Abstract
The Glucagon-like peptide 1 receptor (GLP-1R) is a well-established target for the treatment of type 2 diabetes and GLP-1R agonist-based therapies represent an effective approach which results in several GLP-1 analog drugs. However, the development of nonpeptidic agonist drugs targeting GLP-1R remains unsuccessful. A promising strategy aims to develop orally bioavailable, small-molecule positive allosteric modulators of GLP1-1R. Taking advantage of the recently reported cryo-EM structure of GLP-1R at its active state, we have performed structure-based screening studies which include potential allosteric binding site prediction and in silico screening of drug-like compounds, and conducted in vitro testing and site-specific mutagenesis studies. One compound with low molecular weight was confirmed as a positive allosteric modulator of GLP-1R as it enhances GLP-1′s affinity and efficacy to human GLP-1R in a dose dependent manner. This compound also stimulates insulin secretion synergistically with GLP-1. With the molecular weight of 399, this compound represents one of the smallest known GLP-1R PAMs, and demonstrates other favorable drug-like properties. Site-specific mutagenesis studies confirmed that the binding site of this compound partially overlaps with that of a known antagonist in the transmembrane domain. These results demonstrate that structure-based approach is useful for discovering nonpeptidic allosteric modulators of GLP-1R and the compound reported here is valuable for further drug development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Glucagon-like peptide 1 receptor (GLP-1R) is a member of secretin-like Class B family of G-protein coupled receptors (GPCRs) and plays an essential role in mediating the potentiation of insulin secretion and the suppression of glucagon secretion. Hence, positive modulation of GLP-1R remains an effective strategy for the therapeutic treatment of type 2 diabetes [1], and its incretin peptide GLP-1 and several peptide mimetics including exenatide and liragulitide are successful drugs [2], administered by injection. The therapeutic significance of GLP-1R was underscored by several recent publications of its structure in both inactive and active state [3,4,5,6]. On the other hand, the development of nonpeptidic small molecule agonists of GLP-1R with enhanced bioavailability remains elusive [7,8,9,10,11,12].
Like other Class B GPCRs, GLP-1R has an extracellular N-terminal domain and a seven transmembrane domain (7TM). In ligand binding, its N-terminal domain binds to C-terminal residues of the GLP-1 peptide hormone and its 7TM domain interacts with the N-terminal residues of the peptide for signaling. The extended nature of the orthosteric binding site for GLP-1 hindered the development of orally active small molecule agonists of GLP-1R for therapeutic purpose [4]. Another contributing factor is the lack of structural information on active state of the 7TM of GLP-1R until recently [3,4,5,6]. High-throughput screenings have typically been used to identify small molecule agonists with some in covalent modification e.g. Compound 2 and BETP [7, 9]. However, further development of these lead compounds has not been successful. Up to now, no small molecule drugs acting as GLP-1R agonists are available in the market. Therefore, novel approaches are very desirable in developing small molecule drugs targeting GLP-1R for the treatment of type 2 diabetes.
Given the allosteric nature of GPCRs and the discovery of allosteric sites on GLP-1R [5] and other Class B GPCRs [13], developing small-molecule positive allosteric modulators (PAMs) of GLP-1R for therapeutic intervention represents an attractive approach for drug discovery [14,15,16,17]. Early reported PAMs (Fig. 1) of GLP-1R such as Compound 2, BETP and quercetin have certain limitations including innate electrophilicity, weak potency, or stimulus bias [7,8,9,10,11]. Employing screening and medicinal chemistry, another lab has reported the discovery of a novel GLP-1R PAM (VU0453379) which showed greater therapeutic potential of GLP-1R potentiation and CNS penetration [12]. In our recent work [18], we have taken the rational molecular design approach by first constructing a three-dimensional (3D) structural model of the 7TM domain of GLP-1R at its active state, then applying the ligand-based and structure-based drug design techniques to screen the ZINC database [19] for identification of drug-like small molecule PAMs of GLP-1R. One compound from ZINC database was identified as the potential ago-allosteric modulator (M-4).
