Abstract
Recurrent implantation failure (RIF) is a complex and poorly understood clinical disorder characterized by failure to conceive after repeated embryo transfers. Endometrial receptivity (ER) is a prerequisite for implantation, and ER disorders are associated with RIF. However, little is known regarding the molecular mechanisms underlying ER in RIF. In the present study, RNA sequencing data from the mid-secretory endometrium of patients with and without RIF were analyzed to explore the potential long non-coding RNAs (lncRNAs) and messenger RNAs (mRNAs) involved in RIF. The analysis revealed 213 and 1485 differentially expressed mRNAs and lncRNAs, respectively (fold change ≥ 2 and p < 0.05). Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses indicated that these genes were mostly involved in processes related to immunity or inflammation. 5 key genes (TTR, ALB, TF, AFP, and CFTR) and a key module including 14 hub genes (AFP, ALB, APOA1, APOA2, APOB, APOH, FABP1, FGA, FGG, GC, ITIH2, SERPIND1, TF and TTR) were identified in the protein–protein interaction (PPI) network. The 5 key genes were used to further explore the lncRNA-miRNA-mRNA regulatory network. Finally, the drug ML-193 based on the 14 hub genes was identifed through the CMap. After ML-193 treatment, endometrial cell proliferation was increased, the hub genes were mostly down-regulated, and the ER marker HOXA10 was up-regulated. These results offer insights into the regulatory mechanisms of lncRNAs and mRNAs and suggest ML-193 as a therapeutic agent for RIF by enhancing ER.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infertility poses a significant challenge to human reproduction. Approximately 15% of patients undergoing infertility treatment experience recurrent implantation failure (RIF) [1]. Coughlan et al. [2] proposed classical diagnostic criteria for RIF and defined this condition as a failure to achieve a clinical pregnancy after transfer of at least 4 high-quality embryos in a minimum of 3 fresh or frozen cycles in women under the age of 40 years. Successful embryo implantation occurring during the implantation window is a process that requires a reciprocal interaction between blastocyst and the endometrium [3]. The window of implantation usually spans 3 to 6 days within the secretory phase. During this time span, the endometrium attains the temporal differentiation and reaches receptivity status, allowing embryo adhesion and invasion [4]. The changes that occur during this process are regulated by multiple molecules, including noncoding RNA, cytokines, growth factors, and cell adhesion molecules [5, 6]. Genetic polymorphism of the genes involved in embryo implantation, immune responses and endometrial receptivity (ER) are likely to be involved in the pathophysiology of RIF [7]. RIF places substantial psychological and economic burdens on patients and their families as well as healthcare associations. However, the precise etiology and pathogenesis of RIF are complex and remain unclear.
With the recent advances in high-throughput sequencing technology, it is now well-established that non-coding RNAs, such as microRNA (miRNA), circular RNA and long non-coding RNA (lncRNA), play distinct regulatory roles in biological processes [8]. LncRNA, defined as a kind of non-coding RNA longer than 200 bp, has attracted considerable attention because of its aberrant expression in various diseases [9]. Some lncRNAs can act as competitive endogenous RNAs (ceRNAs) to regulate the expression of downstream messenger RNA (mRNA) by sponging shared miRNA activity, thereby sequestering miRNAs and regulating the function of miRNA-targeted mRNAs [10]. Furthermore, the contributions of dysregulated expression of lncRNAs to the occurrence of RIF have been reported. A study showed that the expression of lncRNA H19 was down-regulated in the endometrium of patients with RIF and acted as a sponge that regulated integrin β3 through its antagonistic effect on let-7, which might affect ER and embryo implantation [11]. Nevertheless, with respect to the clinical value of lncRNA in RIF, evidence regarding the differences in lncRNA expression between patients with and without RIF is still limited.
Therefore, the aim of our study was to identify mRNA and lncRNA related regulatory networks in RIF through RNA sequencing (RNA-seq) and bioinformatics analysis. And drug ML-193 was predicted based on the 14 hub genes identified in the protein–protein interaction (PPI) network, the effect of ML-193 on endometrial cells was partly verified. ML-193 should be further investigated as a potential drug for the treatment of RIF patients in the future.
Materials and Methods
Patients Selection and Sample Collection
The inclusion and exclusion criteria of patients who provided mid-secretory phase endometrium are shown in Table 1. In total, 18 tissue specimens from the mid-secretory phase endometrium (RIF group, n = 10; control group, n = 8) and 10 normal late proliferative endometrial tissue specimens were included in this study. The 18 endometrial samples were obtained during the mid-secretory phase as confirmed by histological diagnoses. Among these specimens, 6 (RIF group, n = 3; control group, n = 3) were randomly selected for RNA-seq, and the remaining 12 (RIF group, n = 7; control group, n = 5) underwent RT-qPCR. The eligibility criteria for the 10 patients who provided the late proliferative endometrial tissue was consistent with our previous study [12]. The 10 endometrium samples were used for primary cell culture. The CCK8 was used in 4 specimens and RT-qPCR was performed in 6.
This study was approved by the Research Ethics Committee of the First Hospital of Lanzhou University (Approval No. LDYYLL2019-45). Informed consent was obtained from all participants.
RNA Isolation and Quality Control
Total RNA was extracted from cells and tissues using TRIzol reagent (Invitrogen, Gibco-BRL, Bethesda, MD, USA) in accordance with the manufacturer’s instructions. The concentration and quality of the extracted RNA were then determined on using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). RNA integrity and genomic DNA contamination were further assessed by standard denaturing agarose gel electrophoresis.
