Abstract
Lake Baikal (Siberia) is the most ancient and deepest of all ancient lakes on Earth. It holds a (mostly endemic) diversity of thousands of animal species, including a speciose radiation of ostracods of the genus Cytherissa. Applying molecular tools to this crustacean group reveals that several morphological species are actually species clusters. Based on combined 16S and 28S DNA sequence data from thirteen classic Cytherissa species and one subspecies sensu Mazepova (1990), we recognize 26 different genetic Cytherissa species, 18 with morphological variation and eight truly cryptic species. These results suggest that the actual specific diversity of Cytherissa in Lake Baikal might easily be double of what is presently known. Baikalian endemic species most likely live in the cradle in which they originated and this opens perspectives to infer modes of speciation. Our current distribution data of Cytherissa species provide first indications for both geographic (lakes basins and shores) and ecological (sediment type, water depth) separation. Our present data thus provide the first steps towards future, rigorous testing of focussed hypotheses on the causality of speciation through either allopatric isolation or parapatric ecological clines.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ancient lakes are natural laboratories for evolutionary studies on the tempo and mode of speciation of their endemic fauna and flora (Martens, 1997). There are only a couple of dozens of these lakes on the globe and they are hotspots of biodiversity, because of their high endemicity and their importance for generating diversity in surrounding areas (Schön & Martens, 2012). Non-marine ostracods are not only an important ecological component of ancient lake taxa, but ancient lakes also contribute up to 25% of all known non-marine specific ostracod diversity (Martens et al., 2008).
On a global scale, Lake Baikal is the oldest extant lake with an estimated age of 25–30 myr (Sherbakov, 1999; Müller et al., 2001) as well as the deepest lake with a maximal depth of more than 1600 m (Sherstyankin & Kuimova, 2006). About 2500 animal (morpho) species occur in Lake Baikal, of which 1455 are endemic (Timoshkin, 2001). Concerning non-marine Ostracoda, more than 90% of Baikalian ostracods are endemic to the lake, and the Cytherissa species flock has the highest specific diversity (Mazepova, 1990). This species flock is probably an example of explosive radiation and has originated 5–8 million years ago (Schön & Martens, 2012), at a time when Lake Baikal’s cold, oxygenated abyss was formed (Sherbakov, 1999).
With the availability of molecular tools, the last twenty years have seen an ever-increasing number of studies detecting so-called cryptic diversity (Bickford et al., 2007), i.e. lineages that are morphologically similar but fulfil all criteria to be different genetic species (Vogler & Monaghan, 2007) according to the phylogenetic species concept (Eldredge & Cracraft (1980), but see also overview in Zhang et al., 2013) or the evolutionary genetic species concept (Birky & Barraclough, 2009). There is mounting evidence that cryptic species occur widely and that their presence is, at least in part, linked to specific types of habitat. For example, freshwater taxa show significantly more cryptic diversity than either terrestrial or marine taxa (Poulin & Pérez-Ponce de León, 2017).
Also in non-marine ostracods, cryptic species have been detected, varying between eight in a putative ancient asexual darwinulid species (Schön et al., 2012) to more than 35 in a single Holarctic temporary pool species (Bode et al., 2010). Likewise, cryptic species have been found in endemic Romecytheridea ostracods from Lake Tanganyika, the second most ancient lake in the world (Schön et al., 2014). The discovery of cryptic lineages throughout all metazoan phyla (Beheregaray & Caccone, 2007; Pfenninger & Schwenk, 2007) is not only important for fundamental science and systematics, but has also profound implications for conservation and management (examples in Brown et al., 2007; Elmer et al., 2007; Fontaneto et al., 2008; Gustafsson et al., 2009; Marrone et al., 2010), especially in unique environments such as ancient lakes. Indeed, if genetic diversity is cryptic, it is equally difficult to recognize it and to protect it from extinction.
Here, we use mitochondrial and nuclear DNA sequence data to test for the presence of cryptic species within 14 known morphological Cytherissa species and subspecies. Our samples come from all three basins of Lake Baikal, from both eastern and western shores and from both different water depths and different types of sediments, enabling us to assess the recent distribution patterns of these ostracods. Our research provides preliminary indications on the caudal importance of allopatric isolation (different basins or shores) and parapatric ecological speciation (depth, sediment types) for the past radiation of endemic Cytherissa species in Lake Baikal.
Materials and methods
Sampling
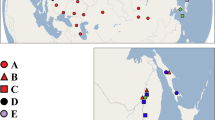
During four expeditions on Lake Baikal, in 1999, 2007, 2009 and 2011, several dozens of samples for ostracods were collected by SCUBA diving, trawling, dredging and with the oceanographic Reineck box-corer, from various locations in Lake Baikal (Fig. 1), including both the eastern and western shore and all three basins and at depths as from 20 m in the littoral photic zone (0–100 m) to deep water habitats of more than 500 m. Ostracods were sorted alive under a light microscope on the research vessels, were fixed in cold 95% pure ethanol for subsequent analyses and separated into preliminary taxonomic groups using the valve outlines of Mazepova (1990) and the hemipenis outlines by Van Mulken et al. (in prep.). We also sampled Cytherissa lacustris, the recent extra-lacustrine spin-off of the Baikalian Cytherissa flock (Schön & Martens, 2012), from Semerwater in the UK (see Table 1 for more details).
Approximate position of sampling stations in Lake Baikal. Labels refer to the sampling stations in Table 1
DNA extraction, PCR and sequencing
For most specimens, valves were removed for scanning electron microscopy (SEM) and the remaining soft parts were used to extract DNA from individual ostracods with a slightly modified protocol of the DNA Easy Blood and Tissue kit (Qiagen), adjusting the elution volumes because of the small size of individual ostracods. We estimated the concentration of the obtained DNA extractions with the Nanodrop and used the eluate with the highest concentration for all subsequent steps of the molecular analysis. With polymerase chain reaction (PCR), we amplified part of the mitochondrial 16S ribosomal region with specific primers (16S-F3 TTAATTCAACATCGAGGTCACAA and 16S-R2 GAGTAAACGGCTGCAGTA) and the D1D2 part of the nuclear Large Subunit (28S) with universal primers (D1D2Fw1 5′-AGCGGAGGAAAAGAAACTA-3′) and (D1D2Rev1 5′-TACTAGAAGGTTCGATTAGTC-3′) (Sonnenberg et al., 2007). Both regions have been successfully sequenced in other studies on non-marine ostracods (Bode et al., 2010; Koenders et al., 2012; Schön et al., 2014), and have also been used for the detection of cryptic species. PCRs were conducted in a T personal Thermoblock (Biometra) with 25 μl volumes of the Qiagen HotStar Mastermix (1.5 mM MgCl2, 200 μM dNTP, Tris·Cl, KCl, (NH4)2SO4, 1.25 U Taq), 0.1 μM of each primer and the following cycling conditions: 15 min at 95°C, followed by 40–42 cycles of 1 min at 95°C, 1 min at 44°C (16S) and 48°C (28S) and 1 min at 72°C, followed by a final extension step of 72°C for 10 min. We used agarose gel electrophoresis and stained gels with GelRed to check if PCR amplifications were successful. Positive amplicons were purified with the GFX PCR DNA and gel band purification kit (GE Healthcare) kit and sequenced in both directions using the PCR primers and the Big Dye kit (ABI) on an ABI 3130x1 capillary DNA sequencer (Life Technologies).
