which, to our knowledge, surpass other correlations reported in the literature, are marked with a ’G’.
The results presented in Tables 3 and 4 are also shown in Figs. 3 and 4, which correspond to Spearmans’s and Pearson’s correlation scores, respectively.
The tests of statistical significance of the results presented in Tables 3 and 4 are shown in Tables 5 and 6, which correspond to Spearman’s and Pearson’s correlation coefficients, respectively. Correlation comparisons, which are considered statistically significant under the assumed confidence level are denoted with ‘ ↑’ if DomESA’s correlation score is higher than the given baseline score, or with ‘ ↓’, if DomESA’s correlation is lower. Correlation comparisons that are considered to be statistically insignificant are denoted by ‘-’.
The method proposed in this paper, DomESA, clearly surpasses both of the ESA baselines. Importantly, the evaluation results of ESA (Medline) are on par with other state-of-the-art methods, yet DomESA provides a substantial improvement. It is worth noting, that the superior results are achieved even though: (1) DomESA has its representation vectors expressed over the same collection of Wikipedia concepts as ESA (Wiki); (2) DomESA does not use resources any more domain-specific than the Medline corpus, which is also used by ESA (Medline). This indicates that the improvement of the results is related to the use of the matrix K.
The intuition behind this can be explained in several ways. Most importantly, however, we feel that the difference between DomESA and the original method is that the original ESA’s notion of relatedness depends on the actual co-occurence of given input texts within specific documents (from either of the corpora ESA is configured with). On the other hand, DomESA leans towards the notion of related input texts appearing in a similar contexts (not necessarily within the same documents), because in the end it is the context (Medline papers) that ‘triggers’ the Wikipedia-based representation.
As we have remarked, in a certain sense, the idea behind DomESA is similar to the one presented in NESA and XESA, only here the orthogonality of Wikipedia articles is being addressed with a Medline-Wikipedia similarity matrix, rather than with Wikipedia-Wikipedia or Medline-Medline similarity matrices. Additionally, the combined use of the two corpora in DomESA adds the domain focus, while the representations of the input are expressed over the space of Wikipedia concepts, just like in the original method.
The statistical significance tests confirm that DomESA provides better results than any other method in the evaluation presented here. W.r.t. the rank correlation, DomESA does significantly outperform each of the other methods on at least one reference dataset, without performing significantly worse than any method on any of the datasets. The same is true regarding the statistical testing of the Pearson’s correlation comparisons. Notably, the methods that provide results close to those of DomESA (in terms of statistical significance) w.r.t. one of the correlation coefficients, perform significantly worse w.r.t. the other correlation coefficient (e.g. ESA setup with Medline corpus).
It is interesting to consider the statistical significance tests of DomESA comparisons with ESA setup with Medline corpus w.r.t. the rank correlation. It turns out, that although DomESA does outperform the ESA baseline on all datasets, only one of the performance margins can be considered significant. This is especially interesting, given that some of these margins are wider than in the case of other methods, e.g. tESA performs better than ESA on the mayo101 reference dataset, but it is the tESA-DomESA comparison that turns out to be statistically significant, rather than the ESA-DomESA comparison. This points to a low correlation between the scores assigned by ESA (Medline) and DomESA. In simple terms, it means that each of these methods ‘gets right’ other parts of the reference dataset (at least partially).
When it comes to comparing DomESA and the original Wikipedia-based ESA it is worth noting, that the domain focus of DomESA, combined with the fact that the method ‘assigns’ Wikipedia concepts to contexts rather than words, translates into specific properties of the distributional representations used in the method. Firstly, it accounts for fuller semantic representations of those input texts that are less likely to appear within the less domain-specific Wikipedia corpus. Secondly, the contexts are less ambiguous, so the representations are ‘channeled’ towards the domain-related articles of Wikipedia. For example, a DomESA’s processing of an input ‘support stockings’ is much more likely (than the one of the original ESA) to generate a representation related to clotting disorders and their treatment rather than to the garment in general. The impact of DomESA’s processing can be illustrated with a specific case of the K matrix truncated at k = 1, which means that each Medline abstract corresponds to one Wikipedia article. This variant of DomESA can be called ‘minimal’, as it will result in the most compact representation vectors (i.e. with the least number of non-zero elements per vector). The direct comparison is presented in Table 7.
