Abstract
Microtubules (made up of α and β-tubulin subunits) play an essential role in the process of mitosis and cell proliferation, thus making them an ideal target for anticancer drugs discovery. Microtubule-targeted drugs, including taxanes and vinca alkaloids, can suppress microtubule dynamics, cause mitotic block and apoptosis, which have been widely used in the treatment of various cancers. There are three unique binding sites (taxanes, vinca alkaloids, and colchicine) in tubulin can be targeted to develop tubulin inhibitors. In this study, we selected the colchicine binding site in tubulin as our target. By performing molecular docking-based virtual screening combined with in vitro tubulin polymerization inhibition assay, we identified two novel and potent tubulin inhibitors (9 and 19). These two compounds arrested cell cycle progression at the G2/M phase and induced apoptosis at sub μM concentrations. In addition, they displayed potent antiproliferative activity with IC50 values in the nM range. Finally, the probable binding modes of 9 and 19 were probed by molecular docking. These two compounds with novel scaffold will shed new light on the lead molecules discovery and the design of new microtubule-targeting agents (MTAs).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Uncontrolled cell division and fast proliferation are fundamental hallmarks of cancer cells. In mammalian, cancer cells divide into two daughter cells were typically by means of mitosis, which is closely related to resembling and disassembling of microtubules [1, 2]. Microtubules, which are key components of the cytoskeleton, play crucial roles in various cellular processes, including the development and maintenance of cell shape, cell signaling, cell division and mitosis [3, 4]. A lot of efforts have led to the identification of several compounds that specifically block the key factors of mitosis [5]. The mechanism of these inhibitors can be largely divided into two categories, the selective inhibitors of mitotic kinases and kinesins, as well as the inhibitors targeting microtubules. Comparing with the inhibitors of kinases, compounds directly targeting microtubules, such as paclitaxel, always showed more potent selectivity and efficiency [6].
Paclitaxel and the vinca alkaloids are the two most successful microtubule-targeted chemotherapeutic drugs, and their most potent actions are mainly due to the suppression of microtubule dynamics, rather than increasing or decreasing microtubule-polymer mass [6]. MTAs can be grouped into two major types (microtubule stabilizers and microtubule destabilizers) based on their mechanism of action (MOA), for example, taxanes belonged to microtubule stabilizers while vinca alkaloids and colchicine belonged to microtubule destabilizers. Both types of MTAs inhibit the necessary dynamics required for microtubule function [7].
There are three main classes of sites on tubulin for microtubule-active drugs: the paclitaxel site, the vinca site, and the colchicine site. Although there are no FDA approved tubulin inhibitors drugs targeting the colchicine binding site currently, this binding site is more amenable to molecules with more favorable physiochemical properties that improve oral bioavailability over taxanes and vinca alkaloids binding sites, for example, have less drug–drug interaction inclination, and are less prone to develop multi-drug resistance [8]. Thus, discovery and design of colchicine site tubulin inhibitors have received extraordinary attention in anticancer drug development.
Virtual screening has been widely and successfully used in identifying modulators of critical targets [9,10,11,12,13,14,15,16,17]. In this study, colchicine binding site in tubulin was selected as the target, and we performed molecular docking-based virtual screening in order to find candidates with novel scaffolds and potent binding affinity of microtubules. After in vitro tubulin polymerization inhibition assay, two novel and potent tubulin inhibitors (9 and 19) were identified. As expected, these two compounds displayed potent anti-proliferative activity on tumor cells (CEM, human lymphocytic leukemia cells; HeLa, human cervical carcinoma cells) with IC50 values in the nM range, arrested cell cycle progression at the G2/M phase and induced apoptosis at sub μM concentrations. Finally, the probable binding modes of 9 and 19 were probed by molecular docking simulations.
