Abstract
Darwinulid ostracods are putative ancient asexuals, and are thus assumed to be unable to purge deleterious mutations from their genomes. Some darwinulids species can be found both above (epigeic) and below ground (hypogeic). We hypothesize that surface populations carry more mutations than their below-ground counterparts, which are buffered from mutagens such as UV-B. Given the age of the investigated area, the Pilbara in Western Australia, we also expect geographic patterning of observed haplotypes. We have used DNA sequence data from the nuclear ITS and the mitochondrial COI region to investigate a (limited) data set on two Australian species, the endemic Vestalenula matildae and V. marmonieri from the Pilbara region. We do not find differences in genetic variability between specimens from subterranean habitats as compared to those from habitats above ground. There was also no congruence between hydrological basins and distribution patterns of the haplotypes identified. Although our data indicate that the two species may have split from each other ca. 70 myr ago, this has not resulted in any clear phylogeographic patterns among the analysed specimens across the regions of the Pilbara.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The paradox of sex is one of the remaining problems in evolutionary biology (Bell, 1982). There are more than 25 hypotheses that attempt to explain why sex is necessary to survive in the long term. The most important ones are the theories on accumulation of mutations (Muller, 1964; Kondrashov, 1988) and slower adaptation (Fisher, 1930; Muller, 1932) to changing environments (Maynard Smith, 1980; Roughgarden, 1991) or to arms races, the so-called Red Queen dynamics (Van Valen, 1973; Hamilton, 1980).
Ancient asexuals are expected to accumulate deleterious mutations at a high frequency as they have lost the ability to purge these from their genomes through meiosis. Muller’s ratchet predicts the irreversible loss of mutation-free genotypes in definite populations of asexuals (Muller, 1964) because once such a mutation-free genotype is lost, for example through drift, it cannot be restored through meiosis. Kondrashov’s hatchet, on the other hand, describes the accumulation of deleterious mutations in the long term if the mutation rate is sufficiently high and effects are at least mildly deleterious (Kondrashov, 1993). Since these processes take time, they should be most prominent in ancient asexuals. If sex and meiosis are absent for millions of years, even the two alleles of the same gene in a diploid organism should independently accumulate mutations (Birky, 1996; Mark Welch & Meselson, 2000), a process that has been called the Meselson effect (Butlin, 2000).
The family Darwinulidae is one of the three examples of putative ancient asexual groups in the animal kingdom besides bdelloid rotifers and some lineages in the oribatid mites (Schön et al., 2008) and, relying on fossil data, may have reproduced without sex for 200 myr or more (Martens et al., 2003).
The family Darwinulidae consists of five genera and about 30 described species (Martens et al., 2008) although diversity was higher in the Palaeozoic, before sex was abandoned (Martens et al., 2003; Schön et al., 2009). Recent species can clearly be distinguished by morphological differences in their valves and/or soft parts (Rossetti & Martens, 1998). Darwinulids are found on all continents (except Antarctica) in different kinds of water bodies such as lakes and rivers, as well as in subterranean (e.g. Artheau, 2007) and (semi-)terrestrial environments (Pinto et al., 2003, 2004) but not in temporary pools (Martens, 1998; Rossetti & Martens, 1998). The majority of the recent species are rare, being restricted to their type locality (e.g. Vestalenula molopoensis) or a certain region (V. cornelia). Some species have an intercontinental distribution: e.g. Darwinula stevensoni, Penthesilenula brasiliensis and P. aotearoa (Rossetti & Martens, 1998).
Most research on darwinulids to date has concentrated on the type species, D. stevensoni. Ecological experiments have shown that D. stevensoni displays the so-called General Purpose Genotype, as the species is able to survive in wide range of salinities and temperatures (Van Doninck et al., 2002). Such a genotype may make it no longer necessary to track environmental changes, because it is already adapted to a wide range of environmental factors. This seems also to be the case for P. brasiliensis, whereas other darwinulid species (P. aotearoa and V. molopoensis) are obviously more specialized in their ecology (Van Doninck et al., 2003).
In contrast to the theoretical genetic predictions mentioned above, sequence data of three nuclear region have shown no (Schön et al., 1998) or extremely low (Schön & Martens, 2003) genetic variability in D. stevensoni, implying that the Meselson effect is absent in this (Schön & Martens, 2003) and other darwinulid ostracods (Schön et al., in preparation). The accumulation of mutations in the nuclear genome of at least a few darwinulids might be counteracted by homogenizing mechanisms such as gene conversion (Schön et al., in preparation; Schön & Martens, 2003).
Genetic investigations of D. stevensoni focusing on phylogenetic and phylogeographic analyses of one African and 14 European populations (Schön et al., 1998; Schön, 2007) have demonstrated some genetic diversity in part of the mitochondrial COI gene. Whereas there is a clear difference between the population from South Africa and the European populations, no obvious phylogeographic patterns could be observed among the latter (Schön, 2007). In P. brasiliensis, several cryptic species have been discovered with COI sequence data, mainly representing intercontinental distributions although also at least four cryptic species were reported locally from Brazil (Schön et al., in preparation).