Analyses of DNA sequence data
We visualized sequencing chromatograms and generated consensus sequences for each specimen with Bioedit (Hall, 1999). Sequence ambiguities were checked by eye and corrected manually, sequences were aligned with MAFFT (Katoh & Standley, 2013) on http://www.ebi.ac.uk and trimmed to equal lengths in BioEdit. Sequence identity was confirmed by BLAST searches in Genbank (Altschul et al., 1997). As outgroup, we used sequence data from Romecytheridea ampla, an ostracod species from Lake Tanganyika belonging to the same subfamily and from which both 16S and 28S sequence data were available (Table 1). We also combined the DNA sequence data from both markers into a congruent dataset with sequence matrix (Vaidya et al., 2011). We trimmed the final alignment for each dataset with the outgroup (Table 1) to equal length and selected the best-fitting evolutionary model in jModeltest 2 (Darriba et al., 2012) using model filtering, the corrected Akaike information criterion (AICc) and 88 different nucleotide substitution models. The parameters of the best-fitting evolutionary models were used in phylogenetic reconstructions with maximum likelihood (ML) (PHYML; Guindon & Gascuel, 2003) and Bayesian approaches (MrBayes 3.2; Ronquist et al., 2012). Not all models selected by jmodeltest2 are implemented in MrBayes and we therefore had to pick the closest ones for Bayesian analyses. For 16S, the TIM1 + I + G model was selected with freqA = 0.2927; freqC = 0.2296; freqG = 0.1968; freqT = 0.2810; [AC] = 1.0000; [AG] = 5.4559; [AT] = 0.6273; [CG] = 0.6273; [CT] = 3.1987; [GT] = 1.0000; p-inv = 0.2280; gamma shape = 0.4750. For 28S, the TPM3uf + G model was selected with the following parameters: freqA = 0.1981; freqC = 0.2286; freqG = 0.3059; freqT = 0.2675; [AC] = 0.0000; [AG] = 3.6775; [AT] = 1.0000; [CG] = 0.0000; [CT] = 3.6775; [GT] = 1.0000; gamma shape = 0.3010. For the combined dataset, the TIM1 + I + G model was selected with freqA = 0.2632; freqC = 0.2145; freqG = 0.2316; freqT = 0.2907; [AC] = 1.0000; [AG] = 5.3221; [AT] = 0.5999; [CG] = 0.5999; [CT] = 3.5948; [GT] = 1.0000; p-inv = 0.4740; gamma shape = 0.3080. In all cases, we constructed ML trees in PHYML with these parameters and 1000 bootstraps. We also constructed ML trees without bootstrap support and outgroups and from haplotype sequences only for the Poisson Tree Processes (PTP) algorithm (Zhang et al., 2013) to delimitate genetic species (see below). For Bayesian approaches, we ran MrBayes with two MCMC chains and 20 million generations, applying the GTR + I+G model for 16S and the combined dataset and the HKY85 + G model for 28S, and sampling trees every 1000 generations. After inspecting the results, we eliminated the first 20000 trees as burn-in and calculated the 50% majority rule consensus tree. All trees were visualized and manipulated with MEGA 6.0 (Tamura et al., 2013) and FigTree (Rambaut, 2017).
Networks
To obtain the best graphic representation of haplotypes and their connectivity at the population level, we also constructed minimum spanning (Bandelt et al., 1999) networks from the 16S and 28S data with popart 1.2 (http://popart.otago.ac.nz) colour-coding the geographic origin (lake basin) as traits.
Delimitating genetic species
We used two different methods for quantitative delimitations of genetic species based on the evolutionary genetic species concept (Birky & Barraclough, 2009), nl. the 4θ (theta) rule (Birky et al., 2010; Birky, 2013) and the PTP algorithm (Zhang et al., 2013). For applying the 4θ rule, we first identified well-supported phylogenetic sister clades from the ML and Bayesian phylogenies with a bootstrap support of more than 75% or a posterior probability of more than 0.8. Within and between the sister clades, we then calculated genetic distances in MEGA 6.0 using the appropriate model for molecular evolution. As with Bayesian analyses, not all models selected by jmodeltest2 are available in MEGA and we chose the closest ones for the calculation of genetic distances. Next, π (nucleotide diversity) and θ (population mutation rate) were calculated taking sampling size of each sister clade into account. Finally, we calculated D (distance between sister clades) and the ratio between θ and D. If the resulting ratio is greater than 4, sister clades are considered to be different evolutionary species (Birky et al., 2010).
We also used a Poisson tree processes (PTP) model to delimit genetic species. This algorithm is based on a shift of the Poisson distributions of substitution rates of branches within and between species in a phylogenetic tree (Zhang et al., 2013) The ML trees of 16S, 28S and the combined 16S/28S dataset were uploaded on the website of bPTP (http://sco.h-its.org/exelixis/web/software/PTP) without outgroups and bootstraps and only representing individual haplotypes. The statistical support of potential genetic species was calculated with the maximal possible number of 500,000 MCM generations and the default burn-in of 10%.
For comparisons, we also applied a third approach for genetic species delimitations, the automatic barcode gap discovery method (ABGD; Puillandre et al., 2012) which calculates genetic distances between all sequences and does not require any phylogenetic information.
Statistical analyses of current distribution data
We summarize current distribution data of all genetic species defined by the congruent molecular datasets regarding ecological (sediment type, water depth), and geographic (different lake basins, different shores) factors. We also compare our ecological distribution data to the much larger dataset of Mazepova (1998) on different sediment types and water depths of morphological Cytherissa species and subspecies.
We then generated a presence–absence matrix for each genetic species from the combined molecular dataset for the four distribution variables, using our geographic and ecological data and the ecological data of Mazepova (1998). This data matrix was used for ordination analyses in PAST (Hammer et al., 2001). More specifically, we conducted a Principal Coordinate Analysis (PCoA) with the jaccard similarity index, and the default transformation exponent of 2. This kind of analyses plots the distribution of genetic Cytherissa species in a coordination system where the axes are linked to the different distribution variables.