With the publication of the active-state structure of GLP-1R [4], we have decided to carry out another round of structure-based screening studies in an effort to identity small molecule PAMs as appropriate lead compounds for further drug development. Through in silico screening and in vitro experiment validation, a compound (C-1) was identified as a PAM of GLP-1R. The site-specific mutagenesis studies confirmed the binding site of C-1 on GLP-1R. The C-1 compound has the molecular weight of 399, which is one of the smallest PAMs of GLP-1R up to now. This compound also demonstrates other drug-like properties. These works further validate the usefulness of the rational design approach in GLP-1R drug discovery and the reported PAM has the potential for further drug development.
Method
The approach presented here includes several steps: (i) In silico structure-based ligand screening; (ii) Experimental validation of the allosteric effects of the top-ranked small molecule compound; and (iii) Site-specific mutagenesis studies to confirm the binding site.
In silico structure-based screening
The cryo-EM structure of rabbit GLP-1R in its active conformation (PDB ID: 5VAI) [4], which has 94% sequence identity with human GLP-1R, was imported in MOE (Molecular Computing Group Inc. version 2018.01), and all hetero atoms, water molecules, Gs protein and the N-terminal domain of the receptor were deleted. Energy minimization was subsequently carried out on the remaining structure using the default settings in MOE. The potential allosteric sites in the 7TM of the GLP-1R was predicted using the SiteFinder module with the default setting in MOE. All the predicted binding sites were manually inspected and the one different from the orthosteric site and with the largest binding volume except for the orthosteric site was chosen as the potential allosteric site in the screening practices below.
For in silico screening, the same cryo-EM structure of rabbit GLP-1R (PDB ID: 5VAI) was imported in the Schrodinger Suite (version 2017). The receptor was cropped to retain only its 7TM and was prepared using Protein Preparation Wizard with default settings. The protein grid was prepared using the Receptor Grid Generation Panel with the default settings and the rotation of hydroxyl group was not allowed. The grid center was kept at (124.2, 132.0, 122.0) with the dimensions of inner grid box as 10 Å × 10 Å × 10 Å and of outer grid box as 30 Å × 30 Å × 30 Å. The prepared GLP-1R grid was then used for ligand docking. The same library of 5,689 drug-like molecules identified in our previous work [18] that have similar ligand properties with the 23 active compounds of GLP-1R were prepared using LigPrep module. The prepared ligands were then docked to the receptor grid using Glide SP mode [20] with default settings. Sixteen different compounds with the highest docking scores were re-docked to the same receptor grid using Glide XP mode and then Induced Fit Docking (IFD) mode. A top-ranked molecule with the smallest molecular weight and the small logP value was chosen for in vitro test.
In vitro testing of potential GLP-1R ago-PAMs
Materials
HEK293 cell stably expressing CRE/CREB luciferase reporter gene (BPS Bioscience #60,515), RPMI medium (Corning #10–040), Krebs Ringer Bicarbonate buffer (Amsbio #KRB-1000), L-Glutamine (Gibco #25,030–081), HEPES (Gibco #15,630–080), Sodium Pyruvate (Gibco #11,360–070), β-mercaptoethanol (MP #806,444), D-Glucose (#G-7528), Fetal Bovine Serum (Fisher #03,600,511), penicillin/streptomycin (Corning #30–002-Cl), Hygromycin B (Alfa Aesar #J60681), Lipofectamine 2000 (Invitrogen #11,668,027), GLP-1R peptide agonist (Sigma #9416), 6 well cell culture plates (Ultra Cruz #sc-204443), 96 well cell culture plates (Sigma #CLS9102), Luciferase cell culture lysis reagent (Promega #E1531), Luciferase assay reagent (Promega #E1501), and Ultra-Sensitive Rat Insulin Kit (Crystal Chem #90,060) were purchased from vendors. Flag-tagged Human GLP-1R and Flag-tagged pCMV-N-Flag negative control vector were purchased from vendors (Sino Biological Inc. #HG13944-NF and #CV061) and INS-1 cells were provided by Dr. Xianxin Hua at Perelman School of Medicine, Pfu DNA polymerase kit (Thermofisher #EP0501) and primers (University of Pennsylvania) were purchased from vendors, plasmid of mutant human GLP-1R S352A and V332W was kindly provided by iHuman Institute, ShanghaiTech University.
