Abstract
Purpose
To identify potential biomarkers and the molecular mechanisms associated with repeated implantation failure (RIF), three microarray datasets, GSE71331 (lncRNA + mRNA), GSE111974 (lncRNA + mRNA), and GSE71332 (miRNA), were retrieved from the Gene Expression Omnibus (GEO) database.
Methods
The differentially expressed mRNAs (DEMs), lncRNAs (DElncRNAs), and miRNAs (DEmiRNAs) between normal control samples (C group) and RIF samples (RIF group) were identified, and then a module partition analysis was performed based on weighted correlation network analysis (WGCNA). Following enrichment analysis of the genes, the lncRNA-miRNA-mRNA interactions (ceRNA) were examined. The mRNAs in the ceRNA network were evaluated using the GSE58144 dataset. Finally, the key RNAs were verified using reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Results
Fifty-three DEmiRNAs, 327 DEMs, and 13 DElncRNAs were identified between the C and RIF groups. According to WGCNA, the magenta module was positively correlated with RIF disease status. The lncRNA-mRNA interaction analysis based on genes in the magenta module revealed the intersecting lncRNAs, including peptidylprolyl isomerase E-like pseudogene (PPIEL) and the testis-specific transcript, y-Linked 14 (TTTY14); these lncRNAs are mainly involved in functions, such as plasma membrane organization. The ceRNA network analysis revealed several interactions, such as TTTY14-miR-6088-semaphorin 5 A (SEMA5A). Finally, SEMA5A and the zinc finger protein 555 (ZNF555) were identified to be significantly upregulated in the RIF group compared with those in the C group in the GSE58144 dataset. The RT-qPCR results aligned with the above results.
Conclusions
Overall, TTTY14, ZNF555, SEMA5A, PPIEL, and miR-6088 could serve as novel biomarkers of RIF.
Highlights
TTTY14, ZNF555, SEMA5A, PPIEL and miR-6088 could be serve as novel biomarkers of RIF.
TTTY14 may participate in RIF progression by regulating the miR-6088/SEMA5A axis.
This study provides new insights for the diagnosis and treatment of RIF.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Repeated implantation failure (RIF) is a disorder in which good-quality embryos fail to implant in the endometrium following several in vitro fertilization cycles [1]. RIF occurs in approximately 10% of women undergoing in vitro fertilization treatment [2]. Although therapies, such as adjuvant drugs (e.g., growth hormone, androgens, and glucocorticoids), have been used to increase pregnancy rates in women with RIF [3, 4], accurately diagnosing and treating women with RIF are difficult tasks.
Some RNAs have been found to be differentially expressed in women with RIF compared to those in normal controls [5]. A previous study revealed that long noncoding RNAs (lncRNAs) participate in regulating endometrial receptivity [5]. A recent study indicated that the lncRNA, TUNAR, is involved in embryo implantation during RIF [6]. According to Li et al., the lncRNA, ENST00000433673, promotes high mRNA expression of ICAM1 and the adhesion of endometrial epithelial cells, which facilitates adhesion and implantation between the embryo and the mother [7]. Diverse endometrial mRNA signatures have also been recognized to contribute to the development of RIF [8]. According to a previous study, microRNAs (miRNAs) participate in endometrial construction in women with RIF and can be used as biomarkers to predict embryo implantation more reliably [9]. A genome-wide study revealed that lncRNAs, miRNAs, and mRNA form a competing endogenous RNA (ceRNA) network during the progression of RIF [10]. In addition, a previous study indicated that ceRNA networks are involved in functions, such as immunological activity, which prepare the endometrium for embryo implantation [11]. Overall, RNAs and the ceRNA regulatory network may play vital roles in the development of RIF. However, the detailed molecular mechanisms and key genes in the ceRNA network that contribute to RIF progression are still unclear.
This multicenter study was retrospective in design. Briefly, bioinformatic analyses were performed based on two lncRNA/mRNA expression profiles and one miRNA expression profile. Genes, including differentially expressed mRNAs (DEMs), differentially expressed lncRNAs (DElncRNAs) and differentially expressed microRNAs (DEmiRNAs) between normal and RIF samples were explored, and then weighted gene co-expression network analysis (WGCNA) was performed. The functions, pathways, and lncRNA-mRNA interactions were investigated based on these genes. Moreover, a ceRNA network was constructed based on the lncRNA-mRNA and miRNA-mRNA interactions. Finally, the expression of the potential genes in the ceRNA network was verified using an additional mRNA dataset. Overall, this study aimed to identify the potential biomarkers and molecular mechanisms associated with RIF.