It can be observed, that the performance of DomESA in terms of Spearman’s rank correlations is barely affected by the change to the parameter k. DomESA produced representations with more non-zero elements, which accounts for DomESA’s greater ability in assigning input texts to Wikipedia contexts (as the method does not require the texts to appear within Wikipedia articles). The difference in the quality in the results (higher correlations), accounts for DomESA providing more accurate representations, even if their size is comparable to those of ESA (which accounts for similar speed of calculations, once the representations are created).
It is also worth noting, that the representations of DomESA are actually compatible with those of ESA, as the semantics of the input texts are being expressed over the same set of concepts (even if the weight distribution is less noisy or more accurate). This also means that DomESA can benefit from some of the same extensions as the original method, such as NESA (Asooja et al. 2015), XESA (Scholl et al. 2010), CL-ESA (Potthast et al. 2008).
When compared to other methods based on vectors extracted directly from Medline (tESA, ESA), DomESA performs slightly better on most datasets in terms of rank correlation (with the exception of the umnsrsRel dataset, for which it provides a second best score), and notably better in terms of linear correlation with human judgment.
Our method also provides better results than the CBOW model trained on a very similar corpus. Although the CBOW model is not an ideal word2vec model for the relatedness approximation problem, our experiment provides a perspective that enables us to compare our results with those obtained by Muneeb et al. (2015), as our results for the CBOW model seem comparable. Our method provides better results, in all of the reference datasets, than any of the methods reported by Muneeb et al. (2015) (word2vec models and GloVe are evaluated). Unfortunately the authors do not mention the rank correlations obtained with their word2vec and GloVe models, but we believe that their results and our CBOW experiment provide enough grounds for a comparison. Obviously, the main disadvantage of DomESA when compared to the word2vec methods is that it operates on vectors of much higher dimensionality and with more non-zero elements, which makes it slower in comparing vectors (i.e. the calculation of cosine similarity is faster on dense and low dimensional vectors, that word2vec operates on).
To the best of our knowledge DomESA provides the highest correlations (both rank and linear) of all the methods reported in the literature in the largest three datasets (umnsrsRel, umsrsSim and mayo101), with the exception of the slightly better ranking performance of tESA in the umnsrsRel dataset. In a very recent evaluation presented by Sajadi et al. (2015), all of the evaluated methods display the relatedness scores below those established by DomESA.
The performance of DomESA in terms of Spearman’s correlation is second to none of the other state-of-the-art methods, which shows the quality of the rankings provided by our method. However, its performance in terms of the Pearson’s correlation scores is also well worth considering. DomESA obtained the highest linear correlations with human judgment among all the evaluated methods, notably improving upon the performance of the original ESA and its derivative (tESA). All things considered, we believe that this is mostly due to the fact that DomESA’s processing incorporates a kind of ‘smoothing’ into its representation of semantics, by relying on ‘fuzzy’ notions of concepts and their similarity, rather than almost binary features (like co-occurrence in a highly weighted document of the corpus). This results in DomESA being less likely to produce outlier results, when compared to other ESA-based methods, which seems to explain this difference in performance. The goal of the processing of CBOW (and other word embedding methods for that matter) is conceptually similar to that of DomESA, as the model ‘learns’ to express words over a set number of dimensions taking into account their contexts. DomESA uses a different computational approach to do a similar thing, i.e. express input texts, given their contexts, over a set of dimensions determined by the Wikipedia corpus. DomESA does not reduce the number of dimensions as CBOW does, but we believe that the similarity of the methods described above explains why Pearson’s correlations obtained with the CBOW model surpass those of the original ESA and tESA. DomESA on the other hand, still outperforms CBOW in terms of Pearson’s correlations, but we believe that the reason for this is twofold: (i) as stated above, DomESA also incorporates some form of semantic smoothing (so CBOW does not have an advantage here); (ii) DomESA’s relatedness approximation is generally more accurate, as the Spearman’s rank correlations seem to suggest.
