Abstract
Glass sponges represent a dominant group of megabenthic deep-sea fauna and play a key role in benthic deep-sea ecosystems. Especially in the Clarion-Clipperton Fracture Zone (CCFZ), a potential deep-sea mining area, they grow on polymetallic nodules or on the surrounding sediment. We investigate hexactinellids from the CCFZ to understand the ecological aspects of deep-sea mining and support the development of future pre-mining risk assessments and monitoring actions. Therefore, this study is published as part of a series of studies, all focusing on deep-sea glass sponges from the CCFZ. Resolving genetic relationships between species is still a fundamental as well as challenging task. Especially understudied groups mostly lack resolution. Combining results derived from taxonomic and phylogenetic data gives deeper insights into glass sponge relationships. Here, we present (1) a set of new primers for sequencing mitochondrial 16S rDNA as well as nuclear 18S and 28S rDNA of glass sponges, (2) first DNA sequencing data for 6 hexactinellid genera and 19 species, as well as (3) the most comprehensive phylogenetic tree of hexactinellid sponges to date including data available from previous studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glass sponges (Porifera, Hexactinellida) represent a dominant group of benthic deep-sea megafauna (Van Soest et al., 2012), especially on polymetallic nodule fields (Amon et al., 2016; Vanreusel et al., 2016; Kaiser et al., 2017). They play an important functional role in benthic deep-sea ecosystems, particularly as they are associated with many other deep-sea invertebrate taxa (Beaulieu, 2001; Bell, 2008; Purser et al., 2016). Although megabenthic glass sponges and glass sponge grounds are globally distributed in the abyssal zone, little is known about their ecology, phylogeny and taxonomy (Hogg et al., 2010, Van Soest et al., 2012). This is probably due to the limited accessibility of deep-sea habitats for sampling, high costs for research vessels and gear deployment, and low commercial interest in glass sponges. Especially in the Clarion-Clipperton Fracture Zone (CCFZ), where global demand and interest for exploration and exploitation of deep-sea minerals are continuously increasing, the CCFZ, which offers the highest known density of polymetallic nodules (Petersen et al., 2016; Wedding et al., 2015; Gollner et al., 2017; Kaiser et al., 2017), has become the world’s largest potential deep-sea mining area. Deep-sea mining of polymetallic nodules and blanketing of the seafloor will cause exhaustive removal of nodules and a subsequent loss of nodule-specific fauna (Thiel, 2001; Vanreusel et al., 2016; Gollner et al., 2017; Kaiser et al., 2017). Its long-term impact on polymetallic nodule field systems is expected to be extreme, probably causing irretrievable ecosystem damage. Recovery of these communities is unlikely as nodule growth rates are estimated to be around 1–10 mm per million years (Petersen et al., 2016). Pre-mining risk assessments and monitoring actions as well as designation of suitable marine protected areas are crucial to manage and minimize the potential mining impact. For this reason, we investigated the phylogeny of deep-sea glass sponges in the CCFZ to increase the actual knowledge on their species occurrence and distribution as well as their biodiversity, ecology and evolution. Furthermore, we put results of this study in a greater context by combining them with data available online (www.ncbi.nlm.nih.gov/genbank). With our result, we will contribute towards future management requirements for deep-sea mining in the CCFZ.
Several studies on the phylogeny of glass sponges were conducted within the last decade, all of them based on continuously increasing datasets of nucleotide sequences (Dohrmann et al., 2008, 2009, 2011, 2012 as well as Reiswig & Dohrmann, 2014). The first study addressing glass sponge phylogeny by Dohrmann et al. (2008) included sequencing data of mitochondrial 16S and nuclear 18S and 28S rDNA from hexactinellid sponges that belong to 3 orders, 9 families, 27 genera and 34 species. Briefly, this study confirmed earlier morphology-based hypotheses (e.g., Mehl, 1992) and showed that glass sponges are divided into two reciprocally monophyletic clades: Hexasterophora and Amphidiscophora. Monophyly of the orders Hexasterophora, Lyssacinosida and Sceptrulophora was not supported. Subsequently, Dohrmann et al. (2009) added data from 11 additional species including individuals of the Aphrocallistidae, Euplectellidae, Farreidae, Rossellidae and other families. Results of this second study gave evidence that the order Lyssacinosida is monophyletic and corroborated that genera such as Hyalonema Gray, 1832, Bathydorus Schulze, 1886 and Rossella Carter, 1872 might be monophyletic. Dohrmann et al. (2011) included data of the mitochondrial cytochrome c oxidase subunit I gene (COI) and data for 7 additional species. Results of that third study proved congruence between COI and rDNA phylogenies but the authors stated also the technical difficulties in sequencing COI from hexactinellid sponges. More lately, studies focusing on the recently described genus Nodastrella Dohrmann, Göcke, Reed & Janussen 2012 (Dohrmann et al, 2012) and the order Sceptrulophora (Reiswig & Dohrmann, 2014) were published.