Materials and methods
Molecular docking-based virtual screening
Molecular docking-based virtual screening was performed using the Autodock 4.2 program [18, 19]. The crystal structure of the tubulin-colchicine complex (PDB ID: 4O2B [20]) was used to construct the docking model. Prior to docking studies, one α and one β tubulin of microtubules as well as crystallographic waters that have interactions with colchicine were retained. The missing hydrogen atoms were added, Gasteiger charges were assigned, and the protein was parameterized with AD4 type by Autodock Tools 1.5.6. Finally, the resulting enzyme structure was used as an input for the Autogrid program. Autogrid performed a pre-calculated atomic affinity grid maps for each atom type in the ligand plus an electrostatics map and a separate de-solvation map presented in the substrate molecule. Grid map with 60 × 60 × 60 points was made according to the conformation of co-crystal ligand, and the grid spacing was set to 0.375 Å. All the SPECS database compounds were prepared using the program LigPrep from the Schrödinger (Schrödinger Release 2015-2: LigPrep, Schrödinger, LLC, New York, NY, 2015) to generate stereoisomers and different protonation states conformers. The OPLS3 force field was used, and the possible ionization states were predicted at pH (7.0 ± 2.0) using Epik. Docking calculations were carried out using the Lamarckian genetic algorithm (LGA). Colchicine was first redocked to tubulin to validate the docking methods, then the prepared SPECS database compounds were docked to tubulin for virtual screening. Pipeline Pilot 8.0 (Pipeline Pilot; Accelrys Software Inc., San Diego, CA) software was used to finish the cluster analysis. The clustering algorithm is a partitioning method in which the original data set is partitioned into ever-smaller regions that define the clusters, and a number of representative objects are chosen from the data set. The corresponding clusters are found by assigning each remaining object to each representative object, selecting the object that is the closest. The representative objects are called the cluster centers, while the other objects are the cluster members.
All the obtained compounds were purchased from commercial supplier (SPECS database). The purity of these compounds was > 95% as declared by the chemical vendor (Table S1).
Tubulin polymerization inhibition assay
Bovine brain tubulin (3 mg/mL, purity > 99%) was purchased from Cytoskeleton, Inc., USA. Tubulin protein was incubated with DMSO (control) or the tested compounds or colchicine (positive control) in general tubulin buffer (100 mM PIPES, 1.0 mM MgCl2, 1 mM EGTA, 1 mM GTP and 5% glycerol) at 0 °C. The reaction was started by warming the samples to 37 °C. Then the absorbance of wavelength at 340 nm (indicated the mass of polymer formation) was detected every 1 min for 20 min by Spectra Max 190 spectrophotometer (Molecular Device).
Tumor cell growth inhibition assay
Cytotoxic effects were examined in the CEM human lymphocytic leukemia cells and HeLa human cervical carcinoma cells by MTT assay. CEM and Hela cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA) and cultured in RPMI 1640 medium supplemented with 10% FBS and 1 × Pen/Strep. To determine the cytotoxic effects of compound 9 and 19, the cells (2 × 105 cells per well) were placed in 100 μL of serum containing media with compounds in 96-well flat-bottomed microtiter plates in triplicate cultures. After three days incubation, the absorbance at 570 nm was recorded on a fully automatic enzyme-labeled meter (Labsystems Inc. Wellscan MK-2, Finland). Compounds were tested in triplicate in at least three independent assays, and the average median inhibitory concentration (IC50) values were reported.
Cell cycle and apoptosis analysis
CEM cells were seeded overnight at 2 × 105 cells/well. Next, DMSO or compounds were added for 24 h. For cell cycle analysis, cells were harvested and washed three times with PBS and at last fixed with 75% ethanol at − 20 °C overnight. The cells were then stained with propidium iodide dye (BD Biosciences) for 20 min in dark conditions at room temperature, followed by subjected to flow cytometry (Cytomics FC 500MPL, Beckman Coulter) detection. For cell apoptosis analysis, harvested cells were measured using AnnexinV-FITC Apoptosis Detection Kit (Vazyme Biotech) according to the manufacturer’s protocol. These results were analyzed by FlowJo V7.6.1.