Results
DNA extraction
We have extracted DNA from more than 100 specimens, and have been successful in obtaining 68 sequences for 16S and 83 sequences of 28S, respectively (Table 1). Developing suitable primers for 16S has been a major obstacle and has involved several rounds of redesigning both forward and reverse primers. Problems with the primers are also the reason why we could not successfully follow the approach of Schön & Martens (2012) in acquiring more COI sequences from the same species and localities, which would have been very useful for further comparisons. Also, the specimens or DNA extractions of Schön & Martens (2012) were no longer available to be included in the current study.
Molecular taxonomy
Combined molecular datasets
Combining both molecular datasets has resulted in phylogenetic trees with some higher support for deeper nodes in the upper part of the tree (Fig. 2) than the phylogenies that were based only on 16S (Fig. S1) or 28S (Fig. S2). The terminal branches in Fig. 2 are generally well supported with bootstrap values of 75% of more and posterior probabilities of more than 0.8. In the combined 16S/28S tree, such well-supported clades consist of sister groups (C. lacustris I and II; C. golyschkinae I and II; C. parallela I and II) but also of clusters of different morphological (sub)species (C. parva and C. sp. 3; C. parallela III and both C. lacustris I and II; C. sernovi insularis I and C. sernovi sernovi). The remaining part of the tree, however, still contains many polytomies, especially at the deeper nodes. With the exception of C. lata I and II and C. tuberculata tuberculata IV and V, respectively, the phylogenetic relationships of the eight other clades remains unresolved.
Congruent phylogeny based on 16S and 28S. This phylogeny has been constructed with Maximum Likelihood and Bayesian methods on DNA sequences of 922 nucleotides each. Statistical support is shown above (PHYML, bootstrap values) and below (MrBayes, posterior probabilities) branches, respectively. C. sp. 1 to C. sp. 4 are new species still awaiting formal description. Roman numbers refer to genetic species according to Table 2. Species printed in bold also show morphological differences. The coloured columns next to the tree show the type of sediment (A), depth (B), basin (C) and shore (D). Missing data are indicated in black
The 16S and 28S DNA sequences come from 13 known morphological species and one subspecies sensu Mazepova (1990) plus four new species that await formal description elsewhere. With the combined molecular data, we identified 26 well-supported phylogenetic clades (Fig. 2). Many of these are congruent with morphological species (C. parva, C. pterygota, C. interposita, C. excelsiformis, C. glomerata and four yet undescribed species (C. spec. 1 to 4) plus one subspecies (C. sernovi sernovi). There are an additional five morphospecies with multiple, well-supported phylogenetic clades or with phylogenetically distant sister clades, both indicating possible cryptic species. Cytherissa tuberculata tuberculata splits into four such clades and C. parallela into three, while two each are found in C. lacustris, C. sernovi insularis, C. golyschkinae, C. sinistrodentata, and C. lata (Fig. 2).
We have used two different methods to test if these phylogenetic clades fulfil the criteria to be considered different evolutionary genetic species. Because of the more limited number of specimens for which DNA sequence data are available from both genomic regions, the number of singletons in the congruent phylogeny (Fig. 2) is larger than in the 16S tree (Fig. S1). Singletons cause potential problems when applying the 4θ rule (see below). Still, this method supports 17 genetic species within morphospecies (Tables 2, S1) plus another eight morphospecies (Fig. 2). The PTP algorithm recognizes all of the clades from the 4θ rule (Table 2) and additionally splits C. parallela I and II into two different genetic species with one singleton each (Table 2). The ABDG method delimitates the same species as PTP (when using the 16S data for ABDG; not shown, data are available from IS on request). We take a conservative approach in delimitating species, using support from all three methods and therefore regard the two clades of C. parallela I and II for now as two genetic species. We can then recognize a total of 26 different genetic species from the combined molecular data. For most of these genetic species, we find variation in valve characters (indicated in bold in Fig. 2), thus providing morphological support for genetic species boundaries. Many of these species are also the same as in Mazepova (1990). We also found four new morphological species (C. sp. 1–4) that are, with the exception of C. sp. 1, also fully supported by the combined molecular data. Other genetic species resemble the species sensu Mazepova (1990) to some extent but show additional valve differences. These species are still awaiting a formal taxonomic description and are for now indicated with Roman numbers after the original species name in Fig. 2 (C. parallela I–III and C. tuberculata tuberculata II–V—see Fig S3, SEM plate). However, in the other instances of genetic species with Roman numbers in Fig. 2, no clear morphological differences are found and these eight remaining lineages are here considered to be true cryptic species.
16S results
Because we could obtain more sequence data from this marker than could be used for the combined dataset, numbers of genetic species are slightly different. If we apply the PTP algorithm or the ABDG method (not shown, data are available from IS on request), we can identify 35 genetic species (Fig. S1). With the 4θ rule, the result is, with 36 genetic species, rather similar, but individual species delimitations are incongruent for morphospecies with several genetic species (Table S3).
When comparing the 16S species boundaries to morphological variability as we did for the combined molecular dataset, we find a total of nine truly cryptic species in the 16S dataset (Fig. S1), one more than with the 16S/28S data.
The structure of the 16S minimum spanning network (Fig. S4) matches the well-supported phylogenetic clades in Fig. S1. We find 58 different haplotypes that are separated from each other by more than 20 mutational steps. Within evolutionary genetic species such as for example C. tuberculata tuberculata III, C. lacustris II or C. golyschkinae IV, we also find haplotypes differing only by small numbers of mutational steps.
28S results
The nuclear ribosomal 28S region shows very little genetic variability amongst Baikalian Cytherissa species. Consequently, the phylogenetic tree is unresolved with very few exceptions (see Fig. S2). There are only 18 haplotypes in the minimum spanning network, with a maximum of four mutation steps (Fig. S5) although more than 80 specimens from have been sequenced from the entire lake. The network shows three very common 28S haplotypes (Fig. S5). The most frequent one is present in more than ten different morphospecies and subspecies. Except for one specimen of C. golyschkinae and C. verrucosa each that are separated by four mutation steps from the next haplotype (Fig. S5), all other single 28S haplotypes are only one or two mutational steps away from the three most common haplotypes or from each other. Because of the limited genetic diversity of 28S, we did not use these DNA sequence data to delimitate genetic species boundaries.