Materials and methods
Microarray data
The keywords “repeated implantation failure” and “Homo sapiens” were used to search all expression profile data uploaded in the past five years (2015–2020) in the Gene Expression Omnibus (GEO) database [12]. Datasets meeting the following criteria were included in our study: (i) sample size > 10; (ii) data unified as the expression profile data of endometrial tissue samples; and (iii) samples classified as RIF and normal control. A total of four datasets, including GSE71331 (lncRNA + mRNA), GSE111974 (lncRNA + mRNA), GSE71332 (miRNA), and GSE58144 (mRNA), were selected. Overall, 108 normal control samples (C group) and 79 RIF samples (RIF group) were used in the current study. Flow chart of the current study is provided in Supplementary Fig. 1. Among the four datasets, GSE71332 was used as the test set, while GSE58144 was used as the validation set as it comprised a relatively single datatype. Detailed information on these datasets is provided in Table 1.
Data preprocessing
Gene expression profile data were downloaded and annotated. For genes associated with different probes, the average values of the different probes were used as the final expression values. A probe with annotation information of “protein_coding” was reserved as an mRNA-compatible probe, while a probe with annotation information of “noncoding RNA” and “pseudogene” was reserved as an lncRNA-corresponding probe. For different miRNAs mapped to the same new ID, the mean value of the different miRNA IDs was considered as the final expression value of this new miRNA ID [13]. This process was performed using the mean algorithm in R software.
Analysis of the differentially expressed genes
The ComBat function [14] in R software (version: 3.34.0) [15] was used to eliminate heterogeneity between the GSE71331 and GSE111974 datasets. Thereafter, the two datasets were combined, and the empirical Bayesian methods in the limma package (3.10.3) of R [16] and t-test were employed to explore the DEMs, DElncRNAs, and DEmiRNAs between the C and RIF groups based on the lncRNA + mRNA and miRNA expression matrix. The Benjamini & Hochberg (BH) adjusted P < 0.05 and |logfold change (FC)| > 0.585 were selected as thresholds. Finally, the results were visualized using a heat map.
Co-expression network analysis
WGCNA (version 1.61) [17] was performed to elucidate the co-expression network. Briefly, the optimal soft threshold (0–1) was selected, and a scale-free network and module partition analysis was performed. The module significance (MS) of each module was calculated, and the correlation between MS and clinical traits in the module was determined. Finally, the gene significance (GS) in the module was investigated.
Function and pathway enrichment analysis
ClusterProfiler (version: 3.16.0) [18] software was used for Gene Ontology (GO)-biological function (GO-BP) [19] and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses [20]. P < 0.05 and count ≥ 2 were selected as cut-off values for the enrichment analysis.
lncRNA-mRNA co-expression investigation
lncRNA-mRNA pairs with r > 0.5 and FDR-adjusted P < 0.05 were selected for further analysis [21]. The results were visualized using the Cytoscape (version 3.4.0) software [22]. The CytoNCA software was used to carry out node topology property analysis in the network [23]. Finally, the degree, betweenness, and closeness of the nodes were obtained.
Enrichment analysis of the lncRNAs
Pathway enrichment analysis of the mRNA (considered as a target gene of lncRNA) was performed using clusterProfiler (version: 3.8.1) in R software [24]. P < 0.05 and count ≥ 2 were considered as cut-off values for significant enrichment results.
ceRNA network construction
The online tool miRWalk (version: 2.0) [25, 26] was used to predict the target genes of miRNAs. If the predicted mRNA existed in at least four of the six databases (including miRWalk2.0 [26], miRDB [27], TargetScan [28], miRanda [29], RNA22 [30], and miRMap [31]), it was considered as a potential mRNA that is regulated by the corresponding mRNA. The miRNAs of key lncRNAs were predicted using the DIANA-LncBase (version: 2.0) database [32]. Finally, the lncRNAs and mRNA that were not only regulated by the same miRNA but had a positive expression relationship were selected to construct the ceRNA network, which was then visualized using Cytoscape software (version: 3.4.0).
