Abstract
The experimental results of the present study show that phenol had a certain inhibitory effect on its utilization by phenol-acclimatized activated sludge (PAAS), which was the inoculum for the microbial fuel cell (MFC) inoculation. In co-metabolic study, utilization of para-nitrophenol (PNP) at low concentrations (< 20 mg L−1) when used with phenol at a fixed concentration (250 mg L−1) favorably proceeded. The behavior was interpreted in terms of carbon catabolite repression (CCR), indicating phenol (250 mg L−1) positively affected consumption of PNP (< 20 mg L−1). The calculated values of degradation rate show the necessity of phenol presence in the system where phenol acted on the inoculum’s ability to withstand the inhibitory effect of PNP. The MFC functionality in electricity generation is also definable by considering CCR applicability and the results show that the negative effect of PNP was repressed by the presence of phenol. For instance, 20 mg L−1 PNP + 250 mg L−1 phenol yielded the highest power density (66.2 mW m−2) and the lowest internal resistance (189 Ω). The PAAS performance was characterized to evaluate cells’ capacity in utilizing inhibitory substrates, and several different models were used. The relevant kinetic parameters are described in terms of PAAS affinity toward the substrate (ks) and the microbe’s sensitivity in responding to the toxic substrate (ki). Luong and Aiba equations were chosen to describe MFC behavior when the PAAS utilized phenol as the sole substrate. While Haldane model was more capable of addressing co-metabolic degradation of PNP.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
A huge amount of organic/inorganic pollutants are produced by industries, while agricultural practices and lifestyles in different societies result in the production of a considerable amount of wastes, which increase in a threatening manner [1, 2]. In handling industrial effluents, a wide range of technological approaches were suggested in the literature, and the first thing to consider is to determine the physical properties of the contaminant(s) of concern and to identify/understand the path of pollutant production [2, 3]. One should be able to predict the consequences of executing a particular treatment focusing on human toxicity and eco-toxicity [4]. Interest in using adsorption technology among different methods, for instance, is rather low because the process limited capacity for pollutant removal, and difficulties are observed in handling problems generated by the solids remaining in the system [5]. Other physicochemical methods, such as photo-degradation, advanced oxidation process, chemical coagulation, and precipitation, experience drawbacks in terms of high operation costs, system’s maintenance requirements, and generation of secondary pollutants probably at higher levels of toxicity [3]. Anaerobic wastewater treatment is explainable on the basis of biological wastewater technologies, and the main concern for replacing the aerobic systems with anaerobic treatment is to control the quality of effluent at different stages, especially for micropollutant degradation and removal [6].
Great attention in recent years were directed toward the bioelectrochemical (BEC) behavior of molecules focusing on devices, such as microbial fuel cells (MFC) in treating organic/inorganic pollutants where the operative design in these reactors relies on the conversion of energy released biochemically to electrical energy [7,8,9]. The BEC concept relies on catalytic functionality of microorganism(s) in performing a series of enzyme-mediated reactions starting from pollutant oxidation [8, 10]. Thus, the release of electrons and H+ ions of pollutants through the microbe’s enzymes plays a decisive role. The released electrons in the oxidative portion of the process interact with the anode electrode, and they enter the electrical circuit while H+ ions pass via an appropriate polymeric membrane and enter the cathode chamber in which both participate in O2 reduction (air cathode) [8, 10]. The process is thermodynamically favorable because of the redox potential values involved in this type of reaction (applicability of Nernst equation) [11]. Microbial cells, which are used as the inoculum for the MFC inoculation, multiply and increase biomass covering the surface of the anode electrode, and these processes facilitate cell acclimatization. Further note in MFC performance is to follow its role in decreasing pollution in terms of the anodic contaminant used by microorganism(s) (chemical oxygen demand ‘COD’ as the resultant report). These organic pollutants are, in fact, growth-inhibitory compounds where inhibition is concentration-dependent. Growth inhibitory behavior of organic contaminants was kinetically modeled, and Haldane, Aiba, and Luong are among popular equations developed based on the applicability of enzyme inhibition concept in describing unstructured growth models in batch experiments [12]. The applicability of the aforementioned kinetic models in a wide range of studies on wastewater treatments was shown well in the literature [13,14,15,16,17].
Irregularity in the microbe’s behavior in the biodegradation process can be seen when the microbe unexpectedly expresses some uncommon response in degrading two organic contaminants simultaneously (co-metabolization phenomena) [18]. Microbe, in addition to having the ability to degrade and utilize growth-supportive substrate (primary substrate), can also respond to second pollutant as a co-metabolic substrate by synthesizing coenzyme/enzyme [18,19,20]. In co-metabolism study, the microbe’s ability to respond to the primary substrate can be improved through an acclimatization process in which the microbe can adjust its metabolic activities, and in this manner, the cells could maintain functionality toward the co-metabolic substrate [18]. For instance, the aerobic growth of Rhodococcus sp.RSP8 on binary substrate systems containing phenol (5–1600 mg L−1) and p-chlorophenol (5–250 mg L−1, p-CP) was explained in terms of co-metabolization phenomena [16]. The inhibitory effect of p-CP as a co-metabolic substrate was stronger than that of the phenol as growth substrate on p-CP and kinetics equations were developed while the Haldane-type kinetic model was suggested for this relationship characterization [16]. The inhibitory effects of trichloroethylene (TCE) as a co-metabolic substrate on phenol degradation by Pseudomonas putida were also reported [14]. The degradation was inhibited at high concentrations of TCE (> 6 mg L−1). The system also experienced biomass loss in this inhibitory condition and recovery of the biomass after one run experiment in the absence of TCE was observed. The researchers found recovery was not as good as recovery of the system when TCE at lower concentrations (< 2 mg L−1) was used [14]. It appears that biomass was spent by the microbial cells in response to the toxicity of TCE in the absence of the phenol as growth substrate. Utilization of non-growth substrate in the presence of the growth substrate at a certain concentration is prevailed readily. However, growth substrate at other substrate levels expresses inhibitory effects on the utilization rate of the non-growth substrate [18].
Para-nitrophenol (PNP) is a phenolic compound, and its toxicity among a wide range of nitroaromatics, has placed it as a priority pollutant in the list of the US Environmental Protection Agency (USEPA) where recommended level of PNP presence in natural water is 10 ng L−1 [19]. The presence of nitroaromatic compounds was reported in many industries involved in producing a wide range of chemicals, such as pesticides, plastics, dyes, explosives, and pharmaceuticals [5]. PNP removal via environmentally friendly technique(s) is highly desirable for instance, attempts were directed to combine aqueous extracts obtained from leaves of Cucumis sativus and Aloe vera with silver ions to make silver nanoparticles [21]. The results of this plant-mediated nanotechnology were highly favorable in terms of PNP degradation and conversion to para-aminophenol (PAP) [21]. A further note on the degradation of PNP has been described on the basis of the test system’s BEC behavior in terms of cathodic reduction of PNP coupled to anodic oxidation of PAP [22]. Analysis was also focused on identifying the involved microbial community which is linked to the particular bacterium and relevant genes being-active in the degradation process [22].