Current distribution of genetic Cytherissa species
Our sampling scheme contains habitats with different ecological (sediment type, water depth) and geographic features (south, central and northern basin; and east and west shores), which could have contributed to different distributions of the genetic Cytherissa species. Because our sample numbers are somewhat limited, we have compared our distribution data to the larger dataset of Mazepova (1998; Table S2). It seems that certain morphological (sub)species have previously been found on more sediment types than in our study (e.g. C. golyschkinae, C. tuberculata tuberculata, C. excelsiformis and C. glomerata; Table S2). Mazepova (1998) also reported a wider depth distribution for these three morphological (sub)species as well as for C. sinistrodentata. For the remaining seven genetic species, our data match the depth distributions of Mazepova (1998) well.
Table 3 summarizes the distribution data of all genetic species from the congruent molecular dataset, arranged in pairs of genetic sister clades to allow easy comparisons. A PCoA analysis of these data shows that most genetic species are well separated from each other (Fig. 3), also the species pairs from Table 3 and the various cryptic species (see above). The first axis with an eigenvalue of 1.5139 explains 35.6% of the overall variation and the second one 22.03%, which are relatively high scores.
Results of the principal coordinate analysis of genetic species defined by the congruent molecular dataset and their ecological and geographic distributions. Identities of genetic species are similar to Fig. 2. Species pairs from Table 3 are indicated by similar colours. Unpaired species are shown in black. If dots are labelled with several species names, these species share the same space in the coordination system. For all species shown, ecological data on sediment type and water depth were taken from Mazepova (1998; see Table S2 for details), and geographical distribution data according to Table 3. Note that the new species C. sp. 4 from Table 3 was not included in the analyses because no data on sediment type and water depth were available for this species from Mazepova (1998). t tuberculata
Discussion
Phylogenetic and network structures
We have sequenced two different genetic markers, namely part of the mitochondrial 16S and part of the nuclear 28S ribosomal region from 18 morphological (sub)species of Cytherissa. The molecular phylogenies from both genomic regions show many polytomies, especially of the deeper nodes, regardless of the methods used for phylogenetic reconstructions (Figs. 2, S1, S2) and when using the two datasets either separately or combined (Figs. 2, S1, S2). In our 16S and combined 16S/28S trees, only the terminal nodes and some deeper nodes (16S/28S) are statistically well supported (Fig. 2), whereas almost the entire 28S phylogeny remains unresolved (Fig. S2). Our phylogenetic results thus resemble those of Schön & Martens (2012), as also in their study, the mitochondrial phylogeny of Baikalian Cytherissa ostracods based on COI had many polytomies, and only the terminal nodes were well supported in the mitochondrial gene (COI), while the nuclear phylogeny (from the ITS1 region) was not resolved at all. Similar incongruences in genetic variability between mitochondrial and nuclear markers have also been reported from other studies on ostracods (Schön et al., 1998, 2010, 2012, 2014; Brandao et al., 2010; Koenders et al., 2012), and on meiofauna in general (Tang et al., 2012), resulting in low phylogenetic resolution and polytomies. One potential causality for this discrepancy is that nuclear ribosomal regions in non-marine ostracods generally seem to evolve at a much slower pace than mitochondrial regions (Schön et al., 2003). Even more relevant here is the detection of explosive speciation in Baikalian Cytherissa (Schön & Martens, 2012), which explains best why our phylogenies are unresolved at the base of the trees.
The topology of our combined tree (Fig. 2) reveals certain similarities with the trees in Schön & Martens (2012), as C. tuberculata is closest to the root of the Cytherissa flock and C. parva forms a well-supported clade with C. sp. 3, apart from the other Cytherissa species. However, in our present results, we cannot detect the four well-supported clades from the COI tree of Schön & Martens (2012). These inconsistencies could be owing to differences in genetic variability between 16S and COI and the fact that our dataset is not fully congruent with the data of Schön & Martens (2012). Because these authors used other specimens, it is also not possible to combine and re-analyse all existing molecular data of Cytherissa. The problem of resolving the Baikalian Cytherissa phylogenies urgently calls for the development of more suitable, large scale molecular markers such as sequencing entire mitogenomes (Schön & Martens, 2016) or large scale genomic data from multiple markers, such as those Meyer et al. (2015) developed for cichlid fish.
Diversity of Baikalian Cytherissa
Our results show that the actual biodiversity of endemic Cytherissa in Lake Baikal is higher than previously thought. Mazepova (1990) recognized a total of 47 species and 10 subspecies of Cytherissa based on valve characters. We have used SEM (see above and Fig. S3) to study differentiation of valve morphology in all specimens and also characterized hemipenis morphology for selected Cytherissa species (Van Mulken et al. in prep.). Both methods provide a much finer resolution of morphological differentiation, as is illustrated by the four (16S/28S combined) to five (16S) new Cytherissa species that we found and that are confirmed with our genetic data. Also, in three Cytherissa morphospecies sensu Mazepova (1990), we can distinguish nine genetic Cytherissa species with clear differences in valve morphologies, that also fulfil the criteria of the evolutionary genetic species concept using the combined DNA sequence data (C. parallela I–III, C. lacustris I & II, C. tuberculata tuberculata II–V; see Fig. S3 for the latter).
We have furthermore detected several truly cryptic species (without any apparent morphological differentiation), which supports the first indications reported by Schön & Martens (2012) for cryptic speciation in Baikalian ostracods. Using the combined molecular data set, we identify eight cryptic species (nine when only using the 16S; see Fig. S1). When applying two different statistical methods to delimitate evolutionary genetic species, the 4θ rule and the PTP algorithm, the overall estimate was for the combined dataset relatively similar with 26 and 27 species, respectively.
However, our combined 16S/28S tree has many well supported clades with singletons (Fig. 2), and the genetic diversity within such clades is zero. Consequently, the ratios would have to be divided by zero and can thus not be calculated (Table S1). The results of the PTP algorithm are probably more robust as this method can also be used for singletons and we found furthermore the same genetic species when applying the ABDG method and this for both 16S and the combined molecular dataset. We could not increase the number of specimens in our study, because of the difficulties to obtain more 16S sequences (see above) and because of low densities of non-marine ostracods in Lake Baikal, especially at greater depths.