As mentioned above, once the representation vectors are calculated, DomESA’s representations yield computational efficiency similar to that of the ESA’s representations, as their size (in terms of non-zero elements) is comparable and the relatedness calculation is essentially the same (cosine similarity between vector representations). However, the creation of the representation vectors in DomESA involves an additional cost of the matrix multiplication step. Also, the process of calculating the matrix K is costly, as it involves N×M document-to-document similarity calculations. Nonetheless, both processes (creation of the representations and matrix calculations) are easily parallelizable and in many usage scenarios they can be treated as a one-time cost. Consequently, DomESA can be expected to scale up well w.r.t. the corpus size – an eventual increase in size can be dealt with seamlessly, with additional computational resources.
It is important to note, that the model proposed here does not fix the problem of corpus dependency of the original method. DomESA provides grounds for relatedness computations for domain-based inputs, so, for example, general domain inputs will be most likely out of the semantic scope of the Medline corpus, which will consequently lead to poor representation vectors. The interoperablility with the original ESA (Wikipedia-based) provides a basis for a workaround solution for comparing domain-focused and general domain inputs, but the problem of deciding which method to use for representing each input is to be solved. More generally, DomESA (and ESA as well) will tend to generate poor representations for inputs that are poorly represented within the domain-focused corpus (e.g. poorly described or newly discovered diseases). It is worth noting, however, that the same can be said of any class of methods, which depend on the semantic coverage of the background knowledge resource – the capacity of the method to leverage the information will naturally be limited by the lack of the information.
Furthermore, our initial experience with DomESA applied to the problem of ontology alignment (Rybiński et al. 2016) seems to point to another issue inherited from ESA. Specifically, it can be observed that the method works well for the basic job of comparing ‘short’ inputs, which consist of 1-2 words/tokens, but tends to generate false positives for longer inputs (especially if they share common relatively ‘high IDF’ words of relatively low semantic value, e.g. ‘recessive’ or ‘autosomal recessive’ in the context of comparing labels of genetic disorders). We believe that this issue may be solved by: (a) involving more information in the relatedness approximation (we have used labels only; synonyms, definitions and ontological neighborhood could also be used to compare concepts), (b) reconsidering the compositional aspect of the method, (c) a combination of approaches (a) and (b).
Approach (a) is quite straightforward – using more information to create the representation vector will result in a statistically better representation, less dependent on the presence of a single common token. Nonetheless, the approach (a) (and consequently (c)) is not a general solution, as it depends on the availability of the additional information, which may be present (or not) in the context of an ontology alignment problem.
A potential solution obtained with an approach (b) is far more appealing in terms of improving the method itself. In the experiments presented here, the concept vectors (the vectors that link the input directly with the documents of the corpus) are created ‘at once’ for the entire input, i.e. a single Tf-Idf vector representing the input is created and we calculate the cosine similarity of this vector with all the Tf-Idf document vectors of the corpus. It is important to note, that many alternative compositional strategies exist, from simple ones (e.g. obtaining representations of individual words and adding them), to more complex approaches, e.g. Kusner et al. (2015). In the latter work, the authors describe an approach for calculating document distances from word embeddings. Their approach accounts not only for similarities (relatedness) between the matching elements (tokens), but also for dissimilarities between the non-matching ones, so it could be a good fit for improving DomESA’s performance with longer texts. We plan to study the applicability of the approach to DomESA in near future.
6 Conclusions
In this paper we have introduced DomESA, a novel extension to the ESA method, designed for domain-oriented use. The method uses a domain-focused background corpus, in addition to Wikipedia articles, to generate higher-quality semantic representations of the input texts. The approach provides semantic relatedness approximation results superior to the original method on all of the benchmarks, which shows its validity.
Our results also indicate that an additional improvement of the results is achieved through the introduction of a ‘fuzzier’ notion of relatedness. In DomESA two inputs can be related due to them appearing in similar contexts, while in the original method they would have to actually co-occur within the same documents. Both this, and the domain focus, are introduced to the DomESA representation vectors in a single computational step.
Moreover, our method, despite the fact that it uses a domain-focused corpus, produces representation vectors expressed over the space of Wikipedia concepts, just like the original ESA set up with Wikipedia. This means, that many of the extensions proposed for the original method can also be applied to DomESA. Those include usage of Wikipedia’s multi-language structure, hyperlinks, classifications and other Wikipedia-specific features. Some of those extensions could be explored in the context of domain-focused semantic relatedness in our future work.