The findings of aforementioned studies have led us to find answers to a new question about the applicability of anaerobic co-metabolism in PNP degradation based on the substrates’ BEC behavior. Efforts in the present study thus were directed toward co-metabolic biodegradation of PNP plus phenol in a laboratory-made MFC reactor (air cathode) using an industrial activated sludge (AS). Kinetic modeling in co-metabolic biodegradation of PNP was evaluated using Haldane, Aiba, Webb, Edwards, and Luong models which were developed based on the growth inhibitory effect of the involved substrates. By applying nonlinear regression concept, data fitting process were carried out and predictive ability of the model was evaluated using different error criteria: R2 (coefficient of determination), χ2 (chi-square), RMSE (root mean square error), and AIC (Akaike Information Criterion). BEC behavior of the MFC reactor was characterized in terms of electrical energy produced bio-chemically. Results expressed in polarization curves were informative and facilitated quality determination of the MFC system in terms of energy output: current density, power density, and Coulombic efficiency were calculated.
2 Materials and methods
2.1 Materials
Phenol, PNP, and mineral salts (Na2HPO4, NaH2PO4.H2O, NH4Cl, KCl) used for the preparation of the synthetic type of wastewater were provided by the local suppliers. Merck reagents used for spectrophotometric determination of COD were purchased locally. The proton exchange membrane (PEM) as Nafion 117 was purchased from the market (DuPont Co., USA) and detail of its treatment is given elsewhere [23]. Removing any organic present in the membrane (immersing in H2O2 solution for 1 h), facilitating hydrogen ions formation from the substrate oxidation (acid washing), and removing residual acid remaining from the previous step (soaking in distilled water, 1 h). Rod-shaped electrodes were graphite in nature with 50 cm2 as the surface area and acquired from the local supplier. Hydration of the electrodes in distilled water for 24 h appeared to be sufficient for the electrode cleaning process [24].
2.2 Acclimation and inoculation processes
A certain volume of the activated sludge (AS) produced in the biological treatment of wastewater of the gas refinery plant located in the southern region of Iran (South Pars Gas Complex), was poured into a large flask (1 L). Large-sized particles and other impurities were separated, and to provide an anaerobic environment, nitrogen sparging through liquid was practiced while the stirring solution was with use of magnetic stirrer. Thick sediment was settled, and mineral salts (4.57 g L−1 Na2HPO4, 2.45 g L−1 NaH2PO4.H2O, 0.31 g L−1 NH4Cl, 0.13 g L−1 KCl), along with phenol as the carbon source for the growth of AS were added to that container (1: 2 v v−1). Phenol at a range of concentrations was used in the acclimation process which was performed at room temperature, where 50 mg L−1 phenol was the first level used, and its decreasing trend was monitored (phenol concentration approached zero level after 24 h). The capacity of AS to consume phenol was characterized by measuring phenol and COD contents at time interval. The slurry at the end of the experiment was let to settle (~ 3 h), and 200 mL from the clear solution at the top of the slurry was removed and replaced with prepared mineral salts plus phenol at a higher concentration (150 mg L−1). The aforementioned process was repeated until 550 mg L−1 of the phenol substrate, where the bacterial mixture was not capable to appropriately degrade phenol, and COD level did not decrease considerably (see Sect. 3.1). The phenol-acclimated activated sludge (PAAS) was transferred to the MFC reactor and the relevant experiments were performed.
2.3 MFC configuration and operation condition
The configuration of a two-chamber MFC as a laboratory-made reactor is shown in Fig. 1, and details of experimental setup along with relevant operations are briefly described as follows. With use of Pyrex glass bottles, the anode and cathode chamber were assembled where the capacity of each compartment was 800 ml, and they were connected to each other via a glass tube having internal diameter and length of 5 and 7 cm, respectively. Electrodes in both compartments were rod-shaped graphite and were 15 cm apart from each other.
The PAAS culture along with the growth medium containing 250 mg L−1 phenol as carbon and energy source were added to the anode chamber (1: 2 v v−1) and by N2 sparging into anolyte (5 min), the oxygen-free environment was provided in the anode compartment. The cathode chamber was filled up with phosphate buffer (100 mM, pH 7.2), and the catholyte was continuously aerated with an aquarium pump. Solutions in both chambers were stirred using magnetic stirrer.
Further note was to ensure the formation of uniform and healthy biofilm thus, open circuit condition (OCC- absence of current) was applied, and when voltage dropped below 100 mV during the process of forming biofilm, the growth medium including phenol was refreshed. Thereafter, by applying closed circuit condition (CCC-at 1 kΩ as the external resistance) capacity of the PAAS biofilm in utilization of phenol was examined (50–550 mg L−1) and the degradation rate in terms of phenol substrate was determined. With the same external resistance (1 kΩ), the biomass capacity for PNP consumption was also quantified (15–60 mg L−1) where no degradation rate could be determined and these findings confirmed the necessity of phenol presence along with PNP, and co-metabolic experiments were carried out as described in the following section. At certain time interval, the sample was taken from the sampling port, which was placed on the top of the anode chamber, and the content of phenol, PNP, and COD was quantified. The performance of MFC reactor in terms of voltage, current, and kinetic modeling was characterized.
2.4 Analytical procedures
The volatile fraction of Mixed liquor volatile suspended solids (MLVSS) is a measure of the microbial cells present in wastewater treatment and the anolyte in the present study is a synthetic type of wastewater, and by measuring MLVSS, one can estimate microbe’s content. In the present study, an appropriate aliquot was taken from the anolyte and the MLVSS was measured according to the standard method as described in the literature [25]. With use of Merck reagents, and following the instructions presented by the manufacturer, and with use of spectrophotometer (JascoV-550 UV–Vis Spectrophotometer), the content of COD was determined. Phenol content of the test solutions was determined spectrophotometrically using Foloin-Ciocalteau reagent [26]. The formation of a blue chromophore, composed of a phosphotungstic-phosphomolybdenum complex is the result of the Folin–Ciocalteau reaction, and the absorbance was measured at 760 nm. PNP concentration also was measured spectrophotometrically, the absorbance of the yellow color developed as the result of PNP reduction at pH > 8 (formation of relevant aminophenol) was read at 400 nm [19]. At the end of treatment, scanning electron microscopy (SEM-AIS2300C, Seron Technology, South Korea) was used to take images from the anode surface, and the following procedure was used to prepare the sample: fixation with glutaraldehyde, washing with a phosphate buffer solution (100 mM, pH 7), the hydration series with aqueous solution of ethanol, and drying at 100 ˚C [27]. Sputter coating process (SC7620 Mini Sputter Coater, Quorum Technologies, United Kingdom) was applied and a thin film of gold was deposited on the sample surface.
