Abstract
Purpose
To determine whether convolutional neural networks (CNN) can be used to accurately ascertain the patient identity (ID) of cleavage and blastocyst stage embryos based on image data alone.
Methods
A CNN model was trained and validated over three replicates on a retrospective cohort of 4889 time-lapse embryo images. The algorithm processed embryo images for each patient and produced a unique identification key that was associated with the patient ID at a timepoint on day 3 (~ 65 hours post-insemination (hpi)) and day 5 (~ 105 hpi) forming our data library. When the algorithm evaluated embryos at a later timepoint on day 3 (~ 70 hpi) and day 5 (~ 110 hpi), it generates another key that was matched with the patient’s unique key available in the library. This approach was tested using 400 patient embryo cohorts on day 3 and day 5 and number of correct embryo identifications with the CNN algorithm was measured.
Results
CNN technology matched the patient identification within random pools of 8 patient embryo cohorts on day 3 with 100% accuracy (n = 400 patients; 3 replicates). For day 5 embryo cohorts, the accuracy within random pools of 8 patients was 100% (n = 400 patients; 3 replicates).
Conclusions
This study describes an artificial intelligence-based approach for embryo identification. This technology offers a robust witnessing step based on unique morphological features of each embryo. This technology can be integrated with existing imaging systems and laboratory protocols to improve specimen tracking.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Background
“To err is human” is a familiar phrase in day-to-day life, but is not an acceptable explanation for errors within an in vitro fertilization (IVF) laboratory [1]. Although difficult to accept and extremely rare, human error does occur within the IVF lab and can result in loss of gametes, or worse, incorrect implantation of mismatched gametes. The daily responsibilities of the IVF lab involve complex procedures requiring precise multi-step protocols, creating many opportunities for error especially when handling increasing volumes of patients throughout the day. These steps include oocyte retrieval (patient identification, dish labeling), sperm collection (patient identification, collection labeling), gamete processing (cumulous cell removal prior to insemination, sperm preparation), insemination (conventional or intracytoplasmic sperm injection), embryo culture, assisted hatching, embryo biopsy, vitrification and warming, and embryo transfer.
The American Society for Reproductive Medicine (ASRM) has identified four key factors that lead to human error which include conscious automaticity, involuntary automaticity, ambiguous accountability, and stress [2]. These factors recognize times when human memory and task orientation lack focus and allow for errors to go unrecognized. Errors can be not only emotionally and medically devastating to patients, but also can be financially, socially, and legally devastating to IVF practices. Of malpractice claims made in the field of Reproductive Endocrinology and Infertility (REI), 38% of them are quoted to be from embryology lab errors [3, 4]. The true rate of occurrence for error is difficult to account for, but one large practice reported a rate of moderate to severe error (defined as an event that negatively affected a cycle or cycle loss due to loss/mishandling of gametes or embryos) as 1 per 1735 procedures or 1 per 429 cycles [5]. The Human Fertilization and Embryology Authority (HFEA) has reported adverse event (includes clinical, near misses, administrative, and laboratory errors) rate of 1% of the 60,000 cycles of IVF treatment in the UK annually. Out of the 465 reported events, 114 of them were laboratory events [6].
To thoroughly track patient specimens, the contents of each dish need to be monitored at each step of the IVF process. National and international organizations such as European Society for Human Reproductive and Embryology (ESHRE), ASRM, and HFEA have proposed best practices that emphasize accuracy of initial labeling with supervision, or a “double witness”, to start the process. ASRM recommends double-witnessing to occur at each of the following steps: patient specimen labeling, egg collection, sperm reception, sperm preparation, mixing sperm and eggs or injecting sperm into eggs, transfer of gametes between tubes/dishes, transfer of embryos into a woman, insemination of a women with sperm prepared in the laboratory, cryopreservation of gametes or embryos, thawing cryopreserved gametes and embryos, the final disposition of gametes or embryos, and transporting gametes or embryos [2]. In addition to the double-witnessing protocols, electronic witnessing systems (EWS), such as barcode identification or radiofrequency identification [7], have also been introduced. These commercial witnessing systems are based on the labeling of all labware used for each case with barcode stickers, or radio frequency identification labels, which can be identified by unique computer-based readers. The risk of sample mismatch due to human error is minimized when using these systems, but as gametes and embryos are moved from one container to another several times during an ART cycle, the possibility of misidentification still exists. To track dish contents instead of individual dishes, manual tagging of oocytes and embryos with polysilicon barcodes has been proposed [8]. This labeling process, however, is an invasive and time-consuming procedure that adds complexity to the IVF workflow. For a gamete and embryo witnessing to be well accepted in the field, it needs to be non-invasive, simple to use, accurate, and easy to incorporate into any IVF laboratory.