RNA-Seq
RNA samples from the RIF and control groups were sent to Cloud-Seq Biotech (Shanghai, China). Briefly, total RNA was purified following removal of ribosomal RNA (rRNA) with a Gen-Seq® rRNA Removal Kit (GenSeq, Inc.). Each rRNA-depleted sample was then subjected to library construction with a GenSeq® Low Input RNA Library Prep Kit (GenSeq, Inc.) per in accordance with the manufacturer’s instructions. Samples were quality-controlled and quantified using the BioAnalyzer 2100 system (Agilent Technologies, Inc., Santa Clara, CA, USA). Library sequencing was performed on an Illumina Novaseq 6000 platform (Illumina, San Diego, CA, USA). Raw data were normalized using edgeR (v3.16.5). Differentially expressed mRNAs and lncRNAs were defined by an FC ≥ 2 and p value < 0.05.
Hierarchical Clustering
Differentially expressed mRNAs and lncRNAs (FC ≥ 2 and p value < 0.05) were calculated with normalized counts. Cluster analysis was generated using the heatmap2 package in the R software. Different endometrial samples were usually clustered in different branches.
GO and KEGG Enrichment Analyses
Gene ontology (GO) analysis and pathway analysis were used to determine the functions of these dysregulated mRNAs and lncRNAs. GO terms were grouped into three categories: biological process (BP), cellular component (CC) and molecular function (MF) (http://www.geneontology.org). Dysregulated mRNAs and lncRNAs were mapped to GO terms in the DAVID (https://david.ncifcrf.gov/). The p value denotes the significance of GO terms enrichment in the genes (p value < 0.05 is recommended). The enrichment score equaled -log10 (p value). The pathway analysis was based on the functional analysis of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (http://www.genome.jp/kegg/). The p value denotes the significance of the pathway correlated to the conditions (p value < 0.05 is recommended).
PPI Network Construction
The PPI pairs between differentially expressed mRNAs were evaluated using the String database (version 11.5; https://cn.string-db.org/) with a medium confidence score of > 0.4. Next, PPI network visualization was performed using Cytoscape. The hub nodes with a high network topology property indicator score were considered to have an important role in the network. The hub nodes in the PPI network were analyzed by CytoNCA plug-in of Cytoscape, and degree centrality, betweenness centrality and closeness centrality were calculated. The “Molecular Complex Detection” (MCODE) plugin of Cytoscape was then employed to identify the hub gene clusters exhibiting a high degree of interaction.
Construction of ceRNA Network
To construct the ceRNA network, differentially expressed mRNAs of interest in the hub nodes of the PPI network were selected. The miRNA-mRNA interaction pairs were obtained from the prediction of TargetScan (https://www.targetscan.org/). Target genes with a context score percentile < 50 were excluded from the TargetScan algorithm. The interactions between miRNAs and differentially expressed lncRNAs were predicted using the miRcode (http://www.mircode.org/) with R software. Next, lncRNA-miRNA-mRNA relationships were constructed based on their shared miRNAs between the miRNA-mRNA and miRNA-lncRNA interactions. The lncRNA-miRNA-mRNA (ceRNA) network was constructed by using Cytoscape 3.9.1.
CMap Analysis
CMap (https://portals.broadinstitute.org/CMap/) is a database of gene expression profiles from cultured human cells treated with bioactive small molecules. It is designed to help scientists to discover functional connections among diseases, genetic perturbation, and drug action. Up-regulated tags of the 14 hub genes were loaded into the CMap online tool, 1046 bioactive molecules were obtained. A norm_cs score ranging from -1 to 1 was used to estimate the closeness between query signatures and compounds: a positive score indicates a promotive effect of the compound on the query signatures, whereas a negative score denotes an inverse effect of the compound on the query signatures. Drugs with negative scores are typically interpreted as having potential therapeutic effects on the biological processes represented by the query signatures.
Cell Culture
10 tissue samples were used for primary cell culture; 4 of them were used for the CCK8 and 6 were used for RT-qPCR. The extraction method of the primary cells was performed as described by Lyu et al. [12].
A Cell Counting Kit-8 (CCK8)
CCK8 assay was used to determine the viability of human embryonic stem cells (hESCs) and Ishikawa (ISK) cells under the treatment of ML-193. Briefly, cells were seeded onto a 96-well plate (hESCs, 1 × 104 cells/well; ISK cells, 5 × 103 cells/well). After 1 day, the cells were treated with ML-193 (0, 5 and 10 mM) for 0 or 24 h. Next, 10 μL of CCK8 solution (NCM, Suzhou, China) was added to each well, and the cells were incubated at 37°C for 2 h. The absorbance at 450 nm was measured using an enzyme-labelling instrument (Infinite M2000 PRO; Tecan, Männedorf, Switzerland). 5 replicate wells were tested for each group.
Real-time Polymerase Chain Reaction (RT-qPCR)
Total RNAs isolated from cells and frozen endometrial tissues were reverse-transcribed to complementary DNA using a high-capacity complementary DNA reverse-transcription kit (Wuhan Servicebio Technology Co., Ltd., Wuhan, China). The expression of lncRNAs in endometrial tissues and the expression of mRNAs in hESCs and ISK cells were measured by RT-qPCR using SYBR Premix Ex Taq (Wuhan Servicebio Technology Co., Ltd.) on an ABI 7900HT instrument (Thermo Fisher Scientific). β-actin was served as an internal control. All primer sequences used in this study were listed in Supplementary Table S1. The 2-(ΔΔCt) method was used to calculate the relative expression of each gene.