2.5 Kinetic modeling
The following Eqs. (1)-(5) are five kinetic models used in the present study to determine degradation rate in the MFC reactor:
where q and qmax are, respectively, specific degradation rate and maximum specific degradation rate (mgsubstrate gVSS−1 h−1); S, ks, ki, and K are initial substrate concentration, half saturation constant, substrate inhibition constant, and Webb constant (mg L−1), respectively. Sm and n represent maximum substrate concentration above which degradation is totally inhibited (mg L−1) and curve parameter, respectively [12, 13].
The values of specific degradation rate (q ‘mgsubstrate gVSS−1 h−1’) are calculated for each initial phenol (50–550 mg L−1) and PNP (15–60 mg L−1) by Eq. (6) as follows:
where x as the biomass content refers to MLVSS (g L−1).
Four error criteria were used to assess the model adjustment to the experimental data, and in this manner, predictive quality of the model was determined. The error criteria are named as follows: R2 (coefficient of determination), χ2 (chi-square), RMSE (root mean square error), and AIC (Akaike Information Criterion). The values of error criteria are calculated using Eqs. (7)–(10):
Akaike Information Criterion [28]:
where yi, \({\widehat{{\text{y}}}}_{{\text{i}}}\), and \(\overline{{\text{y}} }\) are, respectively, the value of response variable experimentally determined, model calculated value for response variable, and the average of observations; n, MSE, and k are number of data points, mean square error, and number of model parameters, respectively.
Reducing the prediction error relies on use of least square technique, as given by Eq. (11):
where ei is the error content.
Focusing on nonlinear nature of the kinetic-based models used in the present study, attempts were directed toward the application of nonlinear regression to minimize the error (ei) associated with the process of kinetics modeling. Gradient descent method is one way used by Levenberg–Marquardt algorithm where iterative efforts are to reach lower and lower value for ei (to minimize the sum of squares of the differences between predicted and measured values) (OriginPro software version 8.6).
2.6 Bioelectrochemical data analyses
Voltage (V) and current (I) were determined considering the Ohm’s law as in Eq. (12) using digital multimeter (MT- 1860 ProsKit Co., Taiwan):
where Rex denotes external resistance (Ω).
Under OCC when no current is in the system the voltage was determined (open circuit voltage ‘OCV’). Under CCC, and by applying external resistance the current was calculated. The following Eqs. (13) and (14) were used for determining current density (CD ‘mA m−2’) and power density (PD ‘mW m−2’):
where A is the surface area of anode (m2).
The polarization curves for phenol (as the sole source of carbon and energy for the PAAS) and PNP + 250 mg L−1 phenol were obtained under CCC environment. In each plot of voltage vs. current density, the internal resistance was calculated from the slop of the linear portion of plot using single-cycle method applying different resistors (100 Ω-30 kΩ). A short time (20–30 min) in each case was needed for the system’s stabilization [10]. The Coulombic efficiency (CE ‘%’) was also calculated using Eq. (15):
where ΔCOD is the difference in COD content between the initial and the final of liquor in the anode chamber. The volume of test liquor is shown by V, 4 is the number of mole of one mole O2 related electrons, 32 is the molecular weight of O2, and the Faraday constant is 96,485 (C mole−1).
3 Results and discussion
3.1 Strategy of MFC operation
Efforts in the first part of the experiments were directed toward acclimatization, which increases the chance of biomass formation with greater tolerance and the ability to withstand toxic compounds. Thus, the response to the presence of growth-inhibitory substrate is to find out how bacterial culture can manage its metabolic activities and express favorable reactions, such as PAAS performance in the degradation of phenol and PNP under anaerobic condition and higher chance was for the PAAS to express bioelectrochemical behavior in a satisfactory manner.
3.1.1 Extent of phenol utilization by the AS during the acclimation process
Microorganisms in the AS used in the present study as the inoculum for the inoculation of MFC, like other ASs are a mixture of aerobic/anaerobic bacteria, and other microorganisms, needed to be anaerobically acclimatized to phenol as carbon and energy source. For determining the microbial composition of activated sludge which is obtainable from different wastewater treatment systems (WWTSs), 16S rRNA gene sequencing technology is the common technique where the presence of Proteobacteria as a major phylum of Gram-negative bacteria in these WWTSs was found to be considerable [29,30,31]. Abundance of Pseudomonas genus which is within Gammaproteobacteria class in these studies is interesting where this genus known as phenol degrader under aerobic and anaerobic conditions, both [32]. The complexity of cell physiology and energy production under anaerobic condition have made bacterial cells respond by developing different regulatory mechanisms and rely on necessary coordination between related cellular parts. Some cellular molecules act as redox signals, and their functionalities are under the effect of environmental stresses (such as oxygen shortage and aromatic substrates) [33, 34]. The signals control genes necessary for the cell adjustment to a new environment (change from oxic to anoxic condition, for instance), and Pseudomonas aeruginosa and Pseudomonas putida among Pseudomonas genus are those species that their performances under oxygen-limited conditions are studied in detail [34, 35].
According to the literature, a brief description of phenol biodegradation under anaerobic condition has been given in Fig. 2 where the formation of hydroxybenzoyl-CoA and removal of aromatic hydroxyl group reductively are the essence of this anaerobic degradation process [36].
According to the literature, a brief description of phenol biodegradation under anaerobic condition has been presented where three enzymatic reactions are highlighted: (1) phenol carboxylase, (2) 4-hydroxy benzoyl CoA ligase (dehydroxylating enzyme), and (3) benzoyl CoA reductase. Products are ultimately formed through ring cleavage
Flask study results of the phenol-acclimatized activated sludge (PAAS) presented in Table 1 show that when the initial phenol concentration increased to a level above 250 mg L−1 the time for observing efficient phenol removal during the acclimation experiments increased by 1½ to 4 times. By increasing the concentration from 250 mg L−1 to 550 mg L−1, the level of COD removal decreased to about 25% (Table 1).