Convolutional neural networks (or CNN), an image-based deep learning neural network, have enormous potential to aid at every step of the IVF process. CNNs process each image as millions of datapoints within a multilayer perceptron capable of analyzing visual imagery. Artificial intelligence (AI) algorithms have been trained, validated, and tested on gamete and embryo images to decipher subtle morphologic markers linked to embryo development such as implantation potential [9]. In many cases, these neural networks have shown to have superior accuracy and consistency in classifying cells when compared to embryologists [10]. This technology has been developed and tested to measure sperm morphology, assess egg quality, perform fertilization assessments, aid in the alignments of oocytes and embryos for micromanipulation, assess embryo quality, and predict developmental outcomes from every stage of preimplantation development [10,11,12,13,14,15,16,17,18]. Deep learning technology allows the computer to examine and process far more features on a cell than can be performed by even the most skillful human eye. Given the wide application of use for artificial intelligence throughout the IVF process, this study aims to assess whether CNNs could be used to find unique features among embryos within a cohort. Using these subtle morphologic differences among embryos affords a non-invasive, streamlined patient identification tool for safe tracking embryos within the IVF laboratory at multiple timepoints. As AI is able to provide objective and standardized analyses of embryo images with remarkable accuracy, we aim to show that AI serves as a powerful tool for embryo witnessing.
Materials and methods
Data collection and handling
Data was collected from patients who underwent a fresh IVF cycle with time-lapse imaging at the Massachusetts General Hospital (MGH) Fertility Center in Boston, MA. After obtaining approval through the Institutional Review Board (IRB#2017P001339), we evaluated a retrospective cohort of 4889 time-lapse imaging videos of embryos collected using a commercial time-lapse imaging system (EmbryoScope, Vitrolife). The imaging system used a Leica × 20 objective that collected images at 10-min intervals under illumination from a single 635-nm LED. Each patients’ cohort of embryos was exported as videos (.avi) using the imaging system software. Videos of individual embryos were broken down into their respective frames to extract images from all timepoints post-insemination. The extracted images from each timepoint were 250 × 250 pixels that were subsequently cropped to 210 × 210 pixels to remove both the timestamps and any potential identifiers present within the frame. Out-of-focus images were included in the datasets and used for both testing and training. Only images of embryos that were completely non-discernable were removed from the study. Since image collection timepoints across all patients were not consistent, we binned them into groups of around 18-min intervals. In total, imaging data from 400 patients were utilized in this study.
Witnessing software development and Unique Identification Score Assignment
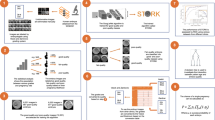
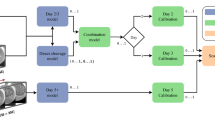
We used pre-developed AI models to build the witnessing software [11, 13]. Two deep convolutional neural network models capable of predicting blastocyst development at the cleavage stage and classifying blastocysts based on their developmental quality were used in combination with a genetic algorithm to generate unique identification scores (UIS) for each embryo within a cohort. Development of the genetic algorithm (GA) has been described in our previous work [12]. The GA scores were used to generate a 12-character ID key for each patient assessed by the witnessing software. These scores were used to determine if the embryos originated from the same patient at a later timepoint (Fig. 1). Embryos were evaluated on day 3 cleavage stage and day 5 blastocyst stage at which time a UIS was generated specific to patient at that timepoint. As a noise mitigation strategy, when evaluating embryo cohorts at a secondary timepoint, scores generated for every available preceding timepoint were also considered for similarity to the initial timepoint.