Apart from the impact of known ESA extensions on DomESA’s performance, there are two other interesting lines of future research related to the method presented in this work. Firstly, we would like to study DomESA’s applicability to the problem of ontology alignment and the improvements it may bring to the process, thus expanding our earlier work (Rybiński et al. 2016). Preliminary results show that DomESA’s forte lies in discovering original correspondences between the ontological concepts. In our future work we would like to build on this quality. Secondly, we would like to study the compositional aspect of the method presented here, with the research question of translating DomESA’s good performance on datasets consisting of pairs of single words or short phrases, to pairs of longer biomedical texts. Both these lines of research converge, at a certain point, on the problem of ontology alignment, in which concepts can often be represented by far more complex structures than short labels.
Notes
We have evaluated the algorithm with the values of k between 1 and 15 and the method seems to work well within this range. In the evaluation presented here we only discuss results for k = 1 and k = 10 for illustrational purposes.
See https://seriousstats.wordpress.com/2012/02/05/comparing-correlations/ for discussion and code.
References
Agirre, E., & Rigau, G. (1996). Word sense disambiguation using conceptual density. In Proceedings of the 16th conference on computational linguistics-volume 1, association for computational linguistics (pp. 16–22).
Asooja, N.A.K., Bordea, G., & Buitelaar, P. (2015). Non-orthogonal explicit semantic analysis. Lexical and Computational Semantics (* SEM 2015).
Barzilay, R., & Elhadad, M. (1997). Using lexical chains for text summarization. In Proceedings of the ACL workshop on intelligent scalable text summarization: July 1997; Madrid, Spain, Association for Computational Linguistics (pp. 10–17).
Dumais, S.T. (2004). Latent semantic analysis. Annual Review of Information Science and Technology, 38(1), 188–230.
Gabrilovich, E., & Markovitch, S. (2007). Computing semantic relatedness using wikipedia-based explicit semantic analysis. In IJCAI, (Vol. 7 pp. 1606–1611).
Guo, X., Liu, R., Shriver, C.D., Hu, H., & Liebman, M.N. (2006). Assessing semantic similarity measures for the characterization of human regulatory pathways. Bioinformatics, 22(8), 967–973.
Haralambous, Y., & Klyuev, V. (2013). Thematically reinforced explicit semantic analysis. International Journal of Computational Linguistics and Applications, 4(1), 79.
Kusner, M.J., Sun, Y., Kolkin, N.I., & Weinberger, K.Q. (2015). From word embeddings to document distances. In Proceedings of the 32nd international conference on machine learning (ICML 2015) (pp. 957–966).
Liu, Y., McInnes, B.T., Pedersen, T., Melton-Meaux, G., & Pakhomov, S. (2012). Semantic relatedness study using second order co-occurrence vectors computed from biomedical corpora, umls and wordnet. In Proceedings of the 2nd ACM SIGHIT international health informatics symposium, ACM (pp. 363–372).
Martinez-Gil, J. (2016). Accurate semantic similarity measurement of biomedical nomenclature by means of fuzzy logic. International Journal of Uncertainty, Fuzziness and Knowledge-Based Systems, 24(02), 291–305.
Mathur, S., & Dinakarpandian, D. (2012). Finding disease similarity based on implicit semantic similarity. Journal of Biomedical Informatics, 45(2), 363–371.
Mikolov, T., Chen, K., Corrado, G.S., & Dean, J. (2013). Efficient estimation of word representations in vector space. In International Conference on Learning Representations. arXiv:1301.3781.
Muneeb, T., Sahu, S.K., & Anand, A. (2015). Evaluating distributed word representations for capturing semantics of biomedical concepts. In ACL-IJCNLP, (Vol. 2015 p. 158).
Pakhomov, S., McInnes, B., Adam, T., Liu, Y., Pedersen, T., & Melton, G.B. (2010). Semantic similarity and relatedness between clinical terms: an experimental study. In AMIA Annual symposium proceedings, american medical informatics association, (Vol. 2010 p. 572).
Pakhomov, S.V., Pedersen, T., McInnes, B., Melton, G.B., Ruggieri, A., & Chute, C.G. (2011). Towards a framework for developing semantic relatedness reference standards. Journal of Biomedical Informatics, 44(2), 251–265.