Statistical Analysis
Lists of differentially expressed mRNAs and lncRNAs were generated by screening with Student’s t-test, using the criteria of an FC of ≥ 2.0 and a p value of < 0.05 between the RIF and control groups. In the GO and KEGG pathway analyses, Fisher’s exact test was used to evaluate the significance of the GO terms or pathway identifier enrichment in the differentially expressed genes. A p value of < 0.05 was considered statistically significant. The statistical analysis was performed with GraphPad Prism software 9.0.0 (GraphPad Software, La Jolla, CA, USA).
Results
Differentially Expressed mRNAs and lncRNAs
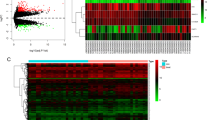
RNA-seq was used to explore the mRNA and lncRNA expression profiles in endometrial tissues from the RIF group (n = 3) and control group (n = 3) (Table 1). The mean ages of the patients in the RIF and control groups were 34.00 ± 3.30 and 30.86 ± 3.39 years, respectively. Differentially expressed mRNAs (Fig. 1a) and lncRNAs (Fig. 1b) were clustered hierarchically according to their expression levels. Volcano plots of the significant differentially expressed mRNAs and lncRNAs were highlighted between the RIF and control groups are shown in Fig. 1c-d. The expression profiles of 213 mRNAs (Supplementary Table S2) and 1485 lncRNAs (Supplementary Table S3) were identified in the RIF and control groups (FC ≥ 2 and p value < 0.05). Among them, 143 mRNAs were up-regulated and 70 mRNAs were down-regulated, while 809 lncRNAs were up-regulated and 676 lncRNAs were down-regulated. Table 2 shows the top 10 up- and down-regulated mRNAs in the RNA-seq study. Table 3 showed the top 10 up- and down-regulated lncRNA transcripts. The classification and chromosome distribution of the dysregulated lncRNAs are shown in Supplementary Fig. 1.
Expression levels of mRNA and lncRNA in RIF. a−b Hierarchical cluster analyses of differentially expressed mRNAs and lncRNAs (fold change ≥ 2 and p value < 0.05) between RIF and control groups. Red and green represent up-regulated and down-regulated expression respectively. c−d The volcano maps of the differentially expressed mRNAs and lncRNAs. X-axis, log2 represents the fold change; Y-axis, expressed as the p value. The two vertical lines represent 2-fold regulation, and the horizontal line represents a p value of 0.05. The red and green dots represent the mRNAs and lncRNAs that are significantly up-regulated and down-regulated, respectively
Gene Ontology and Pathway Analyses
Many lncRNAs act as cis-regulators and that their functions were linked to parental genes with protein-coding ability. GO and KEGG analyses were performed to reveal significant enrichment pathways and to further understand the potential biology of the dysregulated mRNAs and lncRNAs, the GO and KEGG analyses were further performed. GO enrichment analysis was performed for up- and down-regulated mRNAs (FC ≥ 2 and p value < 0.05) between the RIF and control groups, as well as for the lncRNA parental genes. In the GO analysis, the top 10 significant GO terms according to the enrichment score (−log10(p value)) of the dysregulated genes were considered. For up-regulated mRNAs (Fig. 2a), organic anion transport (GO:0015711) was enriched most in BP, the endoplasmic reticulum lumen (GO:0005788) was enriched most in CC, and transporter activity (GO:0005215) was enriched most in MF. For down-regulated mRNAs (Fig. 2b), cell killing (GO:0001906) was the most enriched term in BP, immunological synapse (GO:0001772) was the most enriched term in CC, and IgG binding (GO:0019864) was the most enriched term in MF.
GO analyses of the differentially expressed mRNAs and lncRNAs. a−b The top 10 GO terms of the three different gene functions for up-regulated and down-regulated mRNAs. c−d The top 10 GO terms of the three different gene functions for up-regulated and down-regulated lncRNAs. Red: biological process; Blue: molecular function; Green: cellular component
For up-regulated lncRNAs (Fig. 2c), microtubule cytoskeleton organization (GO:0000226), microtubule-based process (GO:0007017) and cellular localization (GO:0051641) were enriched most in BP; intracellular (GO:0005622), organelle (GO:0043226) and cytoplasm (GO:0005737) were enriched most in CC. And iron binding (GO:0043167), IgG binding (GO:0019864) and anion binding (GO:0043168) were enriched most in MF. For down-regulated lncRNAs (Fig. 2d), flavonoid metabolic processes (GO:0009812), xenobiotic glucuronidation (GO:0052697) and cellular glucuronidation (GO:0052695) were the most enriched terms in BP; cytoplasm (GO:0005737), intracellular (GO:0005622) and endomembrane system (GO:0012505) were the most enriched terms in CC; and glucuronosyltransferase activity (GO:0015020), catalytic activity (GO:0003824) and enzyme binding (GO:0019899) were the most enriched terms in MF.
Further, pathway analyses were performed to test for statistical significance of the differentially expressed mRNA and lncRNA parental genes. The pathway analyses revealed 13 pathways that corresponded to the up-regulated mRNA transcripts, and the most enriched were fat digestion and absorption, Staphylococcus aureus infection, and the peroxisome proliferator activated receptor (PPAR) signaling pathway (Fig. 3a). Among the down-regulated pathways, the chemokine signaling pathway was the most enriched (Fig. 3b). In addition, 35 pathways were enriched for up-regulated lncRNAs, and 26 pathways were enriched for down-regulated lncRNA. Targets of up-regulated lncRNA were mainly enriched in the pathways of platelet activation, inflammatory mediator regulation of TRP channel, and the oxytocin signaling pathway (Fig. 3c). Targets of down-regulated lncRNA were mainly enriched in the pathways of bile secretion, ascorbate and aldarate metabolism, and porphyrin and chlorophyll metabolism (Fig. 3d).