Verification of the identified mRNAs
The expression of the potential genes identified in the ceRNA network was verified using the GSE58144 dataset. The gene expression results between the two groups were visualized using a box diagram with R software. Differences were considered statistically significant at P < 0.05.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The study comprised two groups: women diagnosed with RIF and healthy volunteers. RIF samples were collected from 10 women diagnosed with RIF during the luteal phase at the Assisted Reproductive Medicine Department of Shanghai Ninth People’s Hospital. The participants met the following inclusion criteria: age, 18–38 years; 3 or more implantation failure; and no hormonal preparations used for 3 months prior to sample collection. Women with serious internal and external diseases were excluded from the study. The 10 healthy volunteers were recruited using advertisements posted on social media. The volunteers met the following inclusion criteria: age 18–38 years, proven fertile (have a normal history of childbirth in the past 3 years), and no hormonal or intrauterine contraception used for at least three months prior to sample collection. Women with serious internal and external diseases were excluded from the study. RT-qPCR was performed to determine the key RNAs. Total RNA was extracted using TRIzol reagent, and cDNA was reverse transcribed using Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). After that, cDNA was subjected to RT-qPCR using the SYBR green method (Takara Bio, Inc., Shiga, Japan) and the BioRad IQTM5 Multicolor Realtime PCR Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The following reaction conditions were employed: 95 °C for 10 min, followed by 40 cycles of 15 s at 95 °C, 60 s at 60 °C, and 1 min at 95 °C. The relative mRNA expression of each gene was normalized to that of GAPDH, while the relative miRNA expression was normalized to that of the U6 snRNA. The primer sequences are listed in Table 2. This study was approved by the Ethics Committee of the Shanghai Ninth People’s Hospital, which is affiliated with Shanghai Jiaotong University School of Medicine. The experiment was conducted in triplicate. Statistical significance was determined based on the cut-off value of P < 0.05.
Results
DEMs, DEmiRNAs, and DElncRNAs between the C and RIF groups
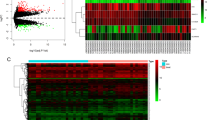
A total of 53 DEmiRNAs (46 upregulated and seven downregulated miRNAs), 327 DEMs (171 upregulated and 156 downregulated mRNAs), and 13 DElncRNAs (eight upregulated and five downregulated lncRNAs) were identified between the C and RIF groups. The volcano plot and heatmap of these genes are shown in Fig. 1.
Volcano plot and heatmap of the differentially expressed lncRNAs (A, B), miRNAs (C, D), and mRNAs (E, F)
The red dot represents upregulated genes; the blue dot represents downregulated genes. For the volcano plot, the X-axis represents the value of log2(FC) while the Y-axis represents the -Log10(p-value); the labeled dots in the volcano plot represent the top 5 upregulated and downregulated genes
WGCNA
WGCNA was performed using all genes. The soft threshold for network construction was 16 (Supplementary Fig. 2), and the fitting degree of the scale-free topological model was 0.9. Using a cutHeight value of 0.3, nine modules were obtained in the current study (Supplementary Fig. 3). The relationship between the expression values of genes in the module and the adjacency correlation between modules was calculated and visualized using a heat map (Supplementary Fig. 4). Based on the correlation between modules and disease status, the black and magenta modules were found to have the highest correlation with disease (Fig. 2A). The correlation between modules and disease status was further investigated according to GS (Fig. 2B). The magenta module was found to be positively correlated with the disease (r = 0.73, P = 3e-11) and was considered the key module for the subsequent analysis.
Results of the correlation analysis for nine modules and disease status. A, Correlation between modules extracted from the datasets and disease; the X-axis represents the different modules while the Y-axis represents the gene significance value. B, Correlation between module and disease status based on the P value
Enrichment investigation for genes in the two modules
Enrichment analysis was performed using the genes in the black and magenta modules. The 315 genes in the black module were found to be mainly enriched in 243 GO-BP functions, such as glomerulus development (GO:0032835, P = 0.0002) (Fig. 3A), and 19 KEGG pathways, such as axon guidance (hsa04360, P = 0.0016) (Fig. 3B). Meanwhile, the 99 genes in the magenta module were mainly enriched in 109 GO-BP functions, such as the regulation of cell growth (GO:0001558, P = 0.0005) (Fig. 3C), and 16 KEGG pathways, such as glioma (hsa05214, P = 0.0034) (Fig. 3D).