Transfection and cell culture
HEK293 cells stably expressing CRE/CREB Reporter (luciferase) were cultured in RPMI medium supplemented with 8% (v/v) fetal bovine serum, 2% (v/v) penicillin/streptomycin, and 100 µg/ml of Hygromycin B. Cells were maintained in an incubator at 37 °C with 5% CO2. Cells were seeded into 6-well cell culture plates one day before transfection. After overnight incubation, one well of cell was transiently transfected with 3.4 µg of human GLP-1R or empty vector, respectively, using lipofectamine 2000. After 4 h of transfection, transfection medium was replaced by RPMI medium supplemented with 5% (v/v) fetal bovine serum and 2% (v/v) penicillin/streptomycin. After 24 h of incubation, cells were trypsinized and seeded into 96 well cell culture plates (5.5 × 104 cells per well) and maintained at 37 °C in 5% CO2 incubator for 24 h. The cells were starved using RPMI with 0.5% sera. After 24 h of starvation, the transfected cells were treated with a compound as indicated.
Luciferase assay
The compound dissolved in 100% DMSO was diluted to indicated concentration in RPMI 1640 (0.5% DMSO included for all cell culture). After 4 h of treatment, cells were harvested by cold luciferase cell culture lysis buffer and kept on shaker for 10 min at 4 °C. Luciferase activity was measured using luciferin substrate and luminescence was read by Wallac 1420 multiplate reader. Luciferase activity of HEK293 reporting cells cultured using 0.5% DMSO and full RPMI medium was used as a vehicle control. Protein concentration of each well was determined by Bradford assay.
Site specific mutagenesis of human GLP-1R
Based on the docking pose of the lead compound, the desired mutation of N406 to A was introduced into N-Flag tag-labeled human GLP-1R using site directed mutagenesis by traditional PCR method using forward primer (5′-TTATACTGCTTTGTCgccAATGAGGTCCAGCTG-3′) and reverse primer (5′-CAGCTGGACCTCATTggcGACAAAGCAGTATAA-3′). The PCR product was then treated with Dpn1 enzyme. The introduction of desired mutation in human GLP-1R plasmid was confirmed by DNA sequencing. The mutants S352A and V332W were kindly provided by ShanghaiTech University. These constructs were used to express mutant GLP-1R in HEK293-CREB luciferase cells.
Glucose stimulated insulin production in INS-1 cells
INS-1 cells were cultured in RPMI 1640 supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate, 10% FBS, 10 mM HEPES, 100 units/ml penicillin, 100 μg/ml streptomycin, and 50 μM β-mercaptoethanol. Cells were maintained in an incubator at 37 °C with 5% CO2. To determine the effect of the GLP-1R agonist compound on insulin production, INS-1 cells were seeded onto 24 well plates. After 48 h of incubation, cells were washed twice with 200 µl of Krebs–Ringer Bicarbonate (KRB) buffer and starved for 2 h in fresh KRB supplemented with 0.1% serum. After 2 h of starvation, the buffer was replaced with 200 µl of KRB containing 0.1% serum, 16.7 mM glucose and 9.7 µM of the compound, or 181 nM of GLP-1 with 0.125% DMSO or 0.125% DMSO alone (vehicle control) and incubated at 37 °C with 5% CO2. After 20 min, the supernatant was collected, centrifuged at 1000 rpm for 5 min at 4 °C, and aliquoted and stored at − 20 °C. These samples were used to determine insulin concentration using insulin detection kit ELISA following the manual.
Data analysis
The concentration-dependent dose response curve was generated using Graph Pad Prism 6.0 for Mac (GraphPad Software Inc., San Diego, CA). The curves were fitted based on sigmoidal dose response with the bottom parameter being kept 0. The EC50 value was calculated from Prism. The statistical difference between different groups was analyzed by 2-way ANOVA module in Prism.
Results
Small-molecule PAMs of GLP-1R identified through in silico screening
The cryo-EM structure of GLP-1R (PDB ID: 5VAI) [4] which showed the active state of the 7TM was adopted for in silico screening. Through SiteFinder analysis in MOE, a number of potential ligand binding sites on this structure were predicted (Supplementary Table S1). Among them, the largest predicted site is the orthosteric GLP-1 binding site; And the second largest site, which is located in the 7TM and far from the orthosteric site, partially overlaps with known allosteric sites in GLP-1R [5] (Supplementary Figure S1). This site was thus regarded as a potential allosteric site and chosen for in silico screening.