A further experiment was to evaluate the catalytic performance of the PAAS as the inoculum for the MFC inoculation where phenol at 250 mg L−1 was added to anode compartment, and voltage generation trend under open circuit condition was followed. Plot of the voltage against the operation time is presented in Fig. 3 where 87 mV as the initial voltage increased to 340 mV within 4½ days. The anolyte was refreshed when the value of voltage under OCC decreased to about 100 mV, i.e., the system’s refreshing was with use of phenol solution having 250 mg L−1 concentration (Fig. 3). The highest OCV value obtained in the present work was 566 mV which is close to 658 mV reported for inoculation of a single chamber MFC with P. aeruginosa [37]. The PAAS’s behavior as the phenol-acclimated microbes under OCC is to express growth and able to multiply where these cells efforts eventually direct their movements toward the anode electrode, and the cells coverage of anode surface is evidenced by the SEM images presented in Fig. 4. Decisive role of anode biofilm in performance of MFC was well described in literature for instance, stacking MFC indicates that efficient formation of biofilm is important and explainable in terms of hydrodynamic property of the anolyte in anode chamber [38]. Actually, water which is the major constituent of the anolyte moves around the anode electrode and the extent of movement depends on the anolyte feeding regime (batch or continuous) [38]. Hydrodynamic characteristics of anolytes and the feeding regime are factors that positively affect the voltage and current density of the MFC which have been studied in some detail by researchers [38].
Recent findings show that P. putida can be categorized as the electroactive bacterium (EAB) and has been placed in EAB group where membrane appendages, such as nanowires and redox mediator molecules are assigned to EABs as cellular tools which efficiently participate in extracellular electron transportation (EET) process (direct and indirect EET) [39]. The presence of Geobacter sp. as electroactive bacteria in different inocula used for the MFC inoculation was reported and the importance of G. Sulfurreducens which obtained from domestic wastewater, lake sediment, and biogas sludge was well shown [40]. Interesting point is a complex relationship which exists between Geobactere and Pseudomonas species which termed syntrophy process where these species both cohabit same environments [41, 42]. Thus, it appears that the presence of Pseudomonas genus in the microbial composition of the PAAS could be a reasonable expectation in the present study.
3.2 MFC performance: PAAS catalytic behavior in degradation of phenol and PNP
Results of biodegradation experiments in the MFC reactor are presented in Fig. 5 where initial concentrations of phenol ranged from 50 to 550 mg/L. The degradation of phenol at 250 mg L−1 initial concentration was complete within 44 h, and less than 1% of COD remained in the test system. The performance of the PAAS in catalytic utilization of phenol at high concentrations (> 250 mg L−1) was inefficient. In fact, 80% of 250 mg L−1 phenol was degraded within 24 h while at this period of time 80% of 550 mg L−1 initial concentration of phenol remained in the system. The later phenol concentration was partially degraded within 157 h. Thus, 250 mg L−1 phenol was chosen as the concentration efficient in degradation by the PAAS in the MFC system. The PAAS culture was not capable of degrading PNP as the sole substrate (15–60 mg L−1). Criddle team has heavily been involved in co-metabolic, and published articles are informative in the fundamental and basics of co-metabolization and Criddle differentiated between growth- and non-growth substrates in providing electron and H+ ions used for the following situations: growth and energy production during the growth phase, cells synthesis, and to support co-metabolism using biomass to decrease maintenance burden on the cells [18]. In order to assess co-metabolic degradation of PNP by the test culture, the MFC experiments were carried out using PNP plus a fixed concentration of phenol (250 mg L−1 as the chosen concentration). Figure 6 illustrates the relevant results. When the initial concentration of PNP was set at 20 mg L−1, the PAAS could utilize 70% of the phenol within 12 h while at the same period of time, only 14% of 20 mg L−1 PNP was degraded. In fact, the extent of the PNP utilization by the PAAS depends on the phenol removal capacity of the culture and as shown in Fig. 6, 76% of the PNP was removed within 96 h while phenol degradation was complete within 48 h. The extent of organic matter removal, describable as COD content, was significant in the experiment (20 mg L−1 PNP + 250 mg L−1 phenol), and a 97% reduction of COD occurred within 96 h. In comparing between the consumption of phenol (250 mg L−1) and PNP (< 30 mg L−1) along with COD reduction by the PAAS, the culture appeared to sense metabolically the selected concentration of phenol as an easily utilizable carbon source. The culture was less capable of degrading PNP at 30 mg L−1 or higher concentrations and removal efficiency remarkably decreased for 30–60 mg L−1 of PNP. PNP at the later concentration range negatively affected phenol utilization, which is definable as a repressor compound. It means that the degradation of PNP plus phenol was incomplete, and 80% of 60 mg L−1 PNP remained in the system after 228 h, and only 70% of the phenol was degraded within the same period of time. The relationship between phenol and PNP can be explained by carbon catabolite repression (CCR), and reverse CCR (rCCR) concepts described as auto-regulatory mechanisms where utilization of a secondary source of carbon/energy which slowly being used by a particular bacterium is under the control of rapidly metabolizable carbon source (diauxy phenomenon discovered by Monod around 1940) [43]. The rCCR as a metabolic strategy found to be operated as almost opposite to CCR [44]. Several studies reported the presence of a secondary substrate as an external carbon source [45]. These studies have also discovered that this source has an inhibitory effect on PNP degradation by some PNP-degrading microbes, such as Arthrobacter sp. [45]. Besides, glucose by some researchers is termed as an external carbon source as the preferred substrate which was non inhibitory for PNP degradation by strains, such as P. putida 1274 [45]. The applicability of CCR and rCCR concepts in the aforementioned study was not considered for their results interpretation. Study on P. putida DLL-E4 showed that several genes and their upregulation/downregulation are heavily involved in the aerobic degradation of PNP by the bacterium which is known as an efficient PNP degrader [46]. The occurrence of CCR and rCCR between phenol and PNP during co-metabolic biodegradation in the anode compartment of MFC using phenol-acclimated activated sludge were not reported in the literature, and data presented in the present work support the applicability of these mechanisms.