Results of the enrichment analysis for genes in the black and magenta modules. A, Gene ontology (GO) biological function (GO-BP) assembled based on genes in the black module. B, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enriched by genes in the black module. C, GO-BP assembled based on genes in the magenta module. D, KEGG pathway enriched by genes in the magenta module. The redder the bubble, the more significant the P value. The larger the bubble, the greater the number of genes enriched in the item
miRNA-mRNA interaction network analysis
Intersection analysis among genes in the magenta module, all DEMs, and all DElncRNAs revealed three intersecting DElncRNAs and 40 intersecting DEMs (Supplementary Fig. 5). An evaluation of the lncRNA-mRNA interaction was performed using these intersecting genes. Based on the results, three lncRNAs (peptidylprolyl isomerase E-like pseudogene (PPIEL), CTAGE family member 7, pseudogene (CTAGE7P), and testis-specific transcript, y-Linked 14 (TTTY14)), 33 mRNAs, and 61 interactions were found in the lncRNA-mRNA interaction network (Fig. 4). Moreover, an enrichment analysis was performed using the three lncRNAs in the lncRNA-mRNA interaction network. These lncRNAs were found to be mainly enriched in GO-BP functions, such as plasma membrane organization (GO:0007009, P = 0.0001, fold enrichment = 27.36), and KEGG pathways, such as pancreatic cancer (hsa05212, P = 0.0024, fold enrichment = 19.32) (Fig. 5).
Enrichment analysis of the three intersecting lncRNAs in the magenta module. A, GO-BP functions assembled based on the three intersecting lncRNAs. B, KEGG pathways enriched by the three intersecting lncRNAs. The X-axis represents the different lncRNAs, while the Y-axis represents the different items of GO-BP functions or KEGG pathways
ceRNA network investigation
Prediction of the interacting DEMs regulated by DEmiRNAs revealed 15 mRNAs and 27 DEmiRNAs, while prediction of the interacting DElncRNAs regulated by DEmiRNAs revealed two lncRNAs and three DEmiRNAs. A ceRNA network was constructed with the lncRNA-miRNA-mRNA interactions, including TTTY14-miR-6088-semaphorin 5 A (SEMA5A) (Fig. 6). As a result, 3 miRNAs, 2 lncRNAs, and 8 mRNAs were found in the network.
Verification of the identified mRNAs
The expression of the eight mRNAs identified in the ceRNA network was verified using the GSE58144 dataset. Only six mRNAs, including barH-like homeobox 1 (BARHL1), calcium/calmodulin-dependent protein kinase II beta (CAMK2B), cyclin-dependent kinase 6 (CDK6), lysosomal-associated membrane protein 3 (LAMP3), SEMA5A, and zinc finger protein 555 (ZNF555), were exported in GSE58144. Gene expression in the C and RIF groups was visualized using a box diagram. All six mRNAs were found to be upregulated in the RIF group compared to the level found in the C group. The mRNA levels of SEMA5A and ZNF555 were significantly upregulated in the RIF group compared to those in the C group (all P < 0.05) (Fig. 7).
RT-qPCR
Finally, we validated the key RNAs, namely SEMA5A, ZNF555, TTTY14, PPIEL, and miR-6088, using RT-qPCR. The levels of SEMA5A, ZNF555, TTTY14, and PPIE were confirmed to be significantly upregulated in the RIF group, while that of miR-6088 was significantly downregulated relative to those in the C group (all P < 0.05; Fig. 8).
Discussion
Some physical habits, such as excessive mental stress, irregular sleep patterns, lack of exercise, or excessive exercise, are reported to be associated with increased RIF in patients [33]. Although the incidence of RIF is high in women following in vitro fertilization treatment, the detailed molecular mechanism remains unclear [34]. In the present bioinformatics study, 53 DEmiRNAs, 327 DEMs, and 13 DElncRNAs were found between the C and RIF groups. WGCNA revealed that the magenta module was positively correlated with RIF disease status. According to the lncRNA-mRNA interaction analysis based on genes in the magenta module, three intersecting lncRNAs, including PPIEL and TTTY14, were found; these lncRNAs were mainly involved in functions, such as plasma membrane organization. Moreover, ceRNA network analysis revealed several interactions, such as TTTY14-miR-6088-SEMA5A. Finally, verification analysis showed that SEMA5A and ZNF555 were significantly upregulated in the RIF group compared to that in the C group. RT-qPCR was performed to verify the key RNAs. Notably, the RT-qPCR results were consistent with the above results.