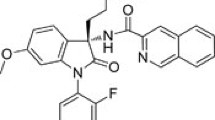
In our previous work [18], a total of 5,689 compounds that have similar ligand properties (molecular weight, xlogP, hydrogen donors, hydrogen acceptor and polar surfac area) to that of 23 known GLP-1R agonists was identified through compound similarity search. Using the Glide SP docking mode from the Schrodinger Suite first, those 5,689 compounds were docked into the predicted allosteric site on the 7TM of GLP-1R. Glide in the Schrodinger Suite is one of the best docking tools to identify potential protein effectors [21]. Based on their docking scores, the top 16 ranked compounds were chosen for re-docking using the Glide XP mode and then the IFD mode. Top ranked poses were visually inspected and as expected they all bound to the proposed allosteric site in GLP-1R (Fig. 2a). Among the top 10 ranked compounds (Table 1), compound C-1 (Fig. 2b) has relatively small molecular weight and the smallest xlogP (octanol/water) [22]. It was purchased and experiemntally tested for its potential activity against GLP-1R.
In vitro activity of the selected top-ranked compound
In vitro activity of the compound C-1 was first studied using human GLP-1R dependent luciferase reporter system. In this screening system, the activation of the human GLP-1R was measured as the amount of luminescence in response to cyclic adenosine monophosphate (cAMP), which in turn was normalized to the amount of protein. Negative control was included in all experiments to evaluate non-specific effect of the compound (if any). From in vitro studies, C-1 was found to activate human GLP-1R and its EC50 value was determined as 21 μM (Fig. 3).
In vitro agonistic activity of compound C-1 in HEK293 cells co-expressing human GLP-1R and a 3x-cAMP response element-luciferase reporter. Dose–response curves of C-1 in the presence and absence of human GLP-1R, respectively (EC50 = 21 μM). HEK293-CREBluciferase cell line transiently expressing human GLP-1R or empty vector was treated with different concentrations of C-1. GLP-1R activation was measured as the amount of luminescence produced, which was normalized by respective protein concentrations. The non-specific effect of C-1 was measured as the amount of luminescence produced in HEK293 cells expressing empty vector without GLP-1R, which was normalized by protein concentration. In all experiments, normalized luminescence was plotted with respect to vehicle control (0.5% DMSO). The dose response curves were generated using sigmoidal dose response parameter from GraphPad Prism 6.0. Data is average of three independent experiments with at least three technical replicates for each treatment conditions and error bars for each concentration were plotted as SEM (n = 3)
Compound C-1 improves GLP-1′s affinity and efficacy to human GLP-1R
Low level [23] and decreased response to GLP-1 have been observed in some Type 2 patients [24,25,26,27]. Therefore, it will be of interest to determine whether compound C-1 can act as a PAM of GLP-1R and enhance the affinity and efficacy of endogenous GLP-1. The activation of GLP-1R by different concentrations of GLP-1 (0.014-145 nM) in combination with C-1 (19.3 µM) was studied by luciferase activity responding to cAMP production using HEK293-CREB cells transiently expressing human GLP-1R. The GLP-1R activity stimulated by GLP-1 in combination with C-1 (19.3 µM) was significantly increased than using GLP-1 alone and the allosteric effect was found to be dose dependent (Fig. 4). Overall, the EC30 of GLP-1 was decreased from 1.5 × 10−10 M for GLP-1 alone to 0.8 × 10−10 M for GLP-1 in combination with C-1 (19.3 µM); And the efficacy of GLP-1 was increased from 74.4 ± 5.3 fold for GLP-1 alone to 92.0 ± 3.5 fold for GLP-1 in the presence of C-1(19.3 µM). These dose response analyses suggested that C-1 clearly acted as a PAM of human GLP-1R.
Potential allosteric effect of C-1 on GLP-1R. The effect of GLP-1 on HEK293 cells expressing human GLP-1R or empty vector in the presence or absence of C-1 (19.3 µM). This concentration was purposely chosen so that GLP-1R was not activated by C-1 alone but by its combination with GLP-1. GLP-1R activation was assessed as luminescence normalized to protein concentration and plotted as luminescence fold change with respect to vehicle control (0.5% DMSO). Data is average of three independent experiments with at least three technical replicates for each conditions and error bars for each concentration were plotted as SEM (n = 3). Statistical analysis was done using 2-way ANOVA (****p < 0.0001; **p < 0.001)
Compound C-1 stimulates insulin secretion
Developing small molecule PAMs of GLP-1R that will stimulate insulin production in pancreatic β cells is the goal of this work. The insulin production activity of C-1 was assessed by in vitro insulin secretion assay in INS-1 cells. The results indicated that like GLP-1, C-1 can stimulate insulin secretion in the presence of 16.7 mM of glucose and the insulin production by GLP-1 and C-1 was more than two-fold compared to vehicle control at both time points (Fig. 5). In addition, no significant difference was observed between the amount of insulin produced by GLP-1 and C-1. However, the amount of insulin produced by GLP-1 in combination with C-1 was more than that produced by GLP-1 and C-1 respectively. These data indicated that C-1 can induce glucose-dependent insulin production in GLP-1R expressed cells as well as improve GLP-1′s efficacy through synergistic effect.