Identification assessment at the cleavage stage
Cleavage stage embryos were examined and imaged at two separate timepoints on day 3 of development at 65 hours post-insemination (hpi) and 70 hpi. These times correspond with when our laboratory assesses embryos for morphologic development, when we transfer embryos to a new dish for extended embryo culture, when we perform laser-assisted hatching or when we transfer cleavage stage embryos. For each embryo, a UIS was generated for each embryo at 65 hpi using the CNN to generate an ID library. At 70 hpi, the embryos were reassessed, and assigned a unique ID key that was matched to patient ID keys available within the library. The absolute error value between the two timepoints (65 hpi and 70 hpi) was calculated for each patient. Minimum error was noted when the UIS generated for a given patient cohort at both timepoints were noted to be of the same patient.
Identification assessment at the blastocyst stage
Blastocyst stage embryos were examined and imaged at two separate timepoints of 105 hpi and 110 hpi on day 5 of development. These times correspond with when the laboratory assesses each patient’s embryos for development and when embryos are moved to a new dish for embryo transfer, trophectoderm biopsy, embryo vitrification, or to an extended culture dish. For each embryo, a UIS was generated for each embryo at 105 hpi using the CNN to generate an ID library. At 110 hpi, embryos were reassessed, and assigned a unique ID key that was matched to patient ID keys available within the library. The absolute error value between the two timepoints (105 hpi and 110 hpi) was calculated for each patient. Minimum error was noted when the UIS generated for a given patient cohort at both timepoints were noted to be of the same patient.
Study design and statistical analysis
The system was tested at two time intervals at day 3 (65 hpi and 70 hpi) and day 5 (105 and 110 hpi). In both scenarios, available images between these timepoints were retrieved for each embryo and were presented to the software to generate a patient ID key using the unique identification score (UIS). In this study, we evaluated the system’s ability to identify patients correctly using an independent set of embryos (400 patients; 2–12 embryos per patient which did not overlap with the training data set used in any previous exercise). To achieve this, each of the 400 patient cohorts was grouped with 7 other randomly selected patient cohort, to form a randomly organized sub-pool from the total pool of available patient data. This allows for evaluation of the AI-based ID-witnessing software in clinically relevant numbers of patients routinely processed at IVF centers around the world. To combat possible bias within the algorithm towards pooled patient cohorts, these sub-pools were randomized for each of replicate analysis (3 replicates) of each of the 400 patients. The absolute error between the ID key of the evaluated patient at the initial timepoint (~ 65 or ~ 105 hpi) and ID keys of all patients within the random sub-pool at secondary timepoint (~ 70 or ~ 110 hpi) was calculated and used by the software automatically to determine the closest matching patient in the sub-pool. When the system identified the patient correctly in the secondary timepoint within the sub-pool of patients, a “pass” was noted. If the system identified the incorrect patient, a “fail” was noted. If the system cannot arrive at any consistent decision when considering all the frames between the initial and secondary timepoint, it is considered as an error and was removed from the analysis. The overall accuracy across all replicates of the 400 patients was calculated for the final assessment of the software.
Results
Of all patients undergoing fresh IVF, 4889 embryos were recorded using time-lapse imaging of sufficient quality for training and validation of the AI CNN. The AI CNN was tested using 8-embryo subsets from 400 patients who underwent fresh IVF on day 3 and day 5 of development over three replicates. The accuracy of the CNN in correctly matching the patient identification within random pools of 8-patient embryo cohorts on day 3 was 100% (n = 400 patients; 3 replicates). The accuracy of the CNN in correctly matching the patient ID of embryo cohorts within random pools of 8 patients on day 5 was 100% (n = 400 patients; 3 replicates).