SEMA5A is involved in axonal guidance during neural development [35]. SEMA5A not only functions in the nervous system, but also in the development of diseases. Dziobek et al. revealed that SEMA5A was upregulated in endometrial cancer and could be used as a supplementary molecular marker for endometrial cancer [36]. In animal models, SEMA5A is regulated by miR-24-1-5p, which participates in endometriosis during the implantation window in rats [37]. Honda et al. performed microarray analysis on neonatal brain exposed to cadmium during gestation and lactation and found abnormal expression of SEMA5A [38]. Moreover, endometrial miRNAs, such as miR-6088, are altered during the window of implantation in patients with RIF [39]. Importantly, embryo implantation requires an optimal endometrium environment, which includes signaling by miRNAs, such as miR-6088 [9]. According to previous studies, miR-6088 is dysregulated in women with a high risk of ovarian cancer [40], and TTTY14 is significantly correlated with overall survival in patients with cancer [41]. Bhat et al. utilized a panel of molecular biology tools to examine the Y chromosome microchimerism in the endometrium using secretory phase samples from fertile and infertile patients with severe (stage IV) ovarian endometriosis and without endometriosis. Based on their result, TTTY14 exhibited a bimodal pattern of expression characterized by low expression in samples from fertile patients and high expression in samples from infertile patients [42]. The lncRNAs in ceRNA have been proven to contribute to RIF progression by regulating the miRNA-mRNA interaction, which can be used as biomarkers for predicting endometrial receptivity [11]. Consistent with the above results, in the present study, SEMA5A was significantly upregulated in the RIF group compared with that in the C group; TTTY14-miR-6088-SEMA5A was one of the ceRNA interactions identified in the current study. Thus, we speculated that the lncRNA, TTTY14, might participate in RIF progression by regulating the miR-6088/SEMA5A axis.
In a previous study, ZNF555 was identified as a putative transcriptional factor highly expressed in human diseases, such as in primary myoblasts, and could serve as a therapeutic target [43]. The lncRNA, PPIEL, is also closely related to non-small cell lung cancer in women [44]. Different DNA methylation statuses of PPIEL were previously revealed to contribute to the development of diseases [45]. Yeung et al. determined whether DNA methylation at birth and in childhood differs by conception using assisted reproductive technologies or ovulation induction compared with conception without fertility treatment. Based on their results, regions in maternally imprinted genes, including IGF1R, PPIEL, and SVOPL, had decreased mean DNA methylation levels among newborns conceived by assisted reproductive technologies [46]. In this study, ZNF555 was one of the two significantly upregulated DEMs in the RIF group relative to the levels in the C group. Meanwhile, the lncRNA, PPIEL, was not only differentially expressed between the C and RIF groups but also enriched in the GO-BP function of plasma membrane organization. More importantly, the RT-qPCR results were consistent with the above results. Thus, we speculated that the mRNA, ZNF555, and lncRNA, PPIEL, might be novel biomarkers for RIF, and PPIEL might participate in the progression of RIF via its plasma membrane organization function.
This study had some limitations, such as its small sample size. Thus, further verification analysis is needed with a larger sample size. In addition, no function assay was performed to validate the conclusion that lncRNA TTTY14 participates in the progression of RIF by regulating miR-6088/SEMA5A axis.
In conclusion, the lncRNA, TTTY14, may participate in the progression of RIF by regulating the miR-6088/SEMA5A axis. Moreover, the mRNA, ZNF555, and lncRNA, PPIEL, could be considered novel biomarkers for RIF, and the lncRNA, PPIEL, might participate in the progression of RIF via its plasma membrane organization function.
Data availability
The data used in this study are publicly available from the GEO database at https://www.ncbi.nlm.nih.gov/geo/, the dataset number are GSE71331, GSE111974, GSE71332, and GSE58144.