3.2.1 Kinetic modeling
A common approach in handling data over substrate inhibition biodegradation as time dependency of substrate utilization by a particular culture is to apply the Monod equation. But that model is less capable of quantifying bacterial growth utilizing toxic substrates, such as phenol and PNP. Based on the substrate inhibition concept, several growth models were developed by the well-recognized researchers and published in the literature [12]. Further point to address is necessity of considering kinetics of reactions catalyzed enzymes or Michaelis–Menten equation with reference to enzyme inhibition subject. By considering behavior of bacterial cells as the receiver of substrate not being growth supportive, mathematical attempts in the present study were directed to describe biodegradation rate of phenol and PNP substrates in the MFC reactor. Determination of specific degradation rate, q (mgsubstrate gVSS−1 h−1), is the first step in studying kinetics of degradation of phenol and PNP. With use of Eq. (6), q values were calculated for phenol (50–550 mg L−1) and for PNP (15–60 mg L−1) concentrations which are shown in Fig. 7 and Fig. 8. Gradual increase of qs is with steady increase of phenol and PNP concentrations at certain range. However, the specific degradation rate decreases at high concentrations of phenol and PNP. Inhibitory effect of phenol on its degradation rate was evident at high concentrations (> 250 mg L−1), and the results are in agreement with the findings on aerobic removal of phenol and p-chlrophenol by Rhodococcus sp.RSP8 as reported by Sinha et al. [16]. Findings on co-metabolic degradation of trichloroethylene (TCE) and phenol by P. putida showed that loss of biomass occurred at high concentrations of TCE (> 6 mg L−1) [14]. In the present study, the low values of qs at high concentrations of PNP can be indicative of biomass loss, i.e., growing cells requirements provided by growth substrate was spent [14, 18].
Fitting the models to experimental data (specific degradation rate vs. initial phenol substrate concentration) where phenol at a range of 50 to 550 mg L−1 was used solely in the MFC reactor. The quality of model prediction which is shown underneath of the figure was determined with use of R2, χ2, RMSE, and AIC
Fitting the models to experimental data (specific degradation rate vs. initial PNP substrate concentration) where PNP at a range of 15 to 60 mg L−1 was co-metabolically degraded in the presence of 250 mg L−1 phenol. The quality of model prediction which is shown underneath of the figure was determined with use of R2, χ2, RMSE, and AIC
In the present study, substrate inhibition models presented in Table 2 were used to describe the experimental data obtained in the form of the degradation rate and results of data fitting process are given underneath of Fig. 7 and Fig. 8. The difference between model predicted value and experimentally measured value characterized as the error term, was determined using Eqs. (7)–(10). The findings of the present study show preference of Aiba equation for describing kinetics of phenol biodegradation by the PAAS culture (R2 = 0.98, χ2 = 0.06, RMSE = 0.24, and AIC = − 11) (Fig. 7). While Haldane model was more capable in addressing co-metabolic degradation of PNP + 250 mg L−1 phenol (R2 = 0.99, χ2 = 6.5E − 4, RMSE = 0.002, and AIC = − 79) (Fig. 8). Ks in case of inhibitory substrate, is definable as half-saturation constant for maximal degradation rate and ki is the inhibition constant as concentration of inhibitor needed to reach half maximal inhibition. The predicted values of kinetics constants are presented in Table 2.
The ks constant of the Aiba kinetic model reported in the present study, is relatively small value and as stated in the literature, ks values obtained under anaerobic conditions are higher than those obtained aerobically [47]. The acclimatization process, which was performed in the present study, was apparently effective and the degradation of phenol alone in the range of 50–250 mg L−1 proceeded favorably. Aerobic treatment of the phenolic wastewater is characteristic of many studies reported in the literature for instance, ks calculated according to Haldane model, was equal to 603.8 mg L−1 using the aerobic sequential batch reactor (SBR) inoculated with the acclimatized activated sludge [48]. In a similar approach, SBR system was used for treating phenolic wastewater and the ks obtained according to Haldane equation was equal to 692 mg L−1 [15]. The high value of ks indicates that substrate is less supportive for the microbe’s performance.
The inhibition constant calculated according to the Aiba model as reported in the present study is high value (Table 2) close to ki value (890 mg L−1) reported for degrading of phenol content of the coke oven wastewater in a shake flask experiment using Pseudomonas citronellolis NS1 [49]. The high ki value shows that microbial culture is less susceptible to the inhibitory substrate and the substrate is less likely to express its inhibitory effect. The low ratio of ks to ki is the result of high value of ki and moderately low value of ks. Thus, it is reasonable to us ks/ki ratio, as suggested in the literature [15], to show the extent of inhibition described by kinetic models. In the present study, with reference to Aiba and Haldane models applicable for phenol biodegradation and for co-metabolic degradation of PNP, the values for ks/ki are as follows: (ks/ki)Ai = 0.06 and (ks/ki)Ha = 0.4. Use of Luong equation gives an opportunity to predict substrate concentration above which culture growth and substrate degradation cease. The values of Sm for phenol and PNP are 605 and 73 mg L−1, respectively (Table 2). The lower value of ks/ki and the higher value of Sm for phenol indicate that the PAAS could degrade phenol readily and the culture was less capable of utilizing PNP.
Further note in biodegradation process is to recognize importance of microorganism(s) types (mixed culture or pure form) and microbe’s living status for instance, P. citronellolis NS1 isolated from coke oven wastewater obtained from the iron and steel plant industry, was able to degrade 1500 mg L−1 of phenol within 90 h [49]. The setup in the aforementioned study was operated aerobically and the biodegradation pattern was modeled according to the suggested by Pamukoglu and Kargi [49]. The acclimatized activated sludge used for phenolic wastewater treatment resulted in ki = 28 mg L−1 using Haldane kinetic model [50].
Aerobic or anaerobic operations of wastewater treatment systems play a determining role in biodegradation studies. Mathematical models have been extensively used to describe behavior of microbe(s) in degradation of inhibitory substrate(s). By considering available published articles, it is time to view things differently and try to gain new knowledge that could ease understanding different approaches used in solving problems usually associated with inhibitory substrate in terms of biodegradation rate.
3.3 MFC behavior: power output in terms of polarization curves
By considering MFC as a power source, then decisive role of this system is explainable as how it is possible to reach a maximum power output. Several factors cooperatively affect MFC performance where cathode modification, substrate type, and concentration as related to the involved microorganism(s), occupy unique position. Enzyme catalyzed reactions which performed by the microbes on the substrate in the anodic region, are behaved in a manner that the biooxidation reactions provide energy not only for the cells survival and multiplication, but also for generation of bioelectricity (as the result of cells attachment on anode electrode and entrance of the released electrons to the electrical circuit as mentioned above). The electrons through electrical circuit eventually reach the cathode electrode. Effect of air cathode modification on power output in MFC was studied in some details by Liu and Liu [51] when nitrogen doped activated carbon prepared to be used as coated material on the air cathode and the power density at 1026 mW m−2 was recorded. The aforementioned value apparently was comparable to platinum when used as a catalyst in the MFC reactor [51]. Further findings were based on an increase in anode to cathode surface area ratio (Aan/Aca) from 0.4 to 1, which decreased internal resistance in this published paper [7]. Although the optimum level of power output in that research was at Aan/Aca = 0.6 [7].