Glucose stimulated insulin production induced by GLP-1 and C-1 in INS-1 cells. INS-1 cells were treated with GLP-1 (0.18 µM) and C-1 (9.7 µM) in the presence of 16.7 mM glucose after 2 h of starvation with KRB buffer. Data is average of three independent experiments and error bars for each concentration were plotted as SEM (n = 3)
Site specific mutagenesis studies confirm the proposed binding site of C-1 in GLP-1R
Inspection of the different binding poses of C-1 in the proposed binding site generated from Glide XP and IFD docking indicated that C-1 forms hydrogen bonds with side chains of residues N406 and S352 (Fig. 2a). Therefore, these two amino acids were chosen for site-specific mutagenesis studies to confirm the proposed binding site. N406A and S352A GLP-1R mutants were either generated or obtained respectively, and further confirmed by sequencing. In addition, V332W was chosen as the negative control for C-1 binding. Residue V332 is not present in the predicted binding site for C-1, but it is one of the residues that was suggested being involved in the binding of Compound 2 [5].
To study the potential change in the allosteric activity of C-1 on GLP-1R mutants N406A, S352A and V332W individually, the effect of GLP-1 (0.014–145 nM) in combination with C-1 (19.3 µM) was compared between WT and mutant GLP-1R transfected HEK-CREB luciferase cell line using luciferase assay. Treatment of the cells with C-1 (19.3 µM) in combination with GLP-1 significantly increased the WT GLP-1R activity, and the allosteric effect was dose dependent, as expected (Fig. 6). Notably, C-1 failed to activate the N406A and S352A mutant GLP-1R activity in the same conditions (Fig. 6). These results suggested that either N406A or S352A mutation abolished the allosteric effect of C-1 and as a result, the GLP-1′s affinity and efficacy was not impacted by the presence of C-1. This is likely due to the fact that both mutations have disrupted the hydrogen bond interactions between N406 or S352 and C-1, which affected the binding of C-1 in the proposed pocket and subsequently abolished its allosteric activity on GLP-1R. Consistently, V332W mutation had no effect on the allosteric activity of C-1, suggesting that C-1 does not bind to other sites other than the proposed one.
Site-specific mutagenesis studies on human GLP-1R. a The effect of GLP-1 in the presence (continuous line) and absence (dotted line) of C-1 (19.3 µM) on HEK293-CREB luciferase cells expressing WT GLP-1R (black) or mutant GLP-1R where N406 was mutated to A (blue). b The effect of GLP-1 in the presence (continuous line) and absence (dotted line) of C-1 (19.3 µM) on HEK293-CREB luciferase cells expressing WT GLP-1R (black) or mutant GLP-1R where S352 was mutated to A (green). c The effect of GLP-1 in the presence (continuous line) and absence (dotted line) of C-1 (19.3 µM) on HEK293-CREB luciferase cells expressing WT GLP-1R (black) or mutant GLP-1R where V332 is mutated to W (red). In all three curves, the effect of GLP-1 in the presence and absence of C-1 has been plotted as the luminescence fold change with respect to vehicle control (0.5% DMSO) and normalized to the respective protein concentrations. Data is average of three independent experiments with at least three technical replicates for each conditions and error bars for each concentration were plotted as SEM (n = 3). Statistical analysis was done using 2-way ANOVA (****p < 0.0001; **p < 0.001). The comparison is done between the data points of curves of luminescence caused due to GLP-1 alone and in combination with C-1 on WT GLP-1R or mutant GLP-1R
Discussion
Targeting the allosteric sites on GLP-1R represents a promising strategy for the development of small molecule drugs that could offer several potential benefits including reduced side effects [16, 28, 29]. However, this strategy has not resulted in the successful discovery of small molecule drugs in the market, which was in part due to the lack of 3D structure information for GLP-1R until very recently [3,4,5,6]. Past small molecule drug discovery efforts were often initiated by high-throughput screening [7,8,9]. In our recent work [18], we attempted to take the rational design approach by first constructing a 3D model of the TM domain of GLP-1R at its active state, then performing in silico structure-based screening. Through in vitro experiments, one compound M-4 was shown to function as a potential ago-allosteric modulator.