Pathway analyses of the differentially expressed mRNAs and lncRNAs. a−b The top 10 for the up-regulated and the down-regulated mRNA enriched signaling pathways. c−d The top 10 pathways for the up-regulated and down-regulated lncRNA target genes. The X-axis shows the enrichment score (-log10(p value)). The size of the dot in the bubble map indicates the number of enriched genes, and the red indicates the p value
Construction of the PPI Network
To explore the functional connection of key genes, a PPI network was constructed from 213 dysregulated mRNAs. Of these mRNAs, 115 of 213 genes were found to be involved in the interactions based on the predictions from the String database (median confidence score of > 0.4). There were 115 nodes and 358 edges in the PPI network (Fig. 4a). According to the ranking of network topology property indicators, we defined hubs as the top 10 nodes ranked separately by degree centrality, betweenness centrality, and closeness centrality were defined as hubs (Table 4). Furthermore, the keyword “reproductive or decidualization or endometrial receptivity or recurrent implantation failure” was entered into the GeneCards (https://www.genecards.org/), and the first 300 results were screened. Among the hub nodes, AFP, ALB, CFTR, TF and TTR emerged as key nodes because of their numerous connections with the results obtained from Genecards.
Construction of a PPI network and identifcation of hubgenes from the PPI network. a Protein–protein interaction (PPI) network of 115 differentially expressed mRNAs between RIF and control groups. The diameter of each circle implies degree centrality. b The subnetwork with 14 hubgenes that extracted from the PPI network with the “Molecular Complex Detection (MCODE)” algorithm
MCODE, a plugin of Cytoscape, was utilized to screen key modules from the PPI network. Application of the criteria of a k-core is 2 revealed 5 subnetworks. Among them, a subnetwork with the highest MCODE score of 13,231 comprising 14 nodes and 86 edges, was selected (Fig. 4b). There were 14 up-regulated genes in this subnetwork (AFP, ALB, APOA1, APOA2, APOB, APOH, FABP1, FGA, FGG, GC, ITIH2, SERPIND1, TF and TTR), and these 14 genes were identified as hub genes.
Construction of the lncRNA-miRNA-mRNA ceRNA Regulatory Network
5 mRNAs of particular interest among the key nodes of the PPI network (AFP, ALB, CFTR, TF and TTR) were selected to further explore the lncRNA-miRNA-mRNA regulatory network. After separately identifying and integrating the 35 miRNA-mRNA and 177 miRNA-lncRNA interactions (Supplementary Table S4), the ceRNA network was constructed. 3 lncRNAs within the ceRNA network were randomly selected to conduct RT-qPCR and exhibited the same change trends as shown in our RNA-seq results (Supplementary Fig. 2). The ceRNA network contained 57 nodes (5 mRNAs, 28 miRNAs and 24 lncRNAs) and 212 edges (Fig. 5). Among the ceRNA relationships, the top 5 lncRNAs with the highest degree centrality were identified, and included ZNRD1-AS1 (degree = 27), TPTEP1 (degree = 23), NPHP3-AS1 (degree = 14), ANKRD18CP (degree = 12), and LINC00284 (degree = 11). The top lncRNAs were the nodes of which indicating important roles in the ceRNA network. The top 5 miRNAs were hsa-miR-27a-3p (degree = 14), hsa-miR-24-3p (degree = 13), hsa-miR-129-5p (degree = 11), hsa-miR-4770 (degree = 10), hsa-miR-613 (degree = 10).
The lncRNA-miRNA-mRNA ceRNA regulatory network. Circular nodes represent mRNAs, triangular nodes represent miRNAs, and rectangular represent lncRNAs. Yellow and blue represent up-regulated and down-regulated expression of mRNAs and lncRNAs in the RNA-seq result respectively. The size of each node implies degree centrality
Identification and Validation Effect of ML-193 on Endometrial Cell Proliferation
In the present study, CMap was employed to identify potential compounds that could disturb the gene expression pattern of the 14 hub genes. The up-regulated tag of the 14 hub genes were submitted to the CMap online tool for analysis. The instances for each compound according to the permuted results of the CMap analysis are listed in Supplementary Table S5. 3 drugs (ML-193, bisphenol-a, and capsazepine) were selected from the top 10 drugs, and only ML-193 showed a proliferative effect as indicated by CCK8 analysis (Supplementary Fig. 3). Thus, ML-193, with the most significant negative enrichment score, was identified as a potential therapeutic compound for RIF. The chemical structure of ML-193 were shown in Fig. 6a.
In addition, the effect of ML-193 on endometrial cells was validated. The results showed that 24 h of treatment with ML-193 promoted proliferation of hESCs and ISK cells (Fig. 6b-c).
Validation of ML-193 Effect on Expression of Hub Genes and HOXA10
14 hub genes were validated to determine whether ML-193 could modulate their expression. Under treatment with ML-193, the expression of mostly 14 hub genes was decreased in hESCs (Fig. 7a) and ISK cells (Fig. 7b).
HOXA10, a biomarker of ER, was also examined. After treatment with ML-193, the expression level of HOXA10 was increased in both hESCs (Fig. 7c) and ISK cells (Fig. 7d).
Discussion
RIF is a complicated clinical disorder in human reproductive medicine, which affects the physical and mental health of couples as well as constituting a huge challenge for clinicians. The present study reports a comprehensive analysis of mRNAs and lncRNAs expression profiles in RIF and constructs PPI and ceRNA networks. According to the above regulatory relationships, 14 hub genes were identified. ML-193, which may be useful for treatment of RIF were identified based on hub genes by CMap. Under treatment with ML-193, hESCs and the ISK cell proliferation increased, hub genes were mostly down-regulated and ER maker HOXA10 was up-regulated. This study provided a basis for exploring the pathogenesis and therapeutic strategies for RIF from the perspective of the regulatory relationship between lncRNAs and mRNAs. Notably, the data indicate that ML-193 could be considered a potential therapeutic agent for RIF by enhancement of ER.