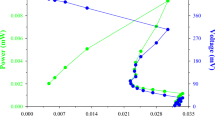
Curve of voltage change with current density is an important electrochemical technique used in evaluating of the performance of involved system of fuel cell and, Fig. 9 shows the relevant results in the present work. A decrease of voltage from the highest point in the polarization curve is expected and explainable in terms of activation loss where microbial enzymes efficiently participate in catalytic pathway in transferring substrate’s electrons to the acceptor molecules. The low capacity of microbes in responding to secondary metabolites which are generated through the enzymatic processes, exert tension to the cells including resistance to electrolyte composition and charge transfer processes. Thus, the values of internal resistance for phenol and PNP + 250 mg L−1 phenol were calculated which are given in Table 3. Response of the PAAS culture to 250 mg L−1 phenol was favorable, but when phenol concentration increased beyond this level and reached 550 mg L−1, Rint increased substantially (181 vs. 692 Ω) (Table 3).
Electron transferring quality of MFC can be quantified in terms of Coulombic efficiency (CE), and with use of Eq. (15), the CE values for the different test concentrations were calculated and are also shown in Table 3. The Coulombic efficiency was highest for the system in which phenol at 250 mg L−1 was used where the internal resistance was lowest among six different concentrations of phenol. Table 3 also shows CE and Rint values for the co-metabolic studies and Considerable similarity in terms of internal resistance and CE between 20 mg L−1 PNP + 250 mg L−1 phenol and 250 mg L−1 phenol was observed. Moreover, a significant decrease in CE content was obtained when the system was treated with 60 mg L−1 PNP + 250 mg L−1 phenol (CE = 3%). The energy loss is higher for the system having larger value of Rint and this value for 60 mg L−1 PNP + 250 mg L−1 phenol was highest in the present study (2711 Ω). The results of power density (PD) vs. current density are shown in Fig. 10. The system showed a notable decrease of nearly 90% in the value of PD when 60 mg L−1 PNP was added to the system compared to the value obtained for 20 mg L−1 PNP in the presence of 250 mg L−1 (66.2 vs. 0.84 mW m−2). PNP at concentrations lower than 30 mg L−1 did not affect negatively on phenol (250 mg L−1) utilization by the PAAS culture and by considering CCR as a regulatory mechanism, the behavior may be interpreted in terms of growth supportive role of phenol (i.e., phenol repressed negative effect of PNP and the bioelectrochemical data were appropriately recorded). PNP at the higher levels (≥ 30 mg L−1) negatively affected the consumption of phenol (250 mg L−1) by the PAAS culture and phenol utilization was repressed by PNP (i.e., growth supportive role of the phenol was interpreted reverse) and the results can be interpreted in terms of rCCR.
4 Conclusions
The chance of inefficient competition between utilization of growth and non-growth substrate by bacterial cells is reduced through co-metabolic degradation of the organa pollutant substrates where the approach is to find how exchange of electrons and H+ ions can take place. The acclimatization process was carried out using a phenol-feeding strategy with a certain range of phenol concentrations. The system relied on the utilization of acclimatized culture for the MFC inoculation. Phenol as the sole growth substrate when used at high concentrations (> 250 mg L−1) showed inhibitory effect on the PAAS behavior in the MFC (self-inhibition). By considering CCR regulatory mechanism, utilization of PNP up to 30 mg L−1 by the inoculum was under influence of 250 mg L−1 phenol consumption, i.e., synthesis of the required enzymes for anaerobic degradation of PNP was allowable. However, PNP at high concentrations (≥ 30 mg L−1) negatively affected phenol utilization and remaining phenol in the system could not act as the growth supportive substrate. It appears rCCR mechanism was better for explaining the obtained results. The inoculum culture appeared to be not involved in the selection of the preferred substrate [44].
Further findings in the present study were to describe the biodegradation kinetics, and among several kinetic models, performances of Luong and Aiba (applied for phenol alone) and Haldane (applied for co-metabolic degradation case as PNP + 250 mg L−1 phenol) favorably explained the involved processes in terms of ks, ki, and relevant ratios. With use of Luong equation, one can estimate the substrate concentration usable to support degradation behavior of the PAAS culture above which the culture growth and substrate degradation ceases.
Genetic studies and characterization of the genes involved in an activated sludge culture under operative conditions in an MFC system are necessary steps for future works.
Data availability
The datasets obtained in this study are available on request to the corresponding author.