The main objectives of this study are to (1) use phylogenetic data to support species identification based on morphological data (published in Kersken et al., 2017, 2018) and (2) provide first insights into the phylogeny of deep-sea glass sponges living on polymetallic nodule fields in the CCFZ. A secondary objective of this work is to (3) produce an expanded and up-to-date phylogenetic framework for glass sponges in general by combining phylogenetic data from this study with data published in previous studies.
Materials and methods
Research project and expedition
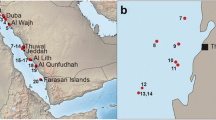
Investigated material was collected during the research expedition SO239 EcoResponse (Assessing the Ecology, Connectivity and Resilience of Polymetallic Nodule Field Systems) by RV Sonne that lasted between 11th Mar. 2015–30th Apr. 2015. The expedition was part of the JPI Oceans action “Ecological Aspects of Deep-Sea Mining” with the Clarion-Clipperton Fracture Zone, northeastern Pacific (8° to 18° N and 90° to 130° W), as working area. Sample sites were located in the following mining exploration areas: BGR licence area (Bundesanstalt für Geowissenschaften und Rohstoffe, Germany); GSR licence area (G-TEC Minerals Resources NV, Belgium); IFREMER licence area (Institut franςais de recherche pour l’exploitation de la mer, France) and IOM licence area (Interoceanmetal, Bulgaria, Cuba, Czech Republic, Poland, Russian Fed. and Slovakia). Furthermore, APEI-3 (Area of Particular Environmental Interest) was included.
Gear deployment and sample handling
All sponges were collected with the Remotely Operated Vehicle ROV Kiel 6000 by the GEOMAR—Helmholtz Centre for Ocean Research in Kiel, Germany. After ascent, sponges were immediately transferred into buckets with pre-cooled seawater and processed in a climate-controlled lab (both with a temperature of + 4°C). For DNA Barcoding, two subsamples were taken of each individual: the first subsample was preserved in pure ethanol (96%) and stored at − 20°C while the second subsample was frozen at − 80°C. Subsequent to subsampling, sponges were transferred into plastic containers filled with denatured ethanol (96%) and stored at room temperature. After the expedition, subsamples were transported to the Senckenberg Biodiversity and Climate Research Centre (BiK-F) in Frankfurt am Main, Germany.
Sponge taxonomy
The light microscope (LM) and a scanning electron microscope (SEM) were used to identify sponge species with help of spicule preparations. Therefore, subsamples were transferred into test tubes and nitric acid (HNO3) was pipetted onto them to dissolve all organic material. Spicules were washed thrice with distilled water to eliminate the nitric acid, and washed thrice with denatured ethanol (96%) afterwards to prepare them for the embedment on the glass slides (LM) as well as the sputter coating on the metal stubs (SEM). Both, LM and SEM were used for species identification, although the LM was primarily used for measuring spicules and the SEM was primarily used for photographing them. The typical literature was used for species identification; for further information, please check Kersken et al. (2017). Furthermore, all sponges collected during the SO239 EcoResponse expedition were deposited and inventoried at the Senckenberg Research Institute and Nature Museum. Identified specimens and their metadata were electronically catalogued with SMF numbers while information is available in the SESAM database (Senckenberg Sammlungsmanagement—www.sesam.senckenberg.de).