Discussion
The system we are presenting was able to uniquely identify day 3 and day 5 embryos with 100% accuracy through 400 patient cohorts over 3 replicates, which serves as a novel application of AI in the embryo identification. Artificial intelligence can be utilized to visually recognize an individual’s embryos and be an additional identification tool for patient’s embryos in the IVF lab. The CNN system described above demonstrated 100% accuracy in matching patient identification with their embryo and an extraordinarily low chance for encountering two patients with the same CNN key. This technology could be utilized to decrease the chance of error when handling human embryos in the IVF lab.
The current gold standard of identification in the embryology lab is double-witnessing. While this system is helpful in reducing error, a major limitation of double-witnessing is the additional time and personnel necessary to complete this protocol. Holmes et al. [19] investigated the time difference between an EWS protocol and a manual double-witnessing protocol in the IVF laboratory and found a three-to-fivefold reduction in witnessing time for procedures with EWS. In settings with high clinical volume, the addition of EWS may help to reduce the time burden of a double-witnessing protocol. Rienzi et al. [20] performed a failure mode and effects analysis (FMEA) in an active IVF clinic to evaluate the utility of EWS in preventing errors in the embryology laboratory. While 99.9% of occasions had no error in the IVF lab, there were still occurrences, albeit rare. The group analyzed the risk of failure after addition of EWS and did see a decline in rates of failure by two thirds. The etiology for these errors were thought to come from one of the following reasons: “heavy clinical work-load and distraction, communication failures between the team, and inadequacy of the labeling system used”[ 20]. Forte et al. [21] performed a patient survey to understand patient concerns about IVF errors and EWS satisfaction and learned that patient concern for error is high as is their satisfaction with utilization of EWS to reduce this risk.
The current technologies include barcode identification, radiofrequency identification, or gamete and embryo direct tagging. Barcode identification is a label placed on the patient’s wrist band while in clinic/treatment and on samples such as embryo culture dish or sperm preparation container. The technology requires a scanner for the barcode and correctly labeled specimen dishes/containers [22, 23]. This technology can also lead to error in terms of printing the labels or incorrectly labeling a specimen. Another EWS is radiofrequency identification (RFID), but this is still under investigation as there is still much to learn about the safety of radiowaves for gametes or preimplantation embryos [23,24,25,26]. Another technology proposed for tracking specimens is direct tagging of oocytes and embryos with polysilicon barcodes. This invasive method of labeling specimens requires reading of the label under the microscope by the embryologist and risks the label being lost during hatching or other embryo handling events [8]. None of these systems replace double-witnessing, each have their own limitations, still have opportunities for error, and can be difficult to implement into an existing IVF lab’s workflow. Adding an element of EWS through AI can aid to this process and help reduce chance for error [7, 27].
One limitation of this study was that all embryos were imaged on the EmbryoScope. Not all laboratories will have access to the same imaging platform or be able to use the same method of imaging during every step of the IVF process. By developing the CNN with only one imaging platform, this may limit the generalizability of our system to images from other imaging platforms. For instance, until recently, AI algorithms developed using the EmbryoScope could not be accurately applied on images captured from an inverted microscope. This is due to a shifting of the data distribution between the algorithm’s training dataset and the dataset from the different imaging system. Additionally, all embryos came from one clinic that is geographically confined to the state of Massachusetts, which may introduce imperceivable bias into the CNN system, further necessitating multicenter assessment of the system.
This study has multiple strengths including the large cohort of embryos that were tested. We included a total of 400 patient cohorts within the test set, with random subsets of embryos also used in testing to overcome potential bias of pooled patient cohorts. The system was able to have 100% accuracy with the entire cohort. We used standard time intervals for the study that are used in the IVF laboratory for morphologic grading of embryos, so the standard care for embryo culture was not interrupted by this process. Additionally, given these are standard timepoints for embryo morphologic assessment during preimplantation development, this AI-ID witnessing system can be easily integrated into any IVF lab workflow.