In the current work, we carried out the structure-based in silico screening studies using the cryo-EM structure of the GLP-1R at its active state [4]. One compound (C-1) was identified and then confirmed as a PAM of GLP-1R using a cAMP response element-based luciferase reporting system, which clearly showed that C-1 activated human GLP-1R in combination with GLP-1 in a dose-dependent way. The allosteric modulating effect of C-1 was further confirmed by insulin secretion experiments. The predicted binding site of C-1 on the TM domain of GLP-1R was subsequently confirmed through site-specific mutagenesis studies (Fig. 6).
The compound (C-1) we identified is structurally and chemically different from those reported in the literature [7,8,9,10,11,12]. Hence it represents a novel PAM of GLP-1R. Further, it has the small molecular weight (< 400), and other favorable drug-like properties e.g. 3.897 for XlogP. In addition, compound C-1 binds at an allosteric site on the TM domain of GLP-1R non-covalently, differently from Compound 2 and BETP. Finally, it was observed that C-1 also induced agonistic activity against GLP-1R independently, although the change is relatively small. Overall, compound C-1 emerges as a great lead compound for further basic drug discovery. Work is in progress to chemically modify C-1 in order to increase its potency.
It should be noted that compound C-1 induced little to none agonistic activities in the absence of GLP-1R expression (Figs. 3 and 4). Considering that HEK293 cells are known to express other functional class B GPCRs e.g. VIPR1 [30], and the fact that no chemical modification has been carried on C-1, this low level of non-specific effect is very intriguing. Nevertheless, undesired stimulation of other class B GPCRs could lead to serious side effects. Further studies are necessary to clearly define the non-specific effects of C-1 and to improve its specificity to GLP-1R if needed.
The discovery of small molecule PAMs of GLP-1R using the rational structure-based approach as demonstrated here further validates the feasibility of this approach in the small molecule drug discovery of other members of the pharmaceutical important Class B family of GPCRs. All of the endogenous ligands for the Class B GPCRs are moderately long peptide hormones which bind to their receptors in a similar manner as GLP-1. The nature of these binding sites makes it challenging for designing small molecule binding to these sites [4, 31]. With existence of the allosteric sites being confirmed for more and more Class B GPCRs, targeting these allosteric sites using structure-based drug design techniques represents a new venue for the development of small molecule drugs targeting these Class B GPCRs.
References
Koole C et al (2013) Recent advances in understanding GLP-1R (glucagon-like peptide-1 receptor) function. Biochem. Soc. Trans. 41:172–179
Prasad-Reddy L, Isaacs D (2015) A clinical review of GLP-1 receptor agonists: efficacy and safety in diabetes and beyond. Drugs Context 4:212283
Jazayeri A et al (2017) Crystal structure of the GLP-1 receptor bound to a peptide agonist. Nature 546:254–258
Zhang Y et al (2017) Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 546:248–253
Song G et al (2017) Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature 546:312–315
Liang YL et al (2018) Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature 555:121–125
Knudsen LB et al (2007) Small-molecule agonists for the glucagon-like peptide 1 receptor. Proc. Natl. Acad. Sci. USA 104:937–942
Chen D et al (2007) A nonpeptidic agonist of glucagon-like peptide 1 receptors with efficacy in diabetic db/db mice. Proc. Natl. Acad. Sci. USA 104:943–948
Sloop KW (2010) Novel small molecule glucagon-like peptide-1 receptor agonist stimulates insulin secretion in rodents and from human islets. Diabetes 59:3099–3107
Wootten D et al (2011) Modulation of the glucagon-like peptide-1 receptor signaling by naturally occurring and synthetic flavonoids. J. Pharmacol. Exp. Ther. 336:540–550
Willard FS, Bueno AB, Sloop KW (2012) Small molecule drug discovery at the glucagon-like peptide-1 receptor. Exp. Diabetes Res. 2012:709893