Molecular work and data analysis
Sequencing of three DNA markers was planned for a total number of 68 specimens: mitochondrial 16S as well as nuclear 18S and 28S ribosomal DNA (rDNA). DNA extraction was performed with two methods: (1) HotSHOT DNA extraction as described by Truett et al. (2000) and (2) glass fiber plate DNA extraction as described by Ivanova et al. (2006). New amplification and sequencing primers were designed in the framework of this study (Table 1), except for two 16S rDNA primers (16S1fw and 16SH_mod) that were already used by Dohrmann et al. (2008). Primer design was mainly based on the data from Dohrmann et al. (2008), Haen et al. (2013) and Vargas et al. (2017). Separate polymerase chain reaction (PCR) protocols were established for amplification (Supplementary Material 1). The PCR mix for each reaction contained 6.5 µl ddH2O, 1 µl 10 × Buffer B, 1 µl MgCl2 (25 mM), 0.1 µl dNTP Mix (2 mM each), 0.1 µl Taq-Polymerase and 0.2 µl of each primer (10 pmol/µl). During the PCR, each reaction tube contained 1 µl of DNA extraction and 9.1 µl of PCR mix (10.1 µl in total). After PCR, DNA extractions of the HotSHOT method were diluted (× 10, × 20, × 40), while extractions after Ivanova et al. 2006 were concentrated (× 3), depending on the intensity of the band when the extraction was run out on an agarose gel (1.4%, 100 V, 30 min). A standard protocol was used for the sequencing reaction (Supplementary Material 2). The sequencing mix for each reaction contained 6.5 µl ddH2O, 1.84 µl 5x Buffer B, 0.16 µl BigDye® Terminator Mix (BigDye® Terminator v3.1, Applied Biosystems, Foster City, USA), 0.5 µl primer and 1.0 µl of the PCR product. During the sequencing reaction, each reaction tube contained 1 µl of pure or diluted PCR product and 9 µl of sequencing mix (10.0 µl in total). DNA sequencing was done in the BiK-F laboratory centre by Sanger sequencing using a capillary sequencer “3730 DNA Analyzer” (Applied Biosystems, Foster City, USA). DNA sequences were analyzed with Geneious v. 4.8.5 (http://www.geneious.com, Kearse et al., 2012). Low quality reads (HQ ≤ 10%) were excluded from the following analyses and ambiguous base calls were corrected manually using the nucleotide code of the International Union of Pure and Applied Chemistry (IUPAC). Consensus sequences of all individuals were checked with a standard nucleotide BLAST (Basic Local Alignment Search Tool) by the National Centre for Biotechnology Information (NCBI) to assure sequenced specimens are glass sponges (Hexactinellida). All assembled nucleotide sequences used in this study were registered and have accession numbers at GenBank (https://www.ncbi.nlm.nih.gov/genbank/) (Supplementary Material 3). Furthermore, additional nucleotide sequences for Hexactinellida were downloaded from GenBank and integrated into the dataset of this study (Supplementary Material 3). For multi-gene analysis, 16S, 18S and 28S rDNA consensus sequences were aligned with MEGA7 (Kumar et al., 2016) using ClustalW (Higgins et al. 1994) and separately tested with jModelTest 2 (Darriba et al., 2012; Guindon & Gascuel, 2003), to find the best common substitution model, which is the General Time Reversible model with separate consideration of invariant sites and a gamma distribution (GTR + I+G) (Tavare 1986). Phylogeny was reconstructed by Bayesian evolutionary analysis with BEAST 2 (Bouckaert et al. 2014) using the model as determined before. Furthermore, the normal distribution of prior and posterior probability was checked with Tracer v 1.6 (Rambaut et al. 2014), while a maximum clade credibility tree was built with FigTree v1.4.3 (Rambaut 2016).
Results
Support of species identification based on morphology
A double-check of species identifications by nucleotide BLASTs (www.blast.ncbi.nlm.nih.gov) gives a low-resolution picture of the sponge species in this study and is sufficient to support but insufficient to replace a species identification based on morphological characters. Several specimens that are difficult to identify were checked: Bolosoma sp. Ijima, 1904 (SMF 11695), Lyssacinosida gen. sp. (SMF 12068), Bathydorus sp. (12064), Lyssacinosida gen. sp. (SMF 12066) and Lyssacinosida gen. sp. (SMF 12080). A more precise identification was possible in the case of Oopsacas sp. Topsent, 1927 (SMF 12068), Acanthascus sp. Schulze, 1886 (SMF 12080) and Lophocalyx sp. Schulze, 1887 (SMF 12066) that were formerly identified as Lyssacinosida gen. sp.