Recently, the ancient asexual status of the Darwinulidae has been debated because Smith et al. (2006) found three males amongst thousands of females in the Japanese species V.cornelia. Whether these males are indeed biologically functional and thus could jeopardize the ancient asexual status of V. cornelia, and by extrapolation the genus Vestalenula as a whole, remains to be demonstrated (Martens & Schön, 2008; Schön et al., 2009; Schurko et al., 2009).
Additional research on darwinulid species other than D. stevensoni is urgently required to test whether the observed genetic and phylogeographic patterns are unique for the type species or apply to more darwinulids. Here, we investigate genetic diversity within two darwinulid species from the Pilbara region in Western Australia, namely V. marmonieri and V. matildae (Martens & Rossetti, 2002). We predict that both species of Vestalenula consist of subterranean populations that are genetically isolated from each other according to hydrological basins, as appears to be the case for amphipods and candonid ostracods from the same Western Australian region (Finston et al., 2007; Karanovic, 2007). If UV is an important mutagene for darwinulid ostracods, we would expect that Vestalenula from habitats above ground have acquired more mutations than in subterranean habitats. The higher temperature fluctuations in surface waters could cause additional environmental stress. The possible influence of both UV and temperature fluctuations will become obvious when comparing intraspecific genetic variabilities of ostracods from the two habitat types.
Materials and methods
Study area and model organisms
The Pilbara region experiences high summer temperatures (average January maximum in Marble Bar 41°C) and warm winters during the day, although nights are cooler inland (average July maximum and minimum 27 and 12°C at Marble Bar, 22 and 8°C at Newman, http://www.bom.gov.au/climate/averages/). Annual rainfall is highly variable between years but averages about 350 mm near the coast and 300 mm inland. Rivers flow for a period of a few weeks after heavy rain: this means they flow about once a year in the southern part of the region and closer to once every second year in the north (Anonymous, 1984). Except after rain, surface water is restricted to pools in river courses that are maintained by groundwater inflow and some springs. Many river pools occur in gorges or at the base of small rock walls.
Both V. marmonieri and V. matildae are endemic to Oceania (Martens & Rossetti, 2002) and occur sympatrically in the study area. Whereas V. marmonieri is found in habitats from above and below ground in Australia and New Caledonia, V. matildae had thus far been reported exclusively from subterranean sites (but see below) in Australia although this was based on three specimens only (Martens & Rossetti, 2002). The two species are readily distinguishable: whereas V. marmonieri belongs to the boteai-group of the genus, which is characterized by a short posteroventral keel on the RV, V. matildae belongs to the danielopoli-group, in which this keel is elongated (see illustrations in Martens & Rossetti, 2002).
We use DNA sequence data from the mitochondrial COI and the nuclear ITS region to investigate phylogeographic patterns and compare genetic diversities between the two habitat types.
Sampling, DNA extraction, PCR amplification and sequencing
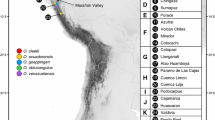
In total, 231 samples from 116 wells and 846 samples from 394 bore holes in Western Australia were collected with modified plankton nets (Eberhard et al., 2007). A further 189 samples from 100 habitats above ground were collected with hand nets. Living samples were sorted and animals stored in 95% EtOH prior to further investigations. DNA was extracted from 23 individual ostracods (see Figure 1 for their geographic distribution and Table 1 for an overview) with the Qiagen Blood and Tissue extraction kit following the manufacture’s protocol.
Distribution of sampled populations in the Pilbara, Western Australia. For population labels, see Table 1
PCR amplification of part of the mitochondrial COI region was conducted with universal primers (Folmer et al., 1994) until the specific primers Vesta COF1 GGAACCTTAATTGGWAATGAY and Vesta COR1 CGGTCTATTGATAKCCKGC could be designed. All PCR amplifications were conducted in a T personal Thermoblock (Biometra) with the following conditions: 25-μl volumes of the Hot Star Master Mix (Qiagen; 1.5 mM MgCl2, 0.1 μM primer, 200 μM dNTP, Tris–Cl, KCl, (NH4)2SO4, 1.25 U Taq) and 15 min at 95°C, 40 cycles with 50 s at 95°C, 50 s at 40°C (Folmer primers) or 44°C (specific primers), 1 min at 72°C and a final extension step for 10 min at 72°C.
ITS1 and ITS2 were first amplified with universal primers ITS1 & ITS4 (White et al., 1990). Whereas PCR products were cloned and sequenced for three individuals as in Schön (2001), the other sequences were obtained with PCR amplification and direct sequencing using the specific primers Vesta ITSF CTTCCCAGGGGAGAAGAGC and Vesta ITSR CGAGGTCCGACAGAAAGAAG. PCR amplifications were conducted as described above for COI, except that annealing temperature varied between 44 and 50°C.
PCR products were checked by agarose gel electrophoresis, stained with Gelred and photographed. PCR products were cleaned [(GFX™ PCR DNA and gel band purification kit (GE Healthcare) according to the manufacture’s protocol)] and sequenced in both directions with the specific PCR primers and the Big Dye kit (ABI) following the manufacture’s protocol on an ABI 3130X.