Inflammatory and immune response were essential for ER to embryo implantation and early placental development [13, 14], and their abnormalities were considered the causes of RIF [15]. In the present study, GO analysis revealed that down-regulated mRNAs were mostly associated with immune responses through several molecular mechanisms, such as cell killing, immunological synapses, and IgG binding (Fig. 2b). Indeed, in preparation for decidualization, immune cells played key roles in immune tolerance and the promotion of pregnancy [16, 17]. KEGG pathway analysis revealed that these genes were related to two critical inflammatory and immune associated signaling pathways, PPAR signaling pathway and inflammatory mediator regulation of transient receptor potential channels. PPAR signaling pathway ranked third in the KEGG analysis of up-regulated mRNAs (Fig. 3c). Activation of the PPAR signaling pathway has been thought to affect female reproduction by interfering with biological functions such as immune tolerance, inflammatory responses and trophoblast invasion [18, 19]. Inflammatory mediator regulation of transient receptor potential channels was found in the up-regulated lncRNAs (Fig. 3a). Mei et al. [20] found genes related to inflammatory mediator regulation of transient receptor potential (TRP) channels had a lower expression level in the chorion-attached endometrium compared to the non-chorion-attached endometrium, this may be related pregnancy failure by dysregulation of maternal immune tolerance. Altogether, our data suggested dysregulated mRNAs and lncRNA may be through involvement in inflammatory and immune responses, to affect ER and embryo implantation.
PPI and lncRNA-miRNA-mRNA ceRNA networks provided the evidence of the functional mechanism of the dysregulated mRNAs and lncRNAs in RIF. The PPI network construction revealed interactions between the dysregulated mRNAs (FC ≥ 2 and p < 0.05). The 14 hub genes of particular focus in PPI network were AFP, ALB, APOA1, APOA2, APOB, APOH, FABP1, FGA, FGG, GC, ITIH2, SERPIND1, TF and TTR (Fig. 4b). Among these genes, APOA1 had higher expression in the mid-secretary phase endometrium, which might be a cause for RIF [21]. TTR, ALB and TF were related to ER. TTR, the carrier for thyroxine and retinol, was a protein marker in the receptive stage of the human endometrium [22, 23], ALB and TF may be involved in the preparation of ER by regulating iron metabolism and immunological tolerance [24]. FGG associated with endometrial inflammation [25], as we all know, inflammatory abnormalities were considered the causes of RIF [15]. FGA could promote the migration and invasion of endometrial stromal cells [26], but few studies have examined the relationship between FGA and ER. AFP was highly expressed in the endometrium of endometrial cancer patients [27], no report to date has indicated that the expression of AFP was involved in RIF. Other genes (APOA2, APOB, APOH, FABP1, GC, ITIH2 and SERPIND1) have not been reported in the pathophysiology of the endometrium. In summary, abnormal expression patterns of these genes were speculated may contribute to impaired ER and the occurrence of RIF. Accumulating reports were also indicated that lncRNAs, as protein-coding gene modulators, served a vital role in mammals through the ceRNA network [28]. Through back-tracing the 5 initially selected mRNAs (TTR, TF, ALB, AFP, and CFTR), lncRNA-miRNA-mRNA relationships were connected based on their shared miRNAs. Accordingly, a ceRNA network was constructed in the present study to better understand the pivotal combined roles of lncRNAs with miRNAs and mRNAs.
Various treatments were available depending on the etiology of RIF, but molecular and cellular abnormalities pose great challenges to treatment [29]. ER was a recognized cause of RIF, and identifying new molecular therapeutic agents to improve ER is critical. Therefore, CMap analysis of the 14 hub genes in this study was performed to explore the compounds that could be used to treat RIF. Based on genome-wide expression profiling of gene transcripts, CMap proposed a data-driven and reliable method to identify new drugs or reposition existing drugs [30]. ML-193, the top drug in this analysis, was an antagonist to GPR55, and several studies have shown that ML-193 can exhibit anti-inflammatory activity [31,32,33]. For example, Kurano et al. [31] found that ML-193 treatment attenuated the elevation of inflammatory cytokines in plasma or tissues in mice, helping to improve insulin resistance and prevent organ damage. Gajghate et al. [32] found that ML-193 reduced inflammatory cell infiltration and increased the number of immune cells during stroke in the brain tissue of mice, thereby attenuating the inflammatory response. Li et al. [33] found that ML-193 exhibited anti-inflammatory activity in LPS-stimulated human dental pulp cells by inhibiting TLR4/Myd88/NF-κB signaling. However, its role against RIF has not been elucidated. The present study showed that ML-193 could promote the proliferation of hESCs and ISK cells (Fig. 6b-c). Successful pregnancy required a receptive endometrium of adequate thickness, insufficient endometrial thickness was closely associated with pregnancy failure [34]. The number of stromal and epithelial cells was reduced in thin endometrium [35]. In additional, proliferation of hESCs was necessary for decidualization, and a decrease in the proliferation rate inhibited decidual formation, leading to pregnancy failure [36]. Thus, ML-193 was speculated whether improves ER by promoting endometrial thickness and decidualization. And ML-193 treatment further down-regulated the mRNA levels of mostly 14 hub genes (Fig. 7a-b), which were up-regulated in RIF. More importantly, among the 14 hub genes, TTR, ALB and TF, which were previously discussed that may involved in ER, were all down-regulated both in hESCs and ISK under the treatment of ML-193. These results suggested that ML-193 may have an effect on ER, so the effect of ML-193 on ER maker HOXA10 was further explored.