Basic results of molecular work
Specimens genetically characterized for this study include at least 27 species that belong to 14 genera and 6 families: Euplectellidae, Euretidae, Leucopsacidae, Rossellidae, Hyalonematidae and Pheronematidae. Sequencing mitochondrial 16S as well as nuclear 18S and 28S rDNA worked, with at least one genetic marker, for 43 of 68 tested individuals (63%) whereof 30 individuals are hexasterophorid sponges and 13 amphidiscophorid sponges. Sequences of two or more DNA markers are obtained from 19 specimens (28%) and sequences of all three markers from 11 tested specimens (16%). Sequencing 16S rDNA is most successful and reveals sequences of 37 individuals (54%). Furthermore, 18S rDNA sequencing worked for 18 individuals (26%) and 28S rDNA sequencing for 19 individuals (28%). Deep-sea glass sponges of 19 species are sequenced for the first time, 8 species of Amphidiscophora (especially Hyalonema spp.) and 11 species of Hexasterophora (Fig. 1 and Table 2). Furthermore, nucleotide sequences of the six sponge genera Bathyxiphus Schulze, 1899, Chonelasma Schulze, 1886, Corbitella Gray, 1867, Holascus Schulze, 1886, Hyalostylus Schulze, 1886 and Poliopogon Thomson, 1878 are sequenced for the first time with a total of 12 16S, 2 18S and 4 28S rDNA sequences (Table 2). The sample set of the latter genera includes a number of eight species whereof four were only recently described (Kersken et al., 2017, 2018).
Consensus sequences of the 16S rDNA have a length of 281–378 nucleotides with a mean length of 354 nucleotides, sequences of the 18S rDNA of 597–1049 nucleotides with a mean length of 946 nucleotides and sequences of the 28S rDNA of 638–1218 nucleotides with a mean length of 1094 nucleotides (Table 2). The alignments of all three markers (16S rDNA with 384, 18S rDNA with 1090 and 28S rDNA with 1352 nucleotides) were concatenated and the final alignment that was used for the Bayesian evolutionary analysis had a total length of 2826 nucleotides (Supplementary material 4).
Bayesian evolutionary analysis with concatenated DNA sequences
The maximum clade credibility tree is inferred from concatenated 16S, 18S and 28S rDNA sequences (Fig. 1). The complete dataset in this study includes nucleotide sequences of 142 individuals that belong to a minimum of 16 families, 59 genera and 86 species. Sequences of 99 individuals were included in previous studies and were downloaded from GenBank (www.ncbi.nlm.nih.gov/genbank/) (Supplementary material 3) while 43 additional sequences were obtained in the framework of this study. Values of posterior probability (pp) are pp = 1.0 for all subclasses (Amphidiscophora and Hexasterophora), orders (Amphidiscophora, Lyssacinosida and Sceptrulophora) as well as families Aphrocallistidae, Dactylocalycidae, Euplectellidae, Hyalonematidae, Leucopsacidae, Pheronematidae, Rossellidae and Tretodictyidae. Furthermore, monophyly of several genera is fully supported (pp = 1.0), although pp values tend to drop on genus or at least on species level, e.g., in the clade Rossella where they range from pp = 0.1–0.9.
The phylogenetic tree can be subdivided into three major clades: (1) Amphidiscophora, (2) Lyssacinosida and (3) Sceptrulophora. Within the (1) Amphidiscophora, two monophyla are formed, one by the family Hyalonematidae and another one by the family Pheronematidae. Within the Hyalonematidae, sequence data of 12 individuals (all identified to the species level) have been added to the former dataset with 4 individuals (all identified to the genus level). The subgenus Hyalonema (Onconema) Ijima, 1927 forms a paraphylum with four specimens of two species (Hyaloenma (Onconema) agassizi Lendenfeld, 1915 and Hyalonema (Onconema) obtusum Lendenfeld, 1915) and the subgenus Hyalonema (Cyliconemaoida) Dohrmann, 2017 forms a monophylum with five specimens of two species (Hyalonema (Cyliconemaoida) campanula Lendenfeld, 1915 and Hyalonema (Cyliconemaoida) ovuliferum Schulze, 1899). The Pheronematidae form a sister group to the Hyalonematidae. The previous dataset includes nucleotide sequences of six individuals whereof three are identified on species level. Data of Poliopogon have been added for the first time, while this study presents sequence data of the recently described species Poliopogon microuncinata Kersken, Janussen & Martinez Arbizu, 2017. Poliopogon is a sister to Schulzeviella Tabachnick, 1990 and Semperella Gray, 1868. Sericolophus Ijima, 1901 and Pheronema Leidy, 1868, but also another individual of Semperella, form a separate group within the Pheronematidae.