In total, we can identify 26 different genetic species with the combined molecular data set, representing 14 morphological (sub)species sensu Mazepova (1990). Our data thus almost double the previously known diversity of endemic Cytherissa species from Lake Baikal. Our sampling includes all three basins of Lake Baikal, five different sediment types and water depths ranging from shallow habitats (c 20 m) to more than 500 m, covering most of the habitat and geographical diversity of Lake Baikal. Extrapolating our results on morphological and cryptic diversity to the entire Baikalian Cytherissa species flock (26 genetic species in 14 morphospecies sensu Mazepova (1990)) implies that we can expect almost twice as many (cryptic) Cytherissa species from Lake Baikal as previously known, with therefore one hundred species, including cryptic ones, being a more realistic estimate than the 47 morphological (sub)species sensu Mazepova (1990) previously known. Studies on ostracods from other ancient lakes have also reported the presence of cryptic species, thus considerably increasing classic diversity estimates (Schön et al., 2014; Karanovic, 2015). Likewise, cryptic species have also been found in Baikalian amphipods (Vainola & Kamaltynov, 1999) and in Baikalian sponges (Itskovich et al., 2015).
High cryptic diversity and cryptic speciation in ancient lakes somewhat negates the recent findings by Poulin & Pérez-Ponce de León (2017), who attributed the higher cryptic diversity in freshwater as compared to terrestrial and marine habitats, to the greater heterogeneity of freshwater habitats. This hypothesis mostly refers to the patchiness and isolation of the many freshwater pools, lakes and rivers. However, in the case of ancient lakes, their long evolutionary history, large size and unusual depth are probably more important for generating cryptic diversity than providing many heterogeneous habitats.
Based on classic morphological species boundaries, ancient lakes have already been identified as major hotspots for non-marine ostracod diversity as, for example, they contribute 25% of all known freshwater ostracod species (Martens et al., 2008). The increase of the known diversity through the discovery of cryptic species from our and other studies emphasises the importance of ancient lakes as biodiversity hot spots, not only for ostracods. This has major implications for the conservation and protection of these lakes and their unique fauna and flora, even outside the lakes themselves (Schön et al., 2000; Schön & Martens, 2012).
Factors linked to speciation in Baikalian Cytherissa
Ancient lakes are in situ laboratories for evolutionary studies in general, and to investigate the factors that have promoted and caused speciation, giving rise to the impressive endemic diversity of these lakes in particular. Mayr (1942, 1963) regarded geographic isolation as the most important driver for (allopatric) speciation and this view dominated the field for a long time. Meanwhile, also the importance of intrinsic factors for sympatric speciation in ancient lakes has been recognized (see, for example, Schön & Martens, 2004 and Cristescu et al., 2010), with cichlid fish still being the most prominent example (e.g. Muschick et al., 2012). Martens (1994, 1997) furthermore re-iterated the term “parapatric speciation”, describing isolation and gene flow along an ecological or geographical gradient, which is highly applicable to Lake Baikal with its deep, fully oxygenated abyss (down to 1600 m), and its north–south length of more than 600 km (Martin, 1994). Because our study detected at least 26 genetic species of Cytherissa, including some cryptic species, and because our sample scheme included all three basins of Lake Baikal from the eastern and western shore, depths ranging from shallow to deep water habitats and five sediment types, we can make a first attempt to assess how recent Cytherissa species could be ecologically and geographically separated. Because of the limited number of molecular data currently available and the lack of extensive, dated phylogenies, our analyses can only provide the very first steps towards future, rigorous testing of hypotheses on allo- or parapatric speciation of non-marine ostracods in Lake Baikal in general and of selected Cytherissa clades in particular.
Geographic and ecological separation
Geographic separation because of historical vicariance might to some extent have shaped Cytherissa diversity, and possibly, also speciation in Lake Baikal. Our PCoA illustrates that most genetic Cytherissa species are clearly separated by extrinsic factors (Fig. 3), even if we use the (wider) ecological distribution data of Mazepova (1998) for water depths and sediment for morphological species and subspecies without being able to differentiate further according to our genetic species (see Table S2). What is more difficult to assess is the extent to which each factor might have contributed to the current ecological and geographic distribution and to speciation in the past. We find several examples where different (cryptic) Cytherissa species seem to be limited in their geographic distribution to a single Baikalian basin or shore (species pairs: C. lacustris I & II; Cytherissa sernovi insularis II & C. sernovi sernovi, C. lata I & II, C.tuberculata tuberculata IV & V; see Table 3). Distribution patterns potentially resulting from allopatric speciation amongst basins have also been described from Lake Tanganyika for ostracods (Schön et al., 2014) and cichlid fish (Snoeks et al., 1994; Rüber et al., 1999, 2001; Sturmbauer et al., 2001; Nevado et al., 2009, 2011). Keeping the lack of ancestral reconstructions and thus rigorous testing of this hypothesis in mind, our preliminary indications for allopatric speciation in Lake Baikal are still noteworthy as only one other case of supposed allopatric speciation from this lake is documented up to now, namely the case of Eulimnogammarus cyaneus versus E. messerschmidtii (Bedulina et al., 2014).
Other examples in our dataset show that besides geographic separation, also ecological factors like water depths or sediment types might have further contributed to the current disjunct distribution of certain Cytherissa species (e.g. the species pairs C. parallela I & II; C. lata I & II and Cytherissa tuberculata tuberculata IV & V, see Table 3).
For most of the genetic Cytherissa species, our depth ranges match the ones of Mazepova (1998) remarkably well. Exceptions include C. glomerata but also the common morphospecies C. sernovi, C. sinistrodentata and C. tuberculata tuberculata, for which Mazepova (1998) reported much wider depth distributions than we found for our different genetic species constituting these classic morphospecies (Table S2). Whether or not there is indeed a clear separation between the genetic/cryptic species by water depths and/or by sediment type (as in snails from Lake Tanganyika; Michel et al., 1992) still has to be further tested with more extensive sampling and subsequent genetic characterization.
Potential intrinsic factors
We can currently not assess at all to which extent intrinsic factors or adaptive evolution has caused sympatric ostracod speciation. To investigate trophic niches, for example, detailed morphological investigations of the appendages involved in food processing or stable isotope analyses would be needed. Preliminary analyses of soft part morphology in Cytherissa offer no indication for trophic specialization in the relevant head appendages (Danielopol & Tétart, 1990).
To study other intrinsic mechanisms, like for example hybridization or introgression which are common in cichlids from African ancient lakes (Koblmüller et al., 2007; Nevado et al., 2009, 2011; Cristescu et al., 2010; Genner & Turner, 2011; Anseeuw et al., 2012; Meier et al., 2017), is not possible to date as no suitable molecular tools are as yet available for ostracods. It remains therefore uncertain whether the large number of 28S haplotypes shared between different Cytherissa species (Fig. S5) is a first indication for hybridization or is merely a reflection of the low variability of this nuclear region.