Abbreviations
- BARHL1:
-
BarH-like homeobox 1
- CAMK2B:
-
Calcium/calmodulin-dependent protein kinase II beta
- CDK6:
-
Cyclin-dependent kinase 6
- ceRNA:
-
Competing endogenous RNA
- CTAGE7P:
-
CTAGE family member 7, pseudogene
- DElncRNAs:
-
Differentially expressed lncRNAs
- DEmiRNAs:
-
Differentially expressed microRNAs
- DEMs:
-
Differentially expressed mRNAs
- GEO:
-
Gene Expression Omnibus
- GO:
-
Gene Ontology
- GS:
-
Significance
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LAMP3:
-
Lysosomal-associated membrane protein 3
- lncRNA:
-
Long noncoding RNA
- MS:
-
Module significance
- PPIEL:
-
Peptidylprolyl isomerase E-like pseudogene
- PPIEL:
-
Peptidylprolyl isomerase E-like pseudogene
- RIF:
-
Repeated implantation failure
- SEMA5A:
-
Semaphorin 5 A
- TTTY14:
-
Testis-specific transcript, y-Linked 14
- TTTY14:
-
Testis-specific transcript, y-Linked 14
- WGCNA:
-
Weighted correlation network analysis
- ZNF555:
-
Zinc finger protein 555
- ZNF555:
-
Zinc finger protein 555
References
Coughlan C, et al. Recurrent implantation failure: definition and management. Reprod Biomed Online. 2014;28(1):14–38.
Laufer N, Simon A. Recurrent implantation failure: current update and clinical approach to an ongoing challenge. Fertil Steril. 2012;97(5):1019–20.
Pourakbari R, et al. Cell therapy in female infertility-related diseases: emphasis on recurrent miscarriage and repeated implantation failure. Life Sci. 2020;258:118181.
Kuroda K, et al. Analysis of the risk factors and treatment for repeated implantation failure: OPtimization of thyroid function, IMmunity, and Uterine Milieu (OPTIMUM) treatment strategy. Am J Reprod Immunol. 2021;85(5):e13376.
Liu L, et al. Altered circular RNA expression in patients with repeated implantation failure. Cell Physiol Biochem. 2017;44(1):303–13.
Chen Q et al. Real-time in vivo magnetic induction tomography in rabbits: a feasibility study. 2020;32(3).
Li D, et al. Preliminary functional inquiry of lncRNA ENST00000433673 in embryo implantation using bioinformatics analysis. Syst Biology Reproductive Med. 2019;65(2):164–73.
Shi C, et al. Diverse endometrial mRNA signatures during the window of implantation in patients with repeated implantation failure. Hum Fertility. 2018;21(3):183–94.
Paul AB, Sadek ST, Mahesan AM. The role of microRNAs in human embryo implantation: a review. J Assist Reprod Genet. 2019;36(2):179–87.
Xu H, et al. Genome-wide analysis of long noncoding RNAs, microRNAs, and mRNAs forming a competing endogenous RNA network in repeated implantation failure. Gene. 2019;720:144056.
Feng C, et al. Construction of implantation failure related lncRNA-mRNA network and identification of lncRNA biomarkers for predicting endometrial receptivity. Int J Biol Sci. 2018;14(10):1361.
Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110.
Liu C, et al. MicroRNA-1202 plays a vital role in osteoarthritis via KCNQ1OT1 has-mir-1202-ETS1 regulatory pathway. J Orthop Surg Res. 2020;15(1):1–9.
Yang D, Parrish RS, Brock GN. Empirical evaluation of consistency and accuracy of methods to detect differentially expressed genes based on microarray data. Computers in Biology & Medicine. 2014;46(1):1–10.
Leek JT, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–3.
Smyth GK et al. LIMMA: linear models for microarray data. In Bioinformatics and computational biology solutions using R and bioconductor. Statistics for Biology and Health. 2005.
Dai Y, et al. Gene co-expression network analysis reveals key pathways and hub genes in Chinese cabbage (Brassica rapa L.) during vernalization. BMC Genomics. 2021;22(1):236.
Yu G, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–7.
Gene Ontology Consortium. Going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–56.
Kanehisa M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–d361.
Tanzhu G, et al. Molecular subtypes and prognostic signature of pyroptosis-related lncRNAs in glioma patients. Front Oncol. 2022;12:779168–8.
Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303.
Lei X, et al. Clustering and overlapping modules detection in PPI network based on IBFO. Proteomics. 2013;13(2):278–90.
Yu G, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics-a J Integr Biology. 2012;16(5):284–7.
Breuer K, et al. InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Res. 2013;41(Database issue):D1228–33.
Dweep H, Gretz N. miRWalk2. 0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697–7.
Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–d131.
Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105.
Betel D, et al. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90–0.