Within the (2) Lyssacinosida, the families Euplectellidae and Rossellidae form monophyla while the Dactylocalycidae and Leucopsacidae form paraphyla. Among the Rossellidae, nucleotide sequences of eight individuals, one identified on species level, were added to the previous dataset and five of these belong to the genus Caulophacus Schulze, 1886. A specimen identified as Acanthascus sp. forms a small group with another Acanthascus but the Acanthascinae herein are polyphyletic as a Rhabdocalyptus Schulze, 1886 is clustering as sister of Crateromorpha Gray in Carter, 1872. Such a big discrepancy is unusual and the material identified by Collins (1998) should be checked taxonomically. The individual identified as Bathydorus sp. forms a small monophylum with two other specimens of the genus Bathydorus. Furthermore, one individual identified as Lophocalyx sp. forms a small monophylum with another Lophocalyx and is part of a group with Caulophacus (Caulophacella) Lendenfeld, 1915 and Doconesthes Topsent, 1928. Furthermore, the Leucopsacidae form a polyphylum close to the family Rossellidae. Among the Leucopsacidae, data of one Oopsacas, identified to genus level, were added and indicate that Oopsacas is paraphyletic. Among the Euplectellidae, nucleotide sequences of 19 individuals, 18 identified to the species level, were added to the previous dataset. Specimens of the genera Corbitella, Holascus and Hyalostylus were sequenced for the first time. Monophyly of subfamilies cannot be observed although it seems that single genera tend to form monophyla. A polyphylum is formed by the genus Holascus as individuals of Holascus euonyx Lendenfeld, 1915 form a group with Acoelocalyx Topsent, 1910 and Malacosaccus Schulze, 1886 while individuals of Holascus spinosus Kersken, Janussen & Martinez Arbizu, 2018 and Holascus taraxacum (Lendenfeld, 1915) form a small monophylum implemented in a group with Atlantisella Tabachnick, 2002, Euplectella Owen, 1841 and Regadrella Schmidt, 1880. Three specimens of Corbitella form a small monophylum while they are a sister group to Acoelocalyx, Holascus and Malacosaccus. Two specimens of the genus Docosaccus Topsent, 1910 were sequenced in this study and form a small monophylum together with a third individual of Docosaccus that was sampled in the northeastern Pacific and described by Kahn et al. (2013) as well as in Dohrmann et al. (2011). Saccocalyx Schulze, 1896 forms a paraphylum, as one specimen of Hertwigia Schmidt, 1880 is in the same group. Within this study, five additional individuals of Saccocalyx, all identified to species level, were combined with the previous data and show how closely Saccocalyx pedunculatus Schulze, 1896 and Saccocalyx microhexcatin Gong, Li & Qiu, 2015 are related. Furthermore, sequence data of a second Bolosoma, identified on genus level, were added to the previous data and indicate that Bolosoma is monophyletic while being included into a bigger group with Hyalostylus and Rhabdopectella Schmidt, 1880. Three individuals of Hyalostylus, all identified to species level, that belong to two species form a small monophylum within the latter described group.
Within the (3) Sceptrulophora, the family Tretodictyidae forms a monophylum as shown by Dohrmann et al. (2011) but other families that were taken in account to be monophyletic are para- or polyphyletic, e.g., Aphrocallistidae, Farreidae and Euretidae (Reiswig & Dohrmann, 2014). In the framework of this study, the first Bathyxiphus was sequenced and is a sister to Homoieurete Reiswig & Kelly, 2011 (Sceptrulophora incertae sedis) while, the first ever sequenced, Chonelasma is a sister to Sarostegia Topsent, 1904 (Sceptrulophora incertae sedis). Other Euretidae (Conorete, Lefroyella and Verrucocoeloidea) form a small monophylum in a big group together with the Farreidae which are paraphyletic.
Discussion
Discussion of phylogeny
Within the family Hyalonematidae, previously sequenced individuals of Hyalonema are distantly related to individuals sequenced within this study because they were sampled in the northwestern Atlantic (Dohrmann et al., 2008, 2009), while all Hyalonematidae of this study were sampled in the northeastern Pacific. Therefore, we expected different species to occur but we are still surprised to see that all Hyalonema from previous studies group aside. This could indicate that those individuals are representatives of different subgenera and a further taxonomic identification would be interesting. Within the family Pheronematidae, it would be important to add data of a Platylistrum and thus have sequence data of a minimum of one representative of each genus. Within the family Rossellidae, monophyly of Caulophacus (Caulophacus) and Caulophacus (Caulodiscus), two subgenera that were so far represented by single specimens cannot be observed. Now, it would be interesting to sequence further specimens of Caulophacus (Oxydiscus) and Caulophacus (Caulophacella) and check if these are also para- or polyphyletic. Among the Euplectellidae, the Corbitellinae are scattered all over the clade, especially Corbitella where sequences of three specimens group with Holascus, Malacosaccus and Acoelocalyx. Thus, our findings are confirming the results of Dohrmann et al. (2008, 2011). Furthermore, we want to recommend a revision of the genus Saccocalyx. Due to our phylogenetic results and former morphological analyses showing solely marginal differences in spicule morphology by presence/absence of microhexactins (Gong et al., 2015; Kersken et al., 2018), we propose a revision of the genus Saccocalyx to check the genus for monospecificity. Within the family Euretidae, it is surprising that Bathyxiphus and Chonelasma are in the same group than Sarostegia as the latter was previously excluded from the family of Euretidae based on phylogenetic results by Reiswig & Dohrmann (2014) which can be confirmed since we find the same group of Conorete Ijima, 1927, Lefroyella Thomson, 1878 and Verrucocoeloidea Reid, 1969 in a well-supported Euretidae/Farreidae clade.