EWS exist and are currently utilized in some laboratories but are not standardized or routine at this time. This study shows the powerful capabilities and increasing promise of image-based AI systems for use in gamete and zygote identification within the IVF lab. This system demonstrates a robust electronic witnessing platform that focuses on subtle, indiscernible morphologic differences between embryos to ensure accurate and precise identification. With this AI-driven EWS alongside current, standard witnessing practices, human error in gamete or zygote identification can be significantly reduced through a platform that can be easily integrated into clinics with Embryoscope time-lapse imaging capabilities.
Conclusions
This study describes the first artificial intelligence-based approach for embryo tracking and patient specimen identification in the IVF laboratory. This technology offers a robust witnessing step based on unique morphological features that are specific to each individual embryo. This technology can be combined with existing identification/witnessing protocols seamlessly to improve specimen tracking in the ART laboratory and avoid human error.
Data transparency
The datasets and machine learning algorithms used and/or analyzed during the current study are available from the corresponding author on reasonable request and under a data transfer agreement with Mass General Brigham. Data regarding any of the subjects in the study has not been previously published unless specified.
References
To err is human. January 10, 2021]; Available from: https://www.merriam-webster.com/dictionary/to%20err%20is%20human.
de los Santos, M.J. and A. Ruiz, Protocols for tracking and witnessing samples and patients in assisted reproductive technology. Fertil Steril, 2013. 100(6): p. 1499–502.
Letterie G. Outcomes of medical malpractice claims in assisted reproductive technology over a 10-year period from a single carrier. J Assist Reprod Genet. 2017;34(4):459–63.
Rasouli MA, Moutos CP, Phelps JY. Liability for embryo mix-ups in fertility practices in the USA. J Assist Reprod Genet. 2021;38(5):1101–7.
Sakkas D, Pool TB, Barrett CB. Analyzing IVF laboratory error rates: highlight or hide? Reprod Biomed Online. 2015;31(4):447–8.
Adverse incidents in fertility clinics: lessons to learn. 2014 January 28, 2021]; Available from: hfea.gov.uk/media/1146/incidents_report_2014_designed_-_web_final.pdf.
Cimadomo D, et al. Failure mode and effects analysis of witnessing protocols for ensuring traceability during PGD/PGS cycles. Reprod Biomed Online. 2016;33(3):360–9.
Novo S, et al. Direct embryo tagging and identification system by attachment of biofunctionalized polysilicon barcodes to the zona pellucida of mouse embryos. Hum Reprod. 2013;28(6):1519–27.
Fitz VW, et al. Should there be an “AI” in TEAM? Embryologists selection of high implantation potential embryos improves with the aid of an artificial intelligence algorithm. J Assist Reprod Genet. 2021;38(10):2663–70.
Bormann, C.L., et al., Consistency and objectivity of automated embryo assessments using deep neural networks. Fertil Steril, 2020. 113(4) 781–787 e1.
Manoj Kumar Kanakasabapathy, P.T., Charles L Bormann, Raghav Gupta, Rohan Pooniwala, Hemanth Kandula, Irene Souter, Irene Dimitriadis, Hadi Shafiee, Deep learning mediated single time-point image-based prediction of embryo developmental outcome at the cleavage stage. 2006.
Bormann, C.L., et al., Performance of a deep learning based neural network in the selection of human blastocysts for implantation. Elife, 2020. 9.
Thirumalaraju P, et al. Evaluation of deep convolutional neural networks in classifying human embryo images based on their morphological quality. Heliyon. 2021;7(2): e06298.
Bormann CL, et al. Deep learning early warning system for embryo culture conditions and embryologist performance in the ART laboratory. J Assist Reprod Genet. 2021;38(7):1641–6.
A Meyer, J.D., N Kelly, H Kandula, M Kanakasabapathy, P Thirumalaraju, C Bormann, H Shafiee, Can deep convolutional neural network (CNN) be used as a non-invasive method to replace Preimplantation Genetic Testing for Aneuploidy (PGT-A)? . Human Reproduction, 2020. 35 1238.