References
Delforno TP, Belgini DR, Hidalgo K, Centurion VB, Lacerda-Júnior GV, Duarte IC, Varesche M, Oliveira V (2020) Anaerobic reactor applied to laundry wastewater treatment: unveiling the microbial community by gene and genome-centric approaches. Int Biodeterior Biodegrad 149:104916. https://doi.org/10.1016/j.ibiod.2020.104916
Sultana S, Choudhury MR, Bakr AR, Anwar N, Rahaman MS (2018) Effectiveness of electro-oxidation and electro-Fenton processes in removal of organic matter from high-strength brewery wastewater. J Appl Electrochem 48:519–528. https://doi.org/10.1007/s10800-018-1185-3
Wang Y, Jiang X, Yang H, Li W, Mu Y, Shen J (2020) Degradation and mineralization of recalcitrant compounds in bioelectrochemical systems. In: Tiquia-Arashiro SM, Pant D (eds) Microbial electrochemical technologies. CRC Press, Taylor & Francis Group, Boca Raton
Arroyo P, Molinos-Senante M (2018) Selecting appropriate wastewater treatment technologies using a choosing-by-advantages approach. Sci Total Environ 625:819–827. https://doi.org/10.1016/j.scitotenv.2017.12.331
Mei X, Liu J, Guo Z, Li P, Bi S, Wang Y, Yang Y, Shen W, Wang Y, Xiao Y, Yang X, Zhou B, Liu H, Wu S (2019) Simultaneous p-nitrophenol and nitrogen removal in PNP wastewater treatment: comparison of two integrated membrane-aerated bioreactor systems. J Hazard Mater 363:99–108. https://doi.org/10.1016/j.jhazmat.2018.09.072
Komolafe O, Mrozik W, Dolfing J, Acharya K, Vassalle L, Mota CR, Davenport R (2021) Occurrence and removal of micropollutants in full-scale aerobic, anaerobic and facultative wastewater treatment plants in Brazil. J Environ Manag 287:112286. https://doi.org/10.1016/j.jenvman.2021.112286
Moradi HG, Mahdavi MA, Gheshlaghi R, Dehghanian M (2023) Electrochemical evaluation of the effect of anode to cathode surface area ratio on power generation in air-cathode microbial fuel cells. J Appl Electrochem. https://doi.org/10.1007/s10800-023-01929-5
Askari A, Vahabzadeh F, Mardanpour MM (2021) Quantitative determination of linear alkylbenzene sulfonate (LAS) concentration and simultaneous power generation in a microbial fuel cell-based biosensor. J Clean Prod 294:126349. https://doi.org/10.1016/j.jclepro.2021.126349
Indriyani YA, Rusmana I, Anwar S, Djajakirana G, Santosa DA (2023) Bioelectrochemical assessment of a novel electrogenic Bacillus altitudinis AC11. 2 for electricity generation in microbial fuel cell (MFC) system. J Appl Electrochem. https://doi.org/10.1007/s10800-023-02020-9
Baniasadi B, Vahabzadeh F (2021) The performance of a cyanobacterial biomass-based microbial fuel cell (MFC) inoculated with Shewanella oneidensis MR-1. J Environ Chem Eng 9:106338. https://doi.org/10.1016/j.jece.2021.106338
Haynie TD (2008) Biological thermodynamics, 2nd edn. Cambridge University Press, Cambridge. https://doi.org/10.1017/CBO9780511802690
Muloiwa M, Nyende-Byakika S, Dinka M (2020) Comparison of unstructured kinetic bacterial growth models. S Afr J Chem Eng 33:141–150. https://doi.org/10.1016/j.sajce.2020.07.006
Wang Y, Meng F, Li H, Zhao S, Liu Q, Lin Y, Wang G, Wu J (2019) Biodegradation of phenol by Isochrysis galbana screened from eight species of marine microalgae: growth kinetic models, enzyme analysis and biodegradation pathway. J Appl Phycol 31:445–455. https://doi.org/10.1007/s10811-018-1517-z
Chen YM, Lin TF, Huang C, Lin JC (2008) Cometabolic degradation kinetics of TCE and phenol by Pseudomonas putida. Chemosphere 72:1671–1680. https://doi.org/10.1016/j.chemosphere.2008.05.035
Kamali M, Gameiro T, Costa ME, Capela I, Aminabhavi TM (2019) Enhanced biodegradation of phenolic wastewaters with acclimatized activated sludge—a kinetic study. Chem Eng J 378:122186. https://doi.org/10.1016/j.cej.2019.122186
Sinha PK, Sinha A, Das M (2011) Microbial removal of phenol and p-chlorophenol from industrial waste water using Rhodococcus sp. RSP8 and its growth kinetic modeling. J Water Resource Prot 3:634. https://doi.org/10.4236/jwarp.2011.38073
Liu X-p (2009) Kinetics of cometabolic degradation of 2-chlorophenol and phenol by Pseudomonas putida. Water Sci Eng 2:110–120. https://doi.org/10.3882/j.issn.1674-2370.2009.03.011
Criddle CS (1993) The kinetics of cometabolism. Biotechnol Bioeng 41:1048–1056. https://doi.org/10.1002/bit.260411107
Maleki M, Motamedi M, Sedighi M, Zamir SM, Vahabzadeh F (2015) Experimental study and kinetic modeling of cometabolic degradation of phenol and p-nitrophenol by loofa-immobilized Ralstonia eutropha. Biotechnol Bioprocess Eng 20:124–130. https://doi.org/10.1007/s12257-014-0593-4
Hazen TC (2010) Cometabolic bioremediation. In: Timmis NK (ed) Handbook of hydrocarbon and lipid microbiology. Springer, Heidelberg
Riaz M, Sharafat U, Zahid N, Ismail M, Park J, Ahmad B, Rashid N, Fahim M, Imran M, Tabassum A (2022) Synthesis of biogenic silver nanocatalyst and their antibacterial and organic pollutants reduction ability. ACS Omega 7:14723–14734. https://doi.org/10.1021/acsomega.1c07365
Luo J, Xu Y, Wang J, Zhang L, Jiang X, Shen J (2021) Coupled biodegradation of p-nitrophenol and p-aminophenol in bioelectrochemical system: mechanism and microbial functional diversity. J Environ Sci (China) 108:134–144. https://doi.org/10.1016/j.jes.2021.02.017
Bagheri M, Daneshvar R, Mogharei A, Vahabzadeh F (2020) Phenol-acclimated activated sludge and Ralstonia eutropha in a microbial fuel Cell for removal of olive oil from mill wastewater. Korean J Chem Eng 37:1233–1240. https://doi.org/10.1007/s11814-020-0538-x
Mohan SV, Saravanan R, Raghavulu SV, Mohanakrishna G, Sarma P (2008) Bioelectricity production from wastewater treatment in dual chambered microbial fuel cell (MFC) using selectively enriched mixed microflora: effect of catholyte. Bioresour Technol 99:596–603. https://doi.org/10.1016/j.biortech.2006.12.026
Bridgewater L, Association APH, Association AWW, Federation WE (2012) Standard methods for the examination of water and wastewater. American Public Health Association, Washington, DC
Box J (1983) Investigation of the Folin-Ciocalteau phenol reagent for the determination of polyphenolic substances in natural waters. Water Res 17:511–525. https://doi.org/10.1016/0043-1354(83)90111-2
Caizán-Juanarena L, Servin-Balderas I, Chen X, Buisman CJ, ter Heijne A (2019) Electrochemical and microbiological characterization of single carbon granules in a multi-anode microbial fuel cell. J Power Sources 435:126514. https://doi.org/10.1016/j.jpowsour.2019.04.042