Discussion of working methods
Sampling deep-sea hexactinellids for genetic analyses can be problematic due to certain limitations, e.g., the time frame between sample collection and handling which is predetermined by the ascent time of the ROV or the temperature difference from bottom water to surface water that can be huge, accelerating DNA degradation. Still, sampling by ROV is up-to-date and the most effective as well as precise method to sample deep-sea sponges. In the molecular lab, DNA extraction is difficult as many species of glass sponges have a low yield of DNA and their silicate spicules can, unintentionally, form pellets and influence extraction negatively. A second DNA extraction method using a glass fiber plate (Ivanova et al., 2006) was applied to increase the number of successful DNA extractions. Amplification of nuclear 18S and 28S sequences using the primers described by Dohrmann et al. (2008) failed in this study. Nucleotide sequences of the cytochrome oxidase subunit 1 (COI) were obtained from only seven individuals and thus excluded from the analysis. Although a separate touchdown PCR protocol was established only for COI, with annealing temperatures ranging from 56 to 48°C, DNA extraction and amplification were unexpectedly challenging as described by Dohrmann et al. (2011). A state-of-the-art multi-gene analysis with BEAST 2 including linked/unlinked loci was planned but failed in the first attempt due to an insufficient overlap of sequence data among sponge individuals (Table 2). In the second attempt, mitochondrial and nuclear sequence data were concatenated before we started a Bayesian evolutionary analysis with BEAST 2 and performed a multi-gene analysis. However, although all sequence data are treated as originating from one locus, we preferred to perform a multi-gene instead of single-gene analysis.
Conclusions
Combining nucleotide sequences of 43 deep-sea glass sponges with data from several previous studies allowed us to generate the most comprehensive phylogenetic tree of hexactinellid sponges to date. It was possible to include data of 6 new genera and 19 species, particularly expanding our knowledge on Euplectellidae (Hexasterophora) and Hyalonematidae (Amphidiscophora): Bathyxiphus, Chonelasma, Corbitella, Holascus, Hyalostylus and Poliopogon. Data of several genera have been extended or added, now showing monophyly: Bolosoma, Corbitella, Docosaccus and Hyalostylus; or now showing paraphyly: Holascus, Oopsacas and Saccocalyx. The Hyalonematidae illustrate what genetic differences we can expect for hexactinellid sponges of the same genus but different habitats, in this case northeastern Pacific and northwestern Atlantic. Results of this study provide first insights into the phylogeny of hexactinellid sponges from the CCFZ and expand the current knowledge on the phylogeny of hexactinellid sponges in general.
References
Amon, D., A. F. Ziegler, T. G. Dahlgren, A. G. Glover, A. Goineau, A. J. Gooday, H. Wiklund & C. R. Smith, 2016. Insights into the abundance an diversity of abyssal megafauna in a polymetallic-nodule region in the eastern Calrion-Clipperton Zone. Scientific Reports 6: 1–12. https://doi.org/10.1038/srep30492.
Beaulieu, S. E., 2001. Life on glass houses: sponge stalk communities in the deep sea. Marine Biology 138: 803–817. https://doi.org/10.1007/s002270000500.
Bell, J. J., 2008. The functional roles of marine sponges. Estuarine, Coastal and Shelf Science 79: 341–353. https://doi.org/10.1016/j.ecss.2008.05.002.
Bouckaert, R., J. Heled, D. Kühnert, T. Vaughan, C. H. Wu, D. Xie, M. A. Suchard, A. Rambaut & A. J. Drummond, 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology 10: e1003537. https://doi.org/10.1371/journal.pcbi.1003537.
Collins, A. G., 1998. Evaluating multiple alternative hypotheses for the origin of Bilateria: an analysis of 18S rRNA molecular evidence. Proceedings of the National Academy of Sciences of the United States of America 95: 15458–15463. PMCID:PMC28064
Darriba, D., G. L. Taboada, R. Doallo & D. Posada, 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. https://doi.org/10.1038/nmeth.2109.