Our results showed after treatment with ML-193, the expression level of HOXA10 was increased in both hESCs (Fig. 7c) and ISK cells (Fig. 7d). Study has found that the expression of HOXA10 was significantly lower in the RIF patients endometrium [37]. As an imperative contributor to embryo im-plantation, HOXA10 played different roles in stromal and epithelial cells [38, 39]. During embryo implantation, expression of HOXA10 was increased in endometrial stromal cells to govern stromal cell transformation to decidual cells [40]. Decidualization was necessary to create a receptive endometrium [41]. Abnormalities in the decidualization process were closely related to RIF [42]. The endometrial epithelium was the first part of the embryo to come into contact with the mother at implantation [43]. HOXA10 was known to play a vital role in regulating changes in epithelial cell function during the establishment of ER [40]. ITGB3 and E-cadherin, which were related to ER, acted as an adhesion medium between the embryo and epithelial cells [44, 45]. Studies showed up-regulation of HOXA10 increase the expression of ITGB3 and E-cadherin in epithelial cells, thereby improving ER and subsequently increasing the embryo adhesion and implantation [45, 46]. Therefore, ML-193 was speculated to improve ER by affecting HOXA10, which regulated stromal cells decidualization as well as adhesion of embryos and epithelial cells. These findings were expected to provide insight into the development of new therapeutic drugs for RIF. The in vitro cellular models used in the study were a limitation of this study, and the specific mechanism of action of ML-193 on ER required further animal experiments.
Conclusion
RIF is a major limiting factor in the success rates of in-vitro fertilization, it not only seriously threatens women's physical health but also damages mental health. In this study, 14 hub genes were identified through a comprehensive bioinformatic analysis of the relationship between lncRNAs and mRNAs in RIF. Additionally, ML-193 was identified as a potential therapeutic agent for RIF. Furthermore, the 14 hub genes and the ER marker HOXA10 were down- and up-regulated, respectively, under treatment with ML-193. These results indicated that ML-193 showed promise as a potential therapeutic agent for RIF by enhancing ER.
Data Availability
The datasets analyzed during the current study are included within the article and its additional files.
Code Availability
Not applicable.
References
Busnelli A, Reschini M, Cardellicchio L, Vegetti W, Somigliana E, Vercellini P. How common is real repeated implantation failure? An indirect estimate of the prevalence. Reprod Biomed Online. 2020;40(1):91–7. https://doi.org/10.1016/j.rbmo.2019.10.014.
Coughlan C, Ledger W, Wang Q, Liu F, Demirol A, Gurgan T, Cutting R, Ong K, Sallam H, Li TC. Recurrent implantation failure: definition and management. Reprod Biomed Online. 2014;28(1):14–38. https://doi.org/10.1016/j.rbmo.2013.08.011.
Pantos K, Grigoriadis S, Maziotis E, Pistola K, Xystra P, Pantou A, Kokkali G, Pappas A, Lambropoulou M, Sfakianoudis K, Simopoulou M. The role of interleukins in recurrent implantation failure: A comprehensive review of the literature. Int J Mol Sci. 2022;23(4). https://doi.org/10.3390/ijms23042198.
Lessey BA, Young SL. What exactly is endometrial receptivity? Fertil Steril. 2019;111(4):611–7. https://doi.org/10.1016/j.fertnstert.2019.02.009.
Ochoa-Bernal MA, Fazleabas AT. Physiologic events of embryo implantation and decidualization in human and non-human primates. Int J Mol Sci. 2020;21(6). https://doi.org/10.3390/ijms21061973.
Liu H, Huang X, Mor G, Liao A. Epigenetic modifications working in the decidualization and endometrial receptivity. Cell Mol Life Sci. 2020;77(11):2091–101. https://doi.org/10.1007/s00018-019-03395-9.
Mrozikiewicz AE, Ożarowski M, Jędrzejczak P. Biomolecular markers of recurrent implantation failure-a review. Int J Mol Sci. 2021;22(18). https://doi.org/10.3390/ijms221810082.
Ali SA, Peffers MJ, Ormseth MJ, Jurisica I, Kapoor M. The non-coding RNA interactome in joint health and disease. Nat Rev Rheumatol. 2021;17(11):692–705. https://doi.org/10.1038/s41584-021-00687-y.
Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220(2). https://doi.org/10.1083/jcb.202009045.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–8. https://doi.org/10.1016/j.cell.2011.07.014.
Zeng H, Fan X, Liu N. Expression of H19 imprinted gene in patients with repeated implanta-tion failure during the window of implantation. Arch Gynecol Obstet. 2017;296(4):835–9. https://doi.org/10.1007/s00404-017-4482-x.
Lyu M, Gao W, Zhang L, Yang X, Yue F, Li H, Ma X, Liu L. Hsa_circ_0001550 impairs de-cidualization by regulating the proliferation and apoptosis of endometrial stromal cells. Reprod Biomed Online. 2023;46(2):225–33. https://doi.org/10.1016/j.rbmo.2022.10.003.
Liu S, Wei H, Li Y, Huang C, Lian R, Xu J, Chen L, Zeng Y. Downregulation of ILT4+ dendritic cells in recurrent miscarriage and recurrent implantation failure. Am J Re-prod Immunol. 2018;80(4):e12998. https://doi.org/10.1111/aji.12998.
Negishi Y, Shima Y, Takeshita T, Morita R. Harmful and beneficial effects of inflammatory re-sponse on reproduction: sterile and pathogen-associated inflammation. Immunol Med. 2020;44(2):98–115. https://doi.org/10.1080/25785826.2020.1809951.