Gu Y, Wei H-L, Balikhin MM (2018) Nonlinear predictive model selection and model averaging using information criteria. Systs Sci Control Eng 6:319–328. https://doi.org/10.1080/21642583.2018.1496042
Zhu S, Wu H, Wei C, Zhou L, Xie J (2016) Contrasting microbial community composition and function perspective in sections of a full-scale coking wastewater treatment system. Appl Microbiol Biotechnol 100:949–960. https://doi.org/10.1007/s00253-015-7009-z
Karray F, Aloui F, Jemli M, Mhiri N, Loukil S, Bouhdida R, Mouha N, Sayadi S (2020) Pilot-scale petroleum refinery wastewaters treatment systems: Performance and microbial communities’ analysis. Process Saf Environ Prot 141:73–82. https://doi.org/10.1016/j.psep.2020.05.022
Askari A, Vahabzadeh F, Mardanpour MM (2021) The identification and performance assessment of dominant bacterial species during linear alkylbenzene sulfonate (LAS)-biodegradation in a bioelectrochemical system. Bioprocess Biosyst Eng 44:2579–2590. https://doi.org/10.1007/s00449-021-02629-0
Tomei MC, Mosca Angelucci D, Clagnan E, Brusetti L (2021) Anaerobic biodegradation of phenol in wastewater treatment: achievements and limits. Appl Microbiol Biotechnol 105:2195–2224. https://doi.org/10.1007/s00253-021-11182-5
Phale PS, Malhotra H, Shah BA (2020) Degradation strategies and associated regulatory mechanisms/features for aromatic compound metabolism in bacteria. Adv Appl Microbiol 112:1–65. https://doi.org/10.1016/bs.aambs.2020.02.002
Bueno E, Mesa S, Bedmar EJ, Richardson DJ, Delgado MJ (2012) Bacterial adaptation of respiration from oxic to microoxic and anoxic conditions: redox control. Antioxid Redox Signal 16:819–852. https://doi.org/10.1089/ars.2011.4051
Lai B, Yu S, Bernhardt PV, Rabaey K, Virdis B, Kromer JO (2016) Anoxic metabolism and biochemical production in Pseudomonas putida F1 driven by a bioelectrochemical system. Biotechnol Biofuels 9:39. https://doi.org/10.1186/s13068-016-0452-y
Lack A, Fuchs G (1992) Carboxylation of phenylphosphate by phenol carboxylase, an enzyme system of anaerobic phenol metabolism. J Bacteriol 174:3629–3636. https://doi.org/10.1128/jb.174.11.3629-3636.1992
Choudhury P, Ray RN, Bandyopadhyay TK, Tiwari ON, Bhunia B (2021) Kinetics and performance evaluation of microbial fuel cell supplied with dairy wastewater with simultaneous power generation. Int J Hydrogen Energy 46:16815–16822. https://doi.org/10.1016/j.ijhydene.2020.08.024
Jain S, Mungray AK (2021) Comparative study of different hydro-dynamic flow in microbial fuel cell stacks. Chin J Chem Eng 32:423–430. https://doi.org/10.1016/j.cjche.2020.10.016
Qi X, Wang H, Gao X, Zhang L, Wang X, Xu P (2022) Efficient power recovery from aromatic compounds by a novel electroactive bacterium Pseudomonas putida B6–2 in microbial fuel cells. J Environ Chem Eng 10:108536. https://doi.org/10.1016/j.jece.2022.108536
Sun G, de Sacadura RD, Thygesen A, Daniel G, Fernando D, Meyer AS (2016) Inocula selection in microbial fuel cells based on anodic biofilm abundance of Geobacter sulfurreducens. Chin J Chem Eng 24:379–387. https://doi.org/10.1016/j.cjche.2015.11.002
Liu X, Tremblay P-L, Malvankar NS, Nevin KP, Lovley DR, Vargas M (2014) A Geobacter sulfurreducens strain expressing Pseudomonas aeruginosa type IV pili localizes OmcS on pili but is deficient in Fe (III) oxide reduction and current production. Appl Environ Microbiol 80:1219–1224. https://doi.org/10.1128/aem.02938-13
Semenec L, Vergara IA, Laloo AE, Petrovski S, Bond PL, Franks AE (2020) Adaptive evolution of Geobacter sulfurreducens in coculture with Pseudomonas aeruginosa. MBio 11:e02875-e12819. https://doi.org/10.1128/mBio.02875-19
Bruckner R, Titgemeyer F (2002) Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol Lett 209:141–148. https://doi.org/10.1111/j.1574-6968.2002.tb11123.x
Park H, McGill SL, Arnold AD, Carlson RP (2020) Pseudomonad reverse carbon catabolite repression, interspecies metabolite exchange, and consortial division of labor. Cell Mol Life Sci 77:395–413. https://doi.org/10.1007/s00018-019-03377-x
Samuel MS, Sivaramakrishna A, Mehta A (2014) Bioremediation of p-Nitrophenol by Pseudomonas putida 1274 strain. J Environ Health Sci Eng 12:53. https://doi.org/10.1186/2052-336x-12-53
Li S, Tang Y, Tang L, Yan X, Xiao J, Xiang H, Wu Q, Yu R, Jin Y, Yu J, Xu N, Wu C, Wang S, Wang C, Chen Q (2022) Preliminary study on the effect of catabolite repression gene knockout on p-nitrophenol degradation in Pseudomonas putida DLL-E4. PLoS ONE 17:e0278503. https://doi.org/10.1371/journal.pone.0278503
Lin Y-H, Cheng Y-S (2020) Phenol degradation kinetics by free and immobilized Pseudomonas putida BCRC 14365 in batch and continuous-flow bioreactors. Processes 8:721. https://doi.org/10.3390/pr8060721
Hussain A, Dubey SK, Kumar V (2015) Kinetic study for aerobic treatment of phenolic wastewater. Water Resour Ind 11:81–90. https://doi.org/10.1016/j.wri.2015.05.002
Panigrahy N, Barik M, Sahoo NK (2020) Kinetics of phenol biodegradation by an indigenous Pseudomonas citronellolis NS1 isolated from coke oven wastewater. J Hazard Toxic Radioact Waste 24:04020019. https://doi.org/10.1061/(ASCE)HZ.2153-5515.0000502
Duan Z (2011) Microbial degradation of phenol by activated sludge in a batch reactor. Environ Prot Eng 37:53–63
Liu Y, Liu Z-M (2019) Promoted activity of nitrogen-doped activated carbon as a highly efficient oxygen reduction catalyst in microbial fuel cells. J Appl Electrochem 49:119–133. https://doi.org/10.1007/s10800-018-1263-6
Acknowledgements
The authors thank Dr. A. Monazzami as IT coordinator at Chemical Engineering Department/AUT, for sincere assistance in the submission process. First author and F. Vahabzadeh express their gratitude for constructive comments made by the anonymous reviewers, which allow us to revise the manuscript properly.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
MPF and Dr FV wrote the main manuscript text. Dr LD helped to carry out the experimental tests. Dr AA helped to prepare Figures and make suggestion. All authors have read the manuscript and agreed for its submission.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pourmirjafary Firuzabady, M., Askari, A., Davarpanah, L. et al. Study on kinetics of co-metabolic degradation of para-nitrophenol and phenol using microbial fuel cell. J Appl Electrochem (2024). https://doi.org/10.1007/s10800-024-02115-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10800-024-02115-x