Dohrmann, M., D. Janussen, J. Reitner, A. G. Collins & G. Wörheide, 2008. Phylogeny and evolution of glass sponges (Porifera, Hexactinellida). Systematic Biology 57: 388–405. https://doi.org/10.1080/10635150802161088.
Dohrmann, M., A. G. Collins & G. Wörheide, 2009. New insights into the phylogeny of glass sponges (Porifera, Hexactinellida): monophyly of Lyssacinosida and Euplectellinae and the phylogenetic position of Euretidae. Molecular Phylogenetics and Evolution 52: 257–262. https://doi.org/10.1016/j.ympev.2009.01.010.
Dohrmann, M., K. M. Haen, D. V. Lavrov & G. Wörheide, 2011. Molecular phylogeny of glass sponges (Porifera, Hexactinellida): increased taxon sampling and inclusion of the mitochondrial protein-coding gene, cytochrome oxidase subunit I. Hydrobiologia 687: 11–20. https://doi.org/10.1007/s10750-011-0727-z.
Dohrmann, M., C. Göcke, J. Reed & D. Janussen, 2012. Integrative taxonomy justifies a new genus, Nodastrella gen. nov., for North Atlantic “Rossella” species (Porifera: Hexactinellida: Rossellidae). Zootaxa 3383: 1–13. https://doi.org/10.11646/zootaxa.3383.1.1.
Gollner, S., S. Kaiser, L. Menzel, D. O. B. Jones, A. Brown, N. C. Mestre, D. Van Oevelen, L. Menot, A. Colaço, M. Canals, D. Cuvelier, J. M. Durden, A. Gebruk, G. A. Egho, M. Haeckel, Y. Marcon, L. Mevenkamp, T. Morato, C. K. Pham, A. Purser, A. Sanchez-Vidal, A. Vanreusel, A. Vink & P. Martínez Arbizu, 2017. Resilience to benthic deep-sea fauna to mining activities. Marine Environmental Research. https://doi.org/10.1016/j.marenvres.2017.04.010.
Gong, L., L. Xinzheng & Q. Jian-Wen, 2015. Two new species of Hexactinellida (Porifera) from the South China Sea. Zootaxa 4034: 182–192.
Guindon, S. & O. Gascuel, 2003. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Systematic Biology 52: 696–704.
Haen, K. M., W. Pett & D. V. Lavrov, 2013. Eight new mtDNA sequences of glass sponges reveal an extensive usage of + 1 frameshifting in mitochondrial translation. Gene 535: 336–344.
Higgins, D., J. Thompson, T. Gibson, J. D. Thompson, D. G. Higgins & T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680.
Hogg, M. M., O. S. Tendal, K. W. Conway, S. A. Pomponi, R. W. M. Van Soest, J. Gutt, M. Krautter & J. M. Roberts, 2010. Deep-sea sponge grounds: reservoirs of biodiversity. UNEP-WCMC Biodiversity Series 32. UNEP-WCMC, Cambridge, UK.
Ivanova, N., J. deWaard & P. Hebert, 2006. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Molecular Ecology Notes 6: 998–1002. https://doi.org/10.1111/j.1471-8286.2006.01428.x.
Kahn, A. S., J. B. Geller, H. M. Reiswig & K. L. Smith Jr., 2013. Bathydorus laniger and Docosaccus maculatus (Lyssacinosida; Hexactinellida): two new species of glass sponge from the abyssal eastern North Pacific Ocean. Zootaxa 3646: 386–400.
Kaiser, S., C. R. Smith & P. Martínez Arbizu, 2017. Editorial: biodiversity of the Clarion Clipperton Fracture Zone. Marine Biodiversity. https://doi.org/10.1007/s12526-017-0733-0.
Kearse, M., R. Moir, A. Wilson, S. Stones-Havas, M. Cheung, S. Sturrock, S. Buxton, A. Cooper, S. Markowitz, C. Duran, T. Thierer, B. Ashton, P. Meintjes & A. Drummond, 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649.
Kersken, D., D. Janussen & P. Martínez Arbizu, 2017. Deep-sea glass sponges (Hexactinellida) from polymetallic nodule fields in the Clarion-Clipperton Fracture Zone (CCFZ), northeastern Pacific: part I – Amphidiscophora. Marine Biodiversity. https://doi.org/10.1007/s12526-017-0727-y.