Wang Q, Sun Y, Fan R, Wang M, Ren C, Jiang A, Yang T. Role of inflammatory factors in the etiology and treatment of recurrent implantation failure. Reprod Biol. 2022;22(4):100698. https://doi.org/10.1016/j.repbio.2022.100698.
Krstic J, Deutsch A, Fuchs J, Gauster M, Gorsek Sparovec T, Hiden U, Krappinger JC, Moser G, Pansy K, Szmyra M, Gold D, Feichtinger J, Huppertz B. (Dis) similarities between the decidual and tumor microenvironment. Biomedicines. 2022;10(5):1065. https://doi.org/10.3390/biomedicines10051065.
Liu S, Diao L, Huang C, Li Y, Zeng Y, Kwak-Kim JYH. The role of decidual immune cells on human pregnancy. J Reprod Immunol. 2017;124:44–53. https://doi.org/10.1016/j.jri.2017.10.045.
Coulam CB. Intralipid treatment for women with reproductive failures. Am J Reprod Immunol. 2021;85(4):e13290. https://doi.org/10.1111/aji.13290.
Zhao L, Zheng X, Liu J, Zheng R, Yang R, Wang Y, Sun L. PPAR signaling pathway in the first trimester placenta from in vitro fertilization and embryo transfer. Biomed Pharmacother. 2019;118:109251. https://doi.org/10.1016/j.biopha.2019.109251.
Mei X, Xu L, Ren Y, Yu M, Kuang L, Li C, Zhang Y, Lu C, Wang Z, Guo Z, Xie X, Huang D, Zhang M. Transcriptome comparison of chorion-attached and non-chorion-attached en-dometrium in mid-gestation of rabbit. Front Vet Sci. 2022;9:838802. https://doi.org/10.3389/fvets.2022.838802.
Brosens JJ, Hodgetts A, Feroze-Zaidi F, Sherwin JR, Fusi L, Salker MS, Higham J, Rose GL, Kajihara T, Young SL, Lessey BA, Henriet P, Langford PR, Fazleabas AT. Proteomic analysis of endometrium from fertile and infertile patients suggests a role for apolipoprotein A-I in embryo implantation failure and endometriosis. Mol Hum Reprod. 2010;16(4):273–85. https://doi.org/10.1093/molehr/gap108.
Diao H, Xiao S, Cui J, Chun J, Xu Y, Ye X. Progesterone receptor-mediated up-regulation of transthyretin in preimplantation mouse uterus. Fertil Steril. 2010;93(8):2750–3. https://doi.org/10.1016/j.fertnstert.2010.01.009.
Beier HM, Beier-Hellwig K. Molecular and cellular aspects of endometrial receptivity. Hum Reprod Update. 1998;4(5):448–58. https://doi.org/10.1093/humupd/4.5.448.
Lancheros-Buitrago J, Rodriguez-Villamil P, Gregory J, Bastos H, Camacho CA, Caballeros JE, Cazales N, Barros E, José de Jesus Silva M, Pimentel A, Mattos RC. Ceruloplasmin, sero-transferrin and albumin presented different abundance in mares' uterine fluid 5 days after in-semination. Theriogenology. 2020;148:194–200. https://doi.org/10.1016/j.theriogenology.2019.11.011.
Jana B, Kaczmarek MM, Romaniewicz M, Brzozowska M. Profile for mRNA transcript abun-dances in the pig endometrium where inflammation was induced by Escherichia coli. Anim Re-prod Sci. 2021;232:106824. https://doi.org/10.1016/j.anireprosci.2021.106824.
Li H, Cai E, Cheng H, Ye X, Ma R, Zhu H, Chang X. FGA controls VEGFA secretion to pro-mote angiogenesis by activating the VEGFR2-FAK signalling pathway. Front Endocrinol (Lausanne). 2022;13:791860. https://doi.org/10.3389/fendo.2022.791860.
Cheng Y, Yang L, Shi G, Chen P, Li L, Fang H, Chen C. Ninjurin 2 rs118050317 gene polymorphism and endometrial cancer risk. Cancer Cell Int. 2021;21(1):1. https://doi.org/10.1186/s12935-020-01646-5.
Chu Q, Xu T, Zheng W, Chang R, Zhang L. Long noncoding RNA MARL regulates antiviral responses through suppression miR-122-dependent MAVS downregulation in lower vertebrates. PLoS Pathog. 2020;16(7):e1008670. https://doi.org/10.1371/journal.ppat.1008670.
Sheikhansari G, Pourmoghadam Z, Danaii S, Mehdizadeh A, Yousefi M. Etiology and management of recurrent implantation failure: A focus on intra-uterine PBMC-therapy for RIF. J Reprod Immunol. 2020;139:103121. https://doi.org/10.1016/j.jri.2020.103121.
Qu XA, Rajpal DK. Applications of Connectivity Map in drug discovery and development. Drug Discov Today. 2012;17(23–24):1289–98. https://doi.org/10.1016/j.drudis.2012.07.017.
Kurano M, Kobayashi T, Sakai E, Tsukamoto K, Yatomi Y. Lysophosphatidylinositol, especially albumin-bound form, induces inflammatory cytokines in macrophages. FASEB J. 2021;35(6):e21673. https://doi.org/10.1096/fj.202100245R.
Gajghate S, Li H, Rom S. GPR55 inactivation diminishes splenic responses and improves neurological outcomes in the mouse ischemia/reperfusion stroke model. Cells. 2024;13(3):280. https://doi.org/10.3390/cells13030280.
Li WJ, Shen J. Antagonism of G protein-coupled receptor 55 prevents lipopolysaccha-ride-induced damages in human dental pulp cells. Hum Exp Toxicol. 2022;41:9603271221099598. https://doi.org/10.1177/09603271221099598.