Results and discussion
Virtual screening and tubulin polymerization inhibition assay identified 2 tubulin inhibitors
Molecular docking-based virtual screening was used to screen small-molecule tubulin inhibitor candidates from SPECS database (http://www.specs.net/) which contains 212,449 compounds. In order to validate the molecular modeling methodology, the inhibitor colchicine that was co-crystalized with tubulin (PDB code: 4O2B) was first redocked to tubulin protein. As shown in Figure S1, the docking result could well reproduce the crystal structure, which indicated that the method was suitable for docking study of tubulin. The workflow of virtual screening was shown in Fig. 1a. After docking, the top-246 compounds ranked based on the docking score were retained for visual inspection to remove the compounds with unfavored geometry, unfavored chemical fitness with the binding sites or binding outside the defined pocket. In addition, compounds with similar chemical scaffolds were clustered by chemical fingerprints, and characteristic compounds were selected in each cluster according to the following considerations: (1) At least one molecule was retained in each cluster to guarantee the diversity of chemical space. (2) Molecules with simple structure and small molecular weight were preferred. (3) Molecules that could form hydrogen bonds with V181of tubulin and the water around colchicine were preferred. Finally, 36 compounds were selected for experimental validation. Among them, compounds 9 and 19 displayed potent tubulin polymerization inhibition activity with IC50 values of 1.68 μM and 2.33 μM, respectively (Fig. 1b). As a reference compound, combretastatin A-4 (CA-4) was included.
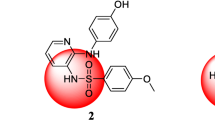
The workflow of tubulin inhibitor discovery and the inhibitory activity of the two hit compounds against tubulin polymerization. a The workflow of tubulin inhibitor discovery. Numerals denote the number of molecules in each stage. b Chemical structures and IC50 curves of compound 9, 19, CA-4 and Colchicine (positive control). Data shown are mean ± SD of three replicates
Growth-inhibitory activity of compounds 9 and 19 in three cancer cell lines
The anti-proliferative activity of the identified two compounds 9 and 19 were evaluated for their growth-inhibitory activity in three human cancer cell lines [lymphoblastic leukemia (CEM) cells, cervical carcinoma (HeLa) cells and colon cancer (HT-29) cells]. As can be seen from Fig. 2, both of this two compounds displayed potent anti-proliferative effects on all the three human cancer cells (CEM, Hela and HT-29), with IC50 values in the nM range.
Inhibition of cell cycle progression and inducement of cell apoptosis in HT-29 human colon cancer cell line
Since MTAs usually arrest cancer cells in G2/M phase and induce cancer cells apoptosis [21]. Meanwhile, given the fact that compound 9 was found to have growth inhibitory effect on human cancer cells, we continued to investigate its actual cellular mechanism of action in human cells. Since MTAs usually induce G2/M cell cycle arrest, the effect of 9 on the cell cycle was studied in HT-29 colon cancer cells for 24 h by flow cytometry. As shown in Fig. 3, an accumulation of cells in G2/M phase was observed after 24 h treatment with compound 9 at the concentrations of 5 nM and 10 nM, respectively. This data indicated an antimitotic mode of action of compound 9.
The effect of compound 9 on cell apoptosis was also examined. Results showed a concentration-dependent increased apoptosis (Fig. 4) upon treatment with compound 9 for 48 h on HT-29 cells. These data together with cell cycle arrest results supported the idea that compound 9 were not only cell-cycle arrestors but also inducers of apoptosis.
Molecular modelling
To gain detailed insights into the binding of this two compounds with tubulin, molecular docking simulation was performed. As shown in Fig. 5a, b, one hydrogen bond that the colchicine carbonyl oxygen makes with the backbone NH of Val181 were established. In addition, there are three water mediated hydrogen bonds that are involved in the interactions of colchicine with tubulin. Although the hydrogen bond established between colchicine and V181 residue does not exist in the interactions of 9 and 19 with tubulin, the water mediated hydrogen bond were maintained. And there are both two water-mediated hydrogen bonds (rendered as red dash lines) established between 9 and tubulin as well as 19 and tubulin, respectively. Besides, hydrophobic interaction was also the main interactions that contributed to the binding affinity between this two compounds and tubulin by further analysis (Fig. S2). In particular, both compounds 9 and 19 were located in the hydrophobic pocket which is composed of residues A180, C241, L242, L248, A250, L255, N258, M259, I318 and K352, except residue A180, the other residues belong to β tubulin. By comparing the binding modes of the two compounds with that of colchicine, we concluded that this two compounds could efficiently occupy the colchicine binding site of tubulin and showed similar binding modes with that of colchicine.