Kersken, D., D. Janussen & P. Martínez Arbizu, 2018. Deep-sea glass sponges (Hexactinellida) from polymetallic nodule fields in the Clarion-Clipperton Fracture Zone (CCFZ), northeastern Pacific: Part II – Hexasterophora. Marine Biodiversity (submitted).
Kumar, S., G. Stecher & K. Tamura, 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology Evolution 33: 1870–1874.
Petersen, S., A. Krätschell, N. Augustin, J. Jamieson, J. R. Hein & M. D. Hannington, 2016. News from the seabed – geological characteristics and resource potential of deep-sea mineral resources. Marine Policy 70: 175–187. https://doi.org/10.1016/j.marpol.2016.03.012.
Purser, A., Y. Marcon, H. J. T. Hoving, M. Vecchione, U. Piatkowski, D. Eason, H. Bluhm & A. Boetius, 2016. Association of deep-sea incirrate octopods with manganese crusts and nodule fields in the Pacific Ocean. Current Biology 26: R1268–R1269.
Rambaut, A., 2016. FigTree v1.4.3 [available on internet at http://tree.bio.ed.ac.uk/software/figtree/].
Rambaut, A., M. A. Suchard, D. Xie & A. J. Drummond, 2014. Tracer v1.6 [available on internet at http://beast.bio.ed.ac.uk/Tracer].
Reiswig, H. M. & M. Dohrmann, 2014. Three new species of glass sponges (Porifera: Hexactinellida) from the West Indies, and molecular phylogenetics of Euretidae and Auloplacidae (Sceptrulophora). Zoological Journal of the Linnean Society 171: 233–253.
Tavare, S., 1986. Some probabilistic and statistical problems in the analysis of DNA sequences. American Mathematical Society: Lectures on Mathematics in the Life Sciences 17: 57–86.
Thiel, H., 2001. Evaluation of the environmental consequences of polymetallic nodule mining based on the results of the TUSCH research association. Deep-Sea Research II 48: 3433–3452.
Truett, G. E., P. Heeger, R. L. Mynatt, A. A. Truett, J. A. Walker & M. L. Warman, 2000. Preparation of PCR quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). BioTechniques 29: 52–54.
Vanreusel, A., A. Hialrio, P. A. Ribeiro, L. Menot & P. Martínez Arbizu, 2016. Threatened by mining, polymetallic nodules are required to preserve abyssal fauna. Nature – Scientific Reports 6: 26808. https://doi.org/10.1038/srep26808
Van Soest, R. V. M., N. Boury-Esnault, J. Vacelet, M. Dohrmann, D. Erpenbeck, N. J. De Voogd, N. Santodomingo, B. Vanhoorne, M. Kelly & J. N. A. Hooper, 2012. Global diversity of sponges (Porifera). PLoS One 7: e35105.
Vargas, S., M. Dohrmann, C. Göcke, D. Janussen & G. Wörheide, 2017. Nuclear and mitochondrial phylogeny of Rossella (Hexactinellida: Lyssacinosida, Rossellidae): a species and a species flock in the Southern Ocean. Polar Biology. https://doi.org/10.1007/s00300-017-2155-7.
Wedding, L. M., S. M. Reiter, C. R. Smith, K. M. Gjerde, J. M. Kittinger, A. M. Friedlander, S. D. Gaines, M. R. Clark, A. M. Thurnherr, S. M. Hardy & L. B. Crowder, 2015. Managing mining of the deep seabed. Science 349: 144–145.
Acknowledgements
The EcoResponse cruise with RV Sonne was financed by the German Ministry of Education and Science (BMBF) as a contribution to the European project JPI-Oceans “Ecological Aspects of Deep-Sea Mining”. The authors acknowledge funding from BMBF under Contract 03F0707E. Furthermore, we want to thank Dr. Barbara Feldmeyer and Dr. Ann-Marie Waldvogel for assistance of molecular lab work and Dr. Martin Dohrmann for information on sponge specific DNA primers and PCR protocols.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Iacopo Bertocci
GenBank: All nucleotide sequences used in this study have accession numbers at GenBank (https://www.ncbi.nlm.nih.gov/genbank/) (Supplementary Material 3).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kersken, D., Kocot, K., Janussen, D. et al. First insights into the phylogeny of deep-sea glass sponges (Hexactinellida) from polymetallic nodule fields in the Clarion-Clipperton Fracture Zone (CCFZ), northeastern Pacific. Hydrobiologia 811, 283–293 (2018). https://doi.org/10.1007/s10750-017-3498-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-017-3498-3