M Kanakasabapathy, C.B., P Thirumalaraju, R Banerjee, H Shafiee, Improving the performance of deep convolutional neural networks (CNN) in embryology using synthetic machine-generated images. Human Reproduction, 2020. 35 1209.
Dimitriadis, C.L.B., M.K. Kanakasabapathy, P. Thirumalaraju, R. Gupta, R. Pooniwala, I. Souter, S.T. Rice, P. Bhowmick, H. Shafiee, Deep convolutional neural networks (CNN) for assessment and selection of normally fertilized human embryos. Fertility and Sterility. 112 272.
Prudhvi Thirumalaraju, M.K.K., Charles L. Bormann, Hemanth Kandula, Sandeep Kota Sai Pavan, Divyank Yarravarapu, Hadi Shafiee, Human sperm morphology analysis using smartphone microscopy and deep learning. Fertility and Sterility, 2019. 112(3) 41.
Holmes R, et al. Comparison of electronic versus manual witnessing of procedures within the in vitro fertilization laboratory: impact on timing and efficiency. F S Rep. 2021;2(2):181–8.
Rienzi L, et al. Failure mode and effects analysis of witnessing protocols for ensuring traceability during IVF. Reprod Biomed Online. 2015;31(4):516–22.
Forte M, et al. Electronic witness system in IVF-patients perspective. J Assist Reprod Genet. 2016;33(9):1215–22.
Hur YS, et al. Development of a security system for assisted reproductive technology (ART). J Assist Reprod Genet. 2015;32(1):155–68.
Perrin RA, Simpson N. RFID and bar codes–critical importance in enhancing safe patient care. J Healthc Inf Manag. 2004;18(4):33–9.
Sato T, et al. Radiofrequency identification tag system improves the efficiency of closed vitrification for cryopreservation and thawing of bovine ovarian tissues. J Assist Reprod Genet. 2019;36(11):2251–7.
Fiocchi S, et al. Temperature increase in the fetus exposed to UHF RFID readers. IEEE Trans Biomed Eng. 2014;61(7):2011–9.
Aitken RJ, et al. Impact of radio frequency electromagnetic radiation on DNA integrity in the male germline. Int J Androl. 2005;28(3):171–9.
Rienzi L, et al. Comprehensive protocol of traceability during IVF: the result of a multicentre failure mode and effect analysis. Hum Reprod. 2017;32(8):1612–20.
Funding
This work was partially supported by the Brigham Precision Medicine Developmental Award (Brigham Precision Medicine Program, Brigham and Women’s Hospital), Innovation Evergreen Fund (Brigham and Women’s Hospital), Partners Innovation Discovery Grant (Partners Healthcare), and R01AI118502, R01AI138800, R61AI40489, and 4U54HL119145-08 (National Institute of Health).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Charles L. Bormann, Manoj Kumar Kanakasabapathy, and Prudhvi Thirumalaraju. The first draft of the manuscript was written by Karissa C. Hammer and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Informed consent was obtained from each individual before participation. Study protocols were approved by the Institutional Review Board (IRB#2017P001339) at Massachusetts General Hospital and Brigham and Women’s Hospital.
Consent for publication
Not applicable.
Conflict of interest
Authors Dr. Hadi Shafiee, Dr. Charles Bormann, Prudhvi Thirumalaraju, and Manoj Kumar Kanakasabapathy wish to disclose a patent, currently licensed by a commercial entity, on the use of AI for embryology (US11321831B2). The rest of the authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Karissa C. Hammer and Victoria S. Jiang are co-first authors.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hammer, K.C., Jiang, V.S., Kanakasabapathy, M.K. et al. Using artificial intelligence to avoid human error in identifying embryos: a retrospective cohort study. J Assist Reprod Genet 39, 2343–2348 (2022). https://doi.org/10.1007/s10815-022-02585-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-022-02585-y