The putative binding modes of 9 and 19 with tubulin. a The binding mode 9. b The binding mode 19. Tubulin is shown as light pink cartoon diagram, and 9, 19 and colchicine are represented as cyan, green and magenta sticks, respectively. The interactive residues are labeled with carbon atoms colored in white. The water mediated hydrogen bonds between colchicine and tubulin are shown as yellow dash lines, while that between 9 and 19 and tubulin are shown as red dash lines. Colchicine binding mode was obtained by the original crystal structure (PDB code: 4O2B)
Conclusion
Owing to the key role in mitosis and other cellular processes, microtubules are regarded as an excellent target for anticancer drug development and have drawn much attention in recent years. Tubulin inhibitors that bind to the colchicine binding site have many advantages over taxanes and vinca alkaloids site binding inhibitors, such as the capacity to overcome multidrug resistance (MDR), favorable pharmacokinetics and well water solubility as well as the high selectivity in killing cancer cells over normal cells. However, up to date, there are no colchicine site binding inhibitors approved by FDA to treat cancer. Thus, it is an urgent need to discovery more colchicine site binding tubulin inhibitors. In this study, by performing molecular docking-base virtual screening, we identified two novel colchicine binding site tubulin inhibitors. Then their anti-proliferative effects on three different human cancer cells were confirmed, and the anti-proliferation mechanism was validated to the cell cycle arrest and cell apoptosis induce. Finally, the binding modes of this two compounds with tubulin were well studied by molecular docking simulation.
References
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100(1):57–70
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674. https://doi.org/10.1016/j.cell.2011.02.013
Jordan A, Hadfield JA, Lawrence NJ, McGown AT (1998) Tubulin as a target for anticancer drugs: agents which interact with the mitotic spindle. Med Res Rev 18(4):259–296
Desai A, Mitchison TJ (1997) Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13:83–117. https://doi.org/10.1146/annurev.cellbio.13.1.83
Olziersky AM, Labidi-Galy SI (2017) Clinical development of anti-mitotic drugs in cancer. Adv Exp Med Biol 1002:125–152. https://doi.org/10.1007/978-3-319-57127-0_6
Jordan MA, Wilson L (2004) Microtubules as a target for anticancer drugs. Nat Rev Cancer 4(4):253–265. https://doi.org/10.1038/nrc1317
Negi AS, Gautam Y, Alam S, Chanda D, Luqman S, Sarkar J, Khan F, Konwar R (2015) Natural antitubulin agents: importance of 3,4,5-trimethoxyphenyl fragment. Bioorg Med Chem 23(3):373–389. https://doi.org/10.1016/j.bmc.2014.12.027
Li L, Jiang S, Li X, Liu Y, Su J, Chen J (2018) Recent advances in trimethoxyphenyl (TMP) based tubulin inhibitors targeting the colchicine binding site. Eur J Med Chem 151:482–494. https://doi.org/10.1016/j.ejmech.2018.04.011
Manglik A, Lin H, Aryal DK, McCorvy JD, Dengler D, Corder G, Levit A, Kling RC, Bernat V, Hubner H, Huang XP, Sassano MF, Giguere PM, Lober S, Da D, Scherrer G, Kobilka BK, Gmeiner P, Roth BL, Shoichet BK (2016) Structure-based discovery of opioid analgesics with reduced side effects. Nature 537(7619):185–190. https://doi.org/10.1038/nature19112
Mao R, Shao J, Zhu K, Zhang Y, Ding H, Zhang C, Shi Z, Jiang H, Sun D, Duan W, Luo C (2017) Potent, selective, and cell active protein arginine methyltransferase 5 (PRMT5) inhibitor developed by structure-based virtual screening and hit optimization. J Med Chem 60(14):6289–6304. https://doi.org/10.1021/acs.jmedchem.7b00587