Sexual selection has often been cited as a major driver in ancient lake speciation, and the best documented examples are of course (again) the cichlid fish (reviewed in Wagner et al., 2012). Sexual selection in ostracods has previously been documented in both freshwater (Martens, 2000) and marine groups (Tsukagoshi, 1988), and is often detectable by wide morphological differences in copulatory structures (hemipenes, prehensile palps) between otherwise closely related species. However, the study by Van Mulken et al. (in prep.) shows that the copulatory appendages in Baikalian Cytherissa species, albeit quite elaborate, are very similar amongst otherwise different species.
Cytherissa lacustris II is found in both Lake Baikal and in the UK (Table 1; Fig. S4; Schön & Martens, 2012) and has different reproductive modes. In Lake Baikal, its “sors” (shallow lagunas associated with the lake) and in Lake Huvsugul (Mazepova, 2006) it is fully sexual, while in the rest of the Holarctic it is obligate asexual (Schön et al., 2000; Schön & Martens, 2012). This variation in reproductive mode indicates that such intrinsic factors might also be relevant for ostracod speciation in ancient lakes (Martens, 1994) and elsewhere, and need to be studied with suitable tools.
Conclusions
To summarize, we found strong evidence from two molecular markers and morphological variation that the Cytherissa diversity from Lake Baikal is probably twice as large as previously known. Our preliminary data also indicate that the 26 genetic species are to some extend separated by ecological (sediment types, water depths) as well as geographic (basin, shore) factors. We argue that these separations might have been causal to allopatric and parapatric speciation along gradients without complete isolation. In the case of Lake Baikal, such gradients include the vast geographic distances among the three basins, between the two shores (east and west) and the large ecological gradient in water depth down to c 1600 m. These hypotheses will need to be rigorously tested in future research.
Other external factors possibly promoting speciation, such as multiple invasions, have already been documented (Schön & Martens, 2012), while also adaptive and intrinsic components are expected to have further contributed to generating the high diversity of Baikalian ostracods and other endemic taxa. We hope that with the increasing availability of various “omics” techniques, also applicable to ostracods (Schön & Martens, 2016), future studies will be able to answer these fundamental questions of evolutionary biology, and in particular on speciation in ancient lakes.
References
Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller & D. J. Lipman, 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402.
Anseeuw, D., B. Nevado, P. Busselen, J. Snoeks & E. Verheyen, 2012. Extensive introgression among ancestral mtDNA lineages: phylogenetic relationships of the Utaka within the Lake Malawi Cichlid Flock. International Journal of Evolutionary Biology 2012: 865603.
Bandelt, H., P. Forster & A. Röhl, 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37–48.
Bedulina, D. S., V. V. Takhteev, S. G. Pogrebnyak, E. B. Govorukhina, E. V. Madyarova, Y. A. Lubyaga, K. P. Vereshchagina, M. A. Timofeyev & T. Luckenbach, 2014. On Eulimnogammarus messerschmidtii, sp. n. (Amphipoda: Gammaridea) from Lake Baikal, Siberia, with redescription of E. cyanoides (Sowinsky) and remarks on taxonomy of the genus Eulimnogammarus. Zootaxa 3838: 518–544.
Beheregaray, L. B. & A. Caccone, 2007. Cryptic biodiversity in a changing world. Journal of Biology 6: 9. doi:10.1186/jbiol60.
Bickford, D., D. J. Lohman, N. S. Sodhi, P. K. L. Ng, R. Meier, K. Winker, K. K. Ingram & A. Das, 2007. Cryptic species as a window on diversity and conservation. Trends in Ecology and Evolution 22: 148–155.
Birky, C. W. J. R., 2013. Species detection and identification in sexual organisms using population genetic theory and DNA sequences. PLoS ONE 8: e52544.
Birky, C. W., J. Adams, M. Gemmel & J. Perry, 2010. Using population genetic theory and DNA sequences for species identification in asexual organisms. PLoS ONE 5: e10609.
Birky Jr., C. W. & T. G. Barraclough, 2009. Asexual speciation. In Schön, I., K. Martens & P. van Dijk (eds), Lost sex, the evolutionary biology of parthenogenesis. Springer, Dordrecht: 201–216.
Bode, S. N. S., D. K. Lamatsch, M. J. F. Martins, O. Schmit, J. Vandekerkhove, F. Mezquita, T. Namiotko, G. Rossetti, I. Schön, R. K. Butlin & K. Martens, 2010. Exceptional cryptic diversity and multiple origins of parthenogenesis in a freshwater ostracod. Molecular Phylogenetics and Evolution 5: 542–552.
Brandao Nunes, S., J. Sauer & I. Schön, 2010. Circumantarctic and eurybathid distribution in Southern Ocean benthos? A genetic test using Macrocyprididae (Crustacea, Ostracoda) as model organism. Molecular Phylogeny and Evolution 159: 219–243.
Brown, D. M., R. A. Brenneman, N. J. Georgiadis, K. Koepfli, J. P. Pollinger, B. Mila, E. L. Louis Jr., G. F. Grether, D. K. Jacobs & R. K. Wayne, 2007. Extensive population genetic structure in the giraffe. BMC Biology 5: 57.
Cristescu, M. E., S. J. Adamowicz, J. J. Vaillant & D. G. Haffner, 2010. Ancient lakes revisited: from the ecology to the genetics of speciation. Molecular Ecology 19: 4837–4851.
Danielopol, D. L. & J. Tétart, 1990. Morphology of Cytherissa and Cyprideis: supplementary data on the appendages and the karyotype. In Danielopol, D. L., P. Carbonel & J. P. Colin (eds), Cytherissa (Ostracoda)—the Drosophila of palaeolimnology, Vol. 47–48, 55–67. Bulletin de l’Institut de Géologie du Bassin d’Acquitaine, Université Bordeaux, Bordeaux.
Darriba, D., G. L. Taboada, R. Doallo & D. Posada, 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772.
Eldredge, N. & J. Cracraft, 1980. Phylogenetic patterns and the evolutionary process. Columbia University Press, New York.
Elmer, K. R., J. A. Davila & S. C. Lougheed, 2007. Cryptic diversity and deep divergence in an upper Amazonian frog, Eleutherodactylus ockendeni. BMC Evolutionary Biology 7: 247.
Fontaneto, D., T. G. Barraclough, K. Chen, C. Ricci & E. A. Herniou, 2008. Molecular evidence for broad-scale distributions in bdelloid rotifers: everything is not everywhere but most things are very widespread. Molecular Ecology 17: 3136–3146.
Genner, M. J. & G. F. Turner, 2011. Ancient hybridization and phenotypic novelty within Lake Malawi’s cichlid fish radiation. Molecular Biology and Evolution 29: 195–206.