Pedersen, T., Pakhomov, S.V.S., Patwardhan, S., & Chute, C.G. (2007). Measures of semantic similarity and relatedness in the biomedical domain. Journal of Biomedical Informatics, 40(3), 288–299.
Pennington, J., Socher, R., & Manning, C.D. (2014). Glove: Global vectors for word representation. In EMNLP, (Vol. 14 pp. 1532–43).
Pesaranghader, A., Rezaei, A., & Pesaranghader, A. (2014). Adapting gloss vector semantic relatedness measure for semantic similarity estimation: an evaluation in the biomedical domain. In Semantic technology (pp. 129–145). New York: Springer.
Pesquita, C., Faria, D., Falcao, A.O., Lord, P., & Couto, F.M. (2009). Semantic similarity in biomedical ontologies. PLoS Computational Biology, 5(7), e1000,443.
Polajnar, T., Aggarwal, N., Asooja, K., & Buitelaar, P. (2013). Improving esa with document similarity. In Advances in information retrieval (pp. 582–593). New York: Springer.
Potthast, M., Stein, B., & Anderka, M. (2008). A wikipedia-based multilingual retrieval model. In European conference on information retrieval, (pp. 522–530). Springer.
Rada, R., Mili, H., Bicknell, E., & Blettner, M. (1989). Development and application of a metric on semantic nets. IEEE Transactions on Systems, Man, and Cybernetics, 19(1), 17–30.
Rybiński, M., & Aldana-Montes, J.F. (2016). TESA: a distributional measure for calculating semantic relatedness. BMC Journal of Biomedical Semantics – accepted for publication.
Rybiński, M., del Mar Roldán-Garcıa, M., Garcıa-Nieto, J., & Aldana-Montes, J.F. (2016). Dismatch results for OAEI. In OM. http://disi.unitn.it/~pavel/om2016/papers/oaei16_paper5.pdf.
Sahay, S., & Ram, A. (2011). Socio-semantic health information access. In AAAI spring symposium: AI and health communication, AAAI.
Sajadi, A., Milios, E.E., Kešelj, V., & Janssen, J.C. (2015). Domain-specific semantic relatedness from wikipedia structure: a case study in biomedical text. In International conference on intelligent text processing and computational linguistics, (pp. 347–360). Springer.
Sánchez, D., & Batet, M. (2011). Semantic similarity estimation in the biomedical domain: an ontology-based information-theoretic perspective. Journal of Biomedical Informatics, 44(5), 749–759.
Scholl, P., Böhnstedt, D, García, R.D., Rensing, C., & Steinmetz, R. (2010). Extended explicit semantic analysis for calculating semantic relatedness of web resources. In Sustaining TEL: from innovation to learning and practice (pp. 324–339). New York: Springer.
Strube, M., & Ponzetto, S.P. (2006). Wikirelate! computing semantic relatedness using wikipedia. In AAAI, (Vol. 6 pp. 1419–1424).
Virginia, G., & Nguyen, H.S. (2015). A semantic text retrieval for Indonesian using tolerance rough sets models. In Transactions on rough sets XIX, (pp. 138–224). Springer.
Zhang, R., Pakhomov, S., McInnes, B.T., & Melton, G.B. (2011). Evaluating measures of redundancy in clinical texts. In AMIA annual symposium proceedings, american medical informatics association, (Vol. 2011 p. 1612).
Zhang, Z., Gentile, A.L., & Ciravegna, F. (2012). Recent advances in methods of lexical semantic relatedness–a survey. Natural Language Engineering, 1(1), 1–69.
Zou, G.Y. (2007). Toward using confidence intervals to compare correlations. Psychological Methods, 12(4), 399.
Acknowledgments
We would like to thank the anonymous referees for their invaluable contributions towards improving the manuscript.
The work presented in this paper was partially supported by grants TIN2014-58304-R (Ministerio de Ciencia e Innovación), P11-TIC-7529 and P12-TIC-1519 (Plan Andaluz de Investigación, Desarrollo e Innovación).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rybiński, M., Aldana Montes, J.F. DomESA: a novel approach for extending domain-oriented lexical relatedness calculations with domain-specific semantics. J Intell Inf Syst 49, 315–331 (2017). https://doi.org/10.1007/s10844-017-0442-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10844-017-0442-y