Ye Y, Zhang B, Mao R, Zhang C, Wang Y, Xing J, Liu YC, Luo X, Ding H, Yang Y, Zhou B, Jiang H, Chen K, Luo C, Zheng M (2017) Discovery and optimization of selective inhibitors of protein arginine methyltransferase 5 by docking-based virtual screening. Org Biomol Chem 15(17):3648–3661. https://doi.org/10.1039/c7ob00070g
Meng F, Cheng S, Ding H, Liu S, Liu Y, Zhu K, Chen S, Lu J, Xie Y, Li L, Liu R, Shi Z, Zhou Y, Liu YC, Zheng M, Jiang H, Lu W, Liu H, Luo C (2015) Discovery and optimization of novel, selective histone methyltransferase SET7 inhibitors by pharmacophore- and docking-based virtual screening. J Med Chem 58(20):8166–8181. https://doi.org/10.1021/acs.jmedchem.5b01154
Zhang J, Liu H, Zhu K, Gong S, Dramsi S, Wang YT, Li J, Chen F, Zhang R, Zhou L, Lan L, Jiang H, Schneewind O, Luo C, Yang CG (2014) Antiinfective therapy with a small molecule inhibitor of Staphylococcus aureus sortase. Proc Natl Acad Sci USA 111(37):13517–13522. https://doi.org/10.1073/pnas.1408601111
Kang JS, Zhang AL, Faheem M, Zhang CJ, Ai N, Buynak JD, Welsh WJ, Oelschlaeger P (2018) Virtual screening and experimental testing of B1 metallo-beta-lactamase Inhibitors. J Chem Inf Model. https://doi.org/10.1021/acs.jcim.8b00133
Mok SWF, Zeng W, Niu Y, Coghi P, Wu Y, Sin WM, Ng SI, Gordillo-Martinez F, Gao JY, Law BYK, Liu L, Yao X, Wong VKW (2018) A method for rapid screening of anilide-containing AMPK modulators based on computational docking and biological validation. Front Pharmacol 9:710. https://doi.org/10.3389/fphar.2018.00710
Zhu K, Jiang C, Tao H, Liu J, Zhang H, Luo C (2018) Identification of a novel selective small-molecule inhibitor of protein arginine methyltransferase 5 (PRMT5) by virtual screening, resynthesis and biological evaluations. Bioorg Med Chem Lett 28(9):1476–1483. https://doi.org/10.1016/j.bmcl.2018.03.087
Wang J, Luo C, Shan C, You Q, Lu J, Elf S, Zhou Y, Wen Y, Vinkenborg JL, Fan J, Kang H, Lin R, Han D, Xie Y, Karpus J, Chen S, Ouyang S, Luan C, Zhang N, Ding H, Merkx M, Liu H, Chen J, Jiang H, He C (2015) Inhibition of human copper trafficking by a small molecule significantly attenuates cancer cell proliferation. Nat Chem 7(12):968–979. https://doi.org/10.1038/nchem.2381
Huey R, Morris GM, Olson AJ, Goodsell DS (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28(6):1145–1152. https://doi.org/10.1002/jcc.20634
Olson GMMDSGRSHRHWEHRKBAJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19(14):24
Prota AE, Danel F, Bachmann F, Bargsten K, Buey RM, Pohlmann J, Reinelt S, Lane H, Steinmetz MO (2014) The novel microtubule-destabilizing drug BAL27862 binds to the colchicine site of tubulin with distinct effects on microtubule organization. J Mol Biol 426(8):1848–1860. https://doi.org/10.1016/j.jmb.2014.02.005
Zhai Y, Kronebusch PJ, Simon PM, Borisy GG (1996) Microtubule dynamics at the G2/M transition: abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J Cell Biol 135(1):201–214
Acknowledgements
This research was supported by a grant from the Chia Tai Tianqing Pharmaceutical Group Co., Ltd. (YWJKJJHKYJJ-Q1115).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, G., Jiao, Y., Huang, C. et al. Identification of novel and potent small-molecule inhibitors of tubulin with antitumor activities by virtual screening and biological evaluations. J Comput Aided Mol Des 33, 659–664 (2019). https://doi.org/10.1007/s10822-019-00206-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00206-y