Analyses of sequence data
Chromatograms were visualized with Chromas Lite 2.01 (http://www.technelysium.com.au/chromas_lite.html), both strands aligned with ClustalX (Larkin et al., 2007) and manually checked for ambiguities. Sequence identity was confirmed by BLAST searches (Altschul et al., 1990) and by translating DNA sequence data into amino acids with MacClade (Maddison & Maddison, 2001). Individual sequences were assembled and used for further analyses. Published sequences of V. molopoensis (Schön et al., 2003; Martens et al., 2005; for Genbank accession numbers, see Table 1) were used as outgroup. All the sequences of V. marmonieri and V. matildae have been submitted to Genbank (see Table 1 for accession numbers).
The optimal models of molecular evolution for the COI and ITS sequences of V. matildae and V. marmonieri were identified with jModeltest (Posada, 2008). The parameters of these models were subsequently applied in PAUP 4.0b.10 (Swofford, 1998) to build neighbour-joining trees and calculate pair-wise distances. For constructing ML trees, PhyML (Guindon & Gascuel, 2003) was run with the parameters identified with jModeltest with bootstraps of 500 replicates for each data set. ML trees were also constructed with TREEPUZZLE (Schmidt et al., 2002) with and without the assumption of a molecular clock to test for equal rates of molecular evolution with the likelihood ratio test. In order to better resolve close phylogenetic relationship and identify haplotypes, parsimonious networks were constructed with tcs 1.21 (Clement et al., 2000) and 95% probability. Absolute rates of molecular evolution for COI and ITS were calculated using the fossil clock of V. molopoensis obtained from phylogenetic reconstructions of the Darwinulidae (Schön et al., 2003).
One-way ANOVA’s were conducted in PAST (Hammer et al., 2001) to check whether differences between categories of pair-wise, genetic distances were statistically significant.
Phylogeographic patterns were investigated with ANeCA (Panchal, 2007) providing an automated version of Nested Clade analyses (Templeton, 1998). Coordinates in degrees, minutes and seconds (see Table 1) were converted to decimal degrees online (http://www.fcc.gov/mb/audio/bickel/DDDMMSS-decimal.html). For each species, the last sampling point of the other Vestalenula species was included as unsampled locality in the datamatrix for the Geodis analyses (Posada et al., 2000) in ANeCA (Panchal, 2007) to allow tests for sampling inadequacies in the inference key.
Results
For COI, jModeltest identified GTR + G with alpha = 0.3750 as the best model. For ITS, the best model was TPM2uf. The ML tree for COI (Fig. 2) shows a clear separation between the two species with a bootstrap support of 100%. Within V. marmonieri, no major groups can be distinguished, and there is no bootstrap support above 60%. Within V. matildae, four clusters can be identified, of which three show bootstrap supports of 78, 82 and 86%, There was no significant difference in the rates of molecular evolution of COI in the three analysed species according to the likelihood ratio test (log L without clock: −909.59, log L with clock: −919.62, degrees of freedom of chi-square distribution = 22, P = 0.58).
The parsimonious networks (Fig. 3a, b) show a finer resolution of the relationships within each species than the ML tree, especially for V. marmonieri. The 15 sequences obtained for this species represent 10 haplotypes (Fig. 3a), which are separated from each other by very few mutational steps, with a maximum of five. There is no clear network structure according to the two habitat types. The most common haplotype of V. marmonieri is shared amongst three sites, two below and one above ground. An additional two haplotypes have been found in both habitat types above and below ground. The network structure for COI sequences from V. matildae (Fig. 3b) is similar. Also, no clear separation between habitat types can be observed. For the latter species, five haplotypes were obtained from the eight sequences, which are again closely connected; the maximal distance between haplotypes is also five mutational steps. Only haplotype 47 from below ground is separated by four mutational steps and thus displays a somewhat larger distance from other specimens. The most common haplotype is shared by three individuals from the same locality below ground. One haplotype is found above as well as below ground.
a Parsimonious network of Western Australian V. marmonieri, based on 394 bp of COI. The line indicates the border between habitats from above and below ground. Haplotypes from above ground are indicated by lighter shades of grey, those from below ground by dark grey. First numbers refer to sampling sites of Table 1 and Fig. 1, while italic numbers are DNA extraction numbers from Table 1. b Parsimonious network of Western Australian V. matildae, based on 385 bp of COI. The line indicates the border between habitats from above and below ground. Haplotypes from above ground are indicated by lighter shades of grey, those from below ground by dark grey. First numbers refer to sampling sites of Table 1 and Fig. 1, while italic numbers are DNA extraction numbers from Table 1
Compared to COI, the ITS region has a much lower variability in both species. Neither V. marmonieri nor V. matildae show any intraspecific differences. Because only one haplotype was identified in V. marmonieri and V. matildae, the parsimonious networks and the ML tree are not shown. Both are available from the first author on request.
When calculating genetic distances of COI within species for different comparisons according to habitat types (Fig. 4), no clear patterns emerge. In