Guindon, S. & O. Gascuel, 2003. PhyML—a simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52: 696–704.
Gustafsson, D. R., D. A. Price & C. Erséus, 2009. Genetic variation in the popular lab worm Lumbriculus variegatus (Annelida: Clitellata: Lumbriculidae) reveals cryptic speciation. Molecular Phylogenetics and Evolution 51: 182–189.
Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symposium Series 41: 95–98.
Hammer, Ø., D. A. T. Harper & P. D. Ryan, 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4(1): 9.
Itskovich, V. B., O. V. Kaluzhnaya, E. Veynberg & D. Erpenbeck, 2015. Endemic Lake Baikal sponges from deep water. 1: potential cryptic speciation and discovery of living species known only from fossils. Zootaxa 3990: 123–137.
Karanovic, I., 2015. Barcoding of ancient lake ostracods (Crustacea) reveals speciation with extremely low distances. PLoS ONE 10: e0121133.
Katoh, K. & D. M. Standley, 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780.
Koblmüller, S., N. Duftner, K. M. Sefc, M. Aibara, M. Stipacek, M. Blanc, B. Egger & C. Sturmbauer, 2007. Reticulate phylogeny of gastropod-shell-breeding cichlids from Lake Tanganyika—the result of repeated introgressive hybridization. BMC Evolutionary Biology 7: 7.
Koenders, A., K. Martens, S. Halse & I. Schön, 2012. Cryptic species of the Eucypris virens species complex (Ostracoda, Crustacea) have invaded Western Australia. Biological Invasions 14: 2187–2201.
Marrone, F., S. Lo Brutto & M. Arculeo, 2010. Molecular evidence for the presence of cryptic evolutionary lineages in the freshwater copepod genus Hemidiaptomus G.O. Sars, 1903 (Calanoida, Diaptomidae). Hydrobiologia 644: 115–125.
Martens, K., 1994. Ostracod speciation in ancient lakes: a review. In Martens, K., B. Goddeeris & G. Coulter (eds), Speciation in Ancient Lakes, Advances in Limnology, Vol. 44. Sinauer Associates, Sunderland: 203–222.
Martens, K., 1997. Speciation in ancient lakes. Trends in Ecology and Evolution 12: 177–182.
Martens, K., 2000. Factors affecting the divergence of mate recognition systems in the Limnocytherinae (Crustacea, Ostracoda). Hydrobiologia 419: 83–101.
Martens, K., I. Schön, C. Meisch & D. J. Horne, 2008. Global diversity of ostracods (Ostracoda, Crustacea) in freshwater. Hydrobiologia 595: 185–193.
Martin, P., 1994. Lake Baikal. In Martens, K., B. Goddeeris & G. Coulter (eds), Speciation in Ancient Lakes. Advances in Limnology. Sinauer Associates, Sunderland.
Mayr, E., 1942. Systematics and the Origin of Species. Columbia University Press, New York.
Mayr, E., 1963. Animal Species and Evolution. The Belknap press, Cambridge.
Mazepova, G., 1990. Rakushokovye ratchki (Ostracoda) Baikala Nauk. Sib. Otdel. Akad. Nauk SSR, Novosibirsk: 1–470.
Mazepova, G. F., 1998. List of Ostracoda species. In Kozhova, O. M. & L. R. Izmest’eva (eds), Lake Baikal Evolution and Biodiversity. Backhuys Publishers, Leiden: 371–377.
Mazepova, G. F., 2006. Ostracoda of Lake Hövsgöl, Mongolia. In Goulden, C. E., T. Sitnikova, J. Gelhaus & B. Boldgiv (eds), The Geology. Biodiversity and Ecology of Lake Hövsgöl (Mongolia). Backhys Publisher, Leiden: 217–232.
Meier, J. I., D. A. Marques, S. Mwaiko, C. E. Wagner, L. Excoffier & O. Seehausen, 2017. Ancient hybridization fuels cichlid fish adaptive radiations. Nature Communications 8: 14363.
Meyer, B. S., M. Matschiner & W. Salzburger, 2015. A tribal level phylogeny of Lake Tanganyika cichlid fishes based on a genomic multi-marker approach. Molecular Phylogenetics and Evolution 83: 56–71.
Michel, E., A. S. Cohen, K. West, M. R. Johnston & P. W. Kat, 1992. Large African lakes as natural laboratories for evolution: examples from the endemic gastropod fauna of Lake Tanganyika. Mitteilungen der Internationalen Vereinigung für Theoretische und Angewandte Limnologie 23: 85–99.
Müller, J., H. Oberhänsli, M. Melles, M. Schwab, V. Rachold & H.-W. Hubberten, 2001. Late Pliocene sedimentation in Lake Baikal: implications for climatic and tectonic change in SE Siberia. Palaeogeography, Palaeoclimatololgy and Palaeoecology 174: 305–326.
Muschick, M., A. Indermaur & W. Salzburger, 2012. Convergent evolution within an adaptive radiation of cichlid fishes. Current Biology 22: 2362–2368.
Nevado, B., S. Koblmüller, C. Sturmbauer, J. Snoeks, J. Usano-Alemany & E. Verheyen, 2009. Complete mitochondrial DNA replacement in a Lake Tanganyika cichlid fish. Molecular Ecology 18: 4240–4255.
Nevado, B., T. Backeljau, M. Hanssens & E. Verheyen, 2011. Repeated unidirectional introgression of nuclear and mitochondrial DNA between four congeneric Tanganyikan cichlids. Molecular Biology and Evolution 28: 2253–2267.
Pfenninger, M. & K. Schwenk, 2007. Cryptic animal species are homogeneously distributed along taxa and biogeographic regions. BMC Evolutionary Biology 7: 121.
Poulin, R. & G. Pérez-Ponce de León, 2017. Global analysis reveals that cryptic diversity is linked with habitat but not mode of life. Journal of Evolutionary Biology 30: 641–649. doi:10.1111/jeb.13034.
Puillandre, N., A. Lambert, S. Brouillet & G. Achaz, 2012. ABGD, automatic barcode gap discovery for primary species delimitation. Molecular Ecology 21: 1864–1877.
Rambaut, A., 2017. Figtree, a graphical viewer of phylogenetic trees. http://tree.bio.ed.ac.uk/software/figtree.
Ronquist, F., M. Teslenko, P. van der Mark, D. Ayres, A. Darling, S. Höhna, B. Larget, L. Liu, M. A. Suchard & J. P. Huelsenbeck, 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542.
Rüber, L., E. Verheyen & A. Meyer, 1999. Replicated evolution of trophic specializations in an endemic cichlid fish lineage from Lake Tanganyika. Proceedings of the National Academy of Sciences USA 96: 10230–10235.