Mouhayar Y, Franasiak JM, Sharara FI. Obstetrical complications of thin endometrium in assisted reproductive technologies: a systematic review. J Assist Reprod Genet. 2019;36(4):607–11. https://doi.org/10.1007/s10815-019-01407-y.
Lv H, Zhao G, Jiang P, Wang H, Wang Z, Yao S, Zhou Z, Wang L, Liu D, Deng W, Dai J, Hu Y. Deciphering the endometrial niche of human thin endometrium at single-cell resolution. Proc Natl Acad Sci U S A. 2022;119(8):e2115912119. https://doi.org/10.1073/pnas.2115912119.
Zhou M, Xu H, Zhang D, Si C, Zhou X, Zhao H, Liu Q, Xu B, Zhang A. Decreased PIBF1/IL6/p-STAT3 during the mid-secretory phase inhibits human endometrial stromal cell proliferation and decidualization. J Adv Res. 2020;30:15–25. https://doi.org/10.1016/j.jare.2020.09.0.
Yang Y, Chen X, Saravelos SH, Liu Y, Huang J, Zhang J, Li TC. HOXA-10 and E-cadherin ex-pression in the endometrium of women with recurrent implantation failure and recurrent mis-carriage. Fertil Steril. 2017;107(1):136–143.e2. https://doi.org/10.1016/j.fertnstert.2016.09.016.
Du H, Taylor HS. The role of hox genes in female reproductive tract development, adult function, and fertility. Cold Spring Harb Perspect Med. 2015;6(1):a023002. https://doi.org/10.1101/cshperspect.a023002.
Blitek A, Kiewisz J, Waclawik A, Kaczmarek MM, Ziecik AJ. Effect of steroids on HOXA10 mRNA and protein expression and prostaglandin production in the porcine endometrium. J Reprod Dev. 2010;56(6):643–8. https://doi.org/10.1262/jrd.10-046k.
Ashary N, Laheri S, Modi D. Homeobox genes in endometrium: from development to decidualization. Int J Dev Biol. 2020;64(1-2-3):227–37. https://doi.org/10.1387/ijdb.190120dm.
Neykova K, Tosto V, Giardina I, Tsibizova V, Vakrilov G. Endometrial receptivity and preg-nancy outcome. J Matern Fetal Neonatal Med. 2022;35(13):2591–605. https://doi.org/10.1080/14767058.2020.1787977.
Zhai J, Ma L, Chang Z, Yu T. Increased expression of prokineticin 2 and its receptor in en-dometrium of recurrent implantation failure patients decreased the expression of MMP9 im-portant for decidualization. Reprod Biol Endocrinol. 2022;20(1):76. https://doi.org/10.1186/s12958-022-00947-w.
Evans J, Hutchison J, Salamonsen LA, Greening DW. Proteomic insights into endometrial receptivity and embryo-endometrial epithelium interaction for implantation reveal critical determinants of fertility. Proteomics. 2020;20(1):e1900250. https://doi.org/10.1002/pmic.201900250.
Germeyer A, Savaris RF, Jauckus J, Lessey B. Endometrial beta3 integrin profile reflects en-dometrial receptivity defects in women with unexplained recurrent pregnancy loss. Reprod Biol Endocrinol. 2014;12:53. https://doi.org/10.1186/1477-7827-12-53.
Bi Y, Huang W, Yuan L, Chen S, Liao S, Fu X, Liu B, Yang Y. HOXA10 improves endometrial receptivity by upregulating E-cadherin†. Biol Reprod. 2022;106(5):992–9. https://doi.org/10.1093/biolre/ioac007.
Zhang M, Zhang Q, Cao Z, Cai X, Liu J, Jiang Y, Zhu Y, Zhou J, Yu L, Zhen X, Hu Y, Yan G, Sun H. MEKK4-mediated phosphorylation of HOXA10 at threonine 362 facilitates embryo adhesion to the endometrial epithelium. Cell Death Discov. 2022;8(1):415. https://doi.org/10.1038/s41420-022-01203-1.
Acknowledgements
The authors would like to thank all the subjects for their generous participation in this study.
Funding
This work was financially supported by the National Natural Science Foundation of China (82360307), the Talent Innovation and Entrepreneurship Project of Lanzhou City (2022-RC-47), Gansu Provincial Science and Technology Program (20JR10TA715, 21JR7RA391), the Medical Innovation and Development Project of Lanzhou University (lzuyxcx-2022-191), Science and Technology Project of Lanzhou (2023-ZD-93), Fund of the First Hospital of Lanzhou University (ldyyyn2022-60), the First Hospital of Lanzhou University Excellence Program (20220060074, 20220060073).
Author information
Authors and Affiliations
Contributions
Lin Liu, Yidan Liu, Yu Tian, Ying Cao and Ting Wang contributed equally to this work. Lin Liu designed and supervised the study. Yidan Liu and Yu Tian conducted the experiments and drafted the manuscript. Ting Wang and Ying Cao collected the samples and analyzed the data. Run Yang, Simin Liu and Shengyan Mi contributed the methodology. Jing Wang and Xiaoling Ma modified the format. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
The study was approved by the Research Ethics Committee of the first Hospital of Lanzhou University. Written informed consents were obtained from all patients prior to study commencement.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, L., Liu, Y., Tian, Y. et al. Identification of Differentially Expressed mRNAs and lncRNAs Contributes to Elucidation of Underlying Pathogenesis and Therapeutic Strategy of Recurrent Implantation Failure. Reprod. Sci. (2024). https://doi.org/10.1007/s43032-024-01630-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s43032-024-01630-8