Morris LC et al (2014) Discovery of (S)-2-cyclopentyl-N-((1-isopropylpyrrolidin2-yl)-9-methyl-1-oxo-2,9-dihydro-1H-py rrido[3,4-b]indole-4-carboxamide (VU0453379): a novel, CNS penetrant glucagon-like peptide 1 receptor (GLP-1R) positive allosteric modulator (PAM). J. Med. Chem. 57:10192–10197
Hollenstein K et al (2013) Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature 499:438–443
May LT, Leach K, Sexton PM, Christopoulos A (2007) Allosteric modulation of G protein-coupled receptors. Annu. Rev. Pharmacol. Toxicol. 47:1–51
Conn PJ, Christopoulos A, Lindsley CW (2009) Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat. Rev. Drug Discov. 8:41–54
Khoury E, Clement S, Laporte SA (2014) Allosteric and biased g protein-coupled receptor signaling regulation: potentials for new therapeutics. Front. Endocrinol. 5:68
Bueno AB (2016) Positive allosteric modulation of the glucagon-like peptide-1 receptor by diverse electrophiles. J. Biol. Chem. 291:10700–10715
Redij T et al (2019) Structural modeling and in silico screening of potential small molecule allosteric agonists of glucagon-like peptide 1 receptor. ACS Omega 4:961–970
Irwin JJ, Shoichet BK (2005) ZINC—a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 45:177–182
Halgren TA et al. (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47(7), 1750–1759.
Repasky MP et al (2012) Docking performance of the glide program as evaluated on the Astex and DUD datasets: a complete set of glide SP results and selected results for a new scoring function integrating WaterMap and glide. J. Comput. Aided Mol. Des. 26:787–799
Wang R, Fu Y, Lai L (1997) A new atom-additive method for calculating partition coefficients. J. Chem. Inf. Comput. Sci. 37:615–621
Lastya A, Saraswati MR, Suastika K (2014) The low level of glucagon-like peptide-1 (glp-1) is a risk factor of type 2 diabetes mellitus. BMC Res. Notes 7(1):849
King AB, Armstrong DU, Chinnapongse S (2003) Comparison of glycemic and lipid response to pioglitazone treatment in Mexican-Americans and non-Hispanic Caucasians with type 2 diabetes. Diabetes Care 26:245–246
Toft-Nielsen MB, Madsbad S, Holst JJ (2001) Determinants of the effectiveness of glucagon-like peptide-1 in type 2 diabetes. J. Clin. Endocrinol. Metab. 86:3853–3860
Knop FK et al (2007) Reduced incretin effect in type 2 diabetes: cause or consequence of the diabetic state? Diabetes 56:1951–1959
Kjems LL, Holst JJ, Volund A, Madsbad S (2003) The influence of GLP-1 on glucose-stimulated insulin secretion: effects on beta-cell sensitivity in type 2 and nondiabetic subjects. Diabetes 52:380–386
Wootten D et al (2017) Allostery and biased agonism at class B G protein-coupled receptors. Chem. Rev. 117:111–138
Pupo AS et al (2016) Recent updates on GPCR biased agonism. Pharmacol. Res. 112:49–57
Atwood BK et al (2011) Expression of G protein-coupled receptors and related proteins in HEK293, AtT20, BV2, and N18 cell lines as revealed by microarray analysis. BMC Genomics 12:14
de Graaf C et al (2011) Structure-based discovery of allosteric modulators of two related class B G-protein-coupled receptors. ChemMedChem 6:2159–2169
Acknowledgements
We thank Dr. Raymond C. Stevens at ShanghaiTech University for sharing with us plasmids of human GLP-1R mutants S352A, V332W and T355A. The first two mutants were used in this work. We thank Dr. James A. McKee for helping prepare Fig. 1 and the figure of the chemical structure of compound C-1. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR001878. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work was supported in part by the Institute for Translational Medicine and Therapeutics' (ITMAT) Transdisciplinary Program in Translational Medicine and Therapeutics at University of Pennsylvania.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Redij, T., Ma, J., Li, Z. et al. Discovery of a potential positive allosteric modulator of glucagon-like peptide 1 receptor through virtual screening and experimental study. J Comput Aided Mol Des 33, 973–981 (2019). https://doi.org/10.1007/s10822-019-00254-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00254-4