Wang G, et al. A nine-gene signature as prognostic biomarker in gastric cancer by bioinformatics analysis. Clin Transl Oncol. 2023;25(11):3296–306.
Charles, et al. miRmap: Comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012;40(22):11673.
Paraskevopoulou M, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–8.
Ni Y, et al. The analysis of fertility quality of life and the influencing factors of patients with repeated implantation failure. Health Qual Life Outcomes. 2021;19(1):32.
Cakmak H, Taylor HS. Implantation failure: molecular mechanisms and clinical treatment. Hum Reprod Update. 2011;17(2):242–53.
Goldberg JL, et al. An oligodendrocyte lineage-specific semaphorin, Sema5A, inhibits axon growth by retinal ganglion cells. J Neurosci. 2004;24(21):4989–99.
Dziobek K, et al. Assessment of the usefulness of the SEMA5A concentration profile changes as a molecular marker in endometrial cancer. Curr Pharm Biotechnol. 2020;21(1):45–51.
Cai H, et al. MicroRNA dysregulation and steroid hormone receptor expression in uterine tissues of rats with endometriosis during the implantation window. Chin Med J. 2018;131(18):2193.
Honda A, et al. Microarray analysis of neonatal brain exposed to cadmium during gestation and lactation. J Toxicol Sci. 2013;38(1):151–3.
Shi C, et al. Endometrial microRNA signature during the window of implantation changed in patients with repeated implantation failure. Chin Med J. 2017;130(5):566.
Pandey R, et al. Circulating miRNA profiling of women at high risk for ovarian cancer. Translational Oncol. 2019;12(5):714–25.
Miao Y, et al. Comprehensive analysis of a novel four-lncRNA signature as a prognostic biomarker for human gastric cancer. Oncotarget. 2017;8(43):75007.
Bhat MA, et al. Genomic evidence of Y chromosome microchimerism in the endometrium during endometriosis and in cases of infertility. Reprod Biol Endocrinol. 2019;17(1):22.
Kim E, et al. ZNF555 protein binds to transcriptional activator site of 4qA allele and ANT1: potential implication in facioscapulohumeral dystrophy. Nucleic Acids Res. 2015;43(17):8227–42.
Heesen M, Klimek M. Nonobstetric anesthesia during pregnancy. Curr Opin Anaesthesiol. 2016;29(3):297–303.
Docherty LE, et al. Genome-wide DNA methylation analysis of patients with imprinting disorders identifies differentially methylated regions associated with novel candidate imprinted genes. J Med Genet. 2014;51(4):229–38.
Yeung EH, et al. Conception by fertility treatment and offspring deoxyribonucleic acid methylation. Fertil Steril. 2021;116(2):493–504.
Acknowledgements
This study was supported by the Natural Science Foundation of Shanghai (Program No. 19411961100) and Comparison of the application effect of PPOS in patients with ovarian reserve dysfunction through randomized controlled tri (Program No. 16411963800).
Author information
Authors and Affiliations
Contributions
LZY and QQH conceived and designed the study, and JY and LZY participated in the acquisition of data. QJC and JY analyzed and interpreted the data. QJC and JY participated in the design of the study and performed statistical analyses. LZY, JY, and QQH conceived the study, participated in its design and coordination and helped draft and revise the manuscript for important intellectual content. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.

Supplementary Material 1: Supplementary Fig. 1.
Flow chart of the current study

Supplementary Material 2: Supplementary Fig. 2.
Weight parameter of adjacency matrix (power) investigation

Supplementary Material 3: Supplementary Fig. 3.
Dynamic tree cut and merged dynamic represent the module before and after the merge

Supplementary Material 4: Supplementary Fig. 4.
Correlation between modules: The X-axis represents modules of different colors, and the Y-axis represents the overall correlation coefficient between genes and the disease state in each module. The change from blue to red indicates that the adjacency correlation value changes from low to high.

Supplementary Material 5: Supplementary Fig. 5.
Venn plot analysis among differentially expressed mRNAs, lncRNAs, and genes in the magenta module
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, L., Ye, J., Chen, Q. et al. lncRNA TTTY14 participates in the progression of repeated implantation failure by regulating the miR-6088/SEMA5A axis. J Assist Reprod Genet 41, 727–737 (2024). https://doi.org/10.1007/s10815-024-03032-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-024-03032-w