Rüber, L., A. Meyer, C. Sturmbauer & E. Verheyen, 2001. Population structure in two sympatric species of the Lake Tanganyika cichlid tribe Eretmodini: evidence for introgression. Molecular Ecology 10: 1207–1225.
Schön, I. & K. Martens, 2004. Adaptive, preadaptive and non-adaptive components of radiations in ancient lakes: a review. Organisms, Diversity and Evolution 4: 137–156.
Schön, I. & K. Martens, 2012. Molecular analyses of ostracod flocks from Lake Baikal and Lake Tanganyika. Hydrobiologia 682: 91–110.
Schön, I. & K. Martens, 2016. Ostracod (Ostracoda, Crustacea) genomics – promises and challenges. Marine Genomics 29: 19–25.
Schön, I., R. K. Butlin, H. I. Griffiths & K. Martens, 1998. Slow molecular evolution in an ancient asexual ostracod. Proceedings of the Royal Society of London Series B 265: 235–242.
Schön, I., E. Verheyen & K. Martens, 2000. Speciation in ancient lake ostracods: comparative analysis of Baikalian Cytherissa and Tanganyikan Cyprideis. Mitteilungen der Internationalen Vereinigung für Theoretische und Angewandte Limnologie 27: 2674–2681.
Schön, I., K. Martens, K. Van Doninck & R. K. Butlin, 2003. Evolution in the slow lane: molecular rates of evolution in sexual and asexual ostracods (Crustacea: Ostracoda). Biological Journal of the Linnean Society 79: 93–100.
Schön, I., K. Martens & S. Halse, 2010. Genetic diversity in Australian ancient asexual Vestalenula (Ostracoda, Darwinulidae)—little variability down-under. Hydrobiologia 641: 59–70.
Schön, I., R. Pinto, S. Halse, A. Smith, K. Martens & C. W. Birky, 2012. Cryptic diversity in putative ancient asexual darwinulids (Crustacea: Ostracoda). PLoS ONE 7: e39844.
Schön, I., C. Poux, E. Verheyen & K. Martens, 2014. High cryptic diversity and persistent lineage segregation in endemic Romecytheridea (Crustacea, Ostracoda) from the ancient Lake Tanganyika. Hydrobiologia 739: 119–131.
Sherbakov, D. Y., 1999. Molecular phylogenetic studies on the origin of biodiversity in Lake Baikal. Trends in Ecology and Evolution 14: 92–95.
Sherstyankin, P. P. & L. N. Kuimova, 2006. Hydrophysical processes in Lake Baikal in its transition from subtropical to modern climates. Hydrobiologia 568(S): 253–257.
Snoeks, J., L. Rüber & E. Verheyen, 1994. The Tanganyika problem: comments on the taxonomy and distribution patterns of its cichlid fauna. Archiv für Hydrobiologie, Beihefte Ergebnisse der Limnologie 44: 355–372.
Sonnenberg, R., A. W. Nolte & D. Tautz, 2007. An evaluation of LSU rDNA D1-D2 sequences for their use in species identification. Frontiers in Zoology 4: 6.
Sturmbauer, C., S. Baric, W. Salzburger, L. Rüber & E. Verheyen, 2001. Lake level fluctuations synchronize genetic divergences of cichlid fishes in African lakes. Molecular Biology and Evolution 18: 144–154.
Tamura, K., G. Stecher, D. Peterson, A. Filipski & S. Kumar, 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729.
Tang, C. Q., F. Leasi, U. Obertegger, A. Kieneke, T. G. Barraclough & D. Fontaneto, 2012. The widely used small subunit 18S rDNA molecule greatly underestimates true diversity in biodiversity surveys of the meiofauna. Proceedings of the National Academy of Sciences of the United States of America 109: 16208–16212.
Timoshkin, O. A., 2001. Lake Baikal: diversity of fauna, problems of its immiscibility and origin, ecology and ‘exotic’ communities. In Timoshkin, O. A. (ed.), Index of animal species inhabiting Lake Baikal and its catchment area. Nauka, Novosibirsk: 74–113.
Tsukagoshi, A., 1988. Reproductive character displacement in the ostracod genus Cythere. Journal of Crustacean Biology 8: 563–575.
Vaidya, G., D. J. Lohman & R. Meier, 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27: 171–180.
Vainola, R. & R. M. Kamaltynov, 1999. Species diversity and speciation in the endemic amphipods of Lake Baikal: molecular evidence. Crustaceana 72: 945–956.
Vogler, A. P. & M. T. Monaghan, 2007. Recent advances in DNA taxonomy. Journal for Zoological Systematics and Evolutionary Research 45: 1–10.
Wagner, C. E., L. J. Harmon & O. Seehausen, 2012. Ecological opportunity and sexual selection together predict adaptive radiation. Nature 487: 366–369.
Zhang, J., P. Kapli, P. Pavlidis & A. Stamatakis, 2013. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29: 2869–2876.
Acknowledgements
This work was funded by the Intra European Marie Curie Fellowship CRYSTAL (Cryptic ostracod species in an Ancient Lake: the Cytherissa flock from Baikal, contract: PIEF-GA-2009-253767) and a SYNTHESYS grant to VP. DS acknowledges the RBFBR grant 15-04-03848 for funding. We also acknowledge the ESF EUROCHORES programme Eurodiversity for funding the MOLARCH project (05_EDIV_FP237-MOLARCH), of which Erik Verheyen (Brussels, Belgium) was the coordinator. We thank Julian Cillis (Brussels, Belgium) for technical assistance with SEM images and Janet Higuchi (Maringa, Brazil) for assembling the figure with the SEM pictures. Zohra Elouaazizi is gratefully acknowledged for assistance with DNA sequencing. We also acknowledge our Russian colleagues, foremost M. Grachev and O. Timoshkin, for their support during sampling and the visits of KM and VP to Irkutsk. We also wish to thank the members of the molecular lab and of the Freshwater Biology team in Brussels for their help and assistance, especially Tasnim Patel and Marie Cours. We also thank two referees for their useful comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Isa Schön and Valentina Pieri share the position as first authors.
Guest editors: Koen Martens, Sidinei M. Thomaz, Diego Fontaneto & Luigi Naselli-Flores / Emerging Trends in Aquatic Ecology II.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Schön, I., Pieri, V., Sherbakov, D.Y. et al. Cryptic diversity and speciation in endemic Cytherissa (Ostracoda, Crustacea) from Lake Baikal. Hydrobiologia 800, 61–79 (2017). https://doi.org/10.1007/s10750-017-3259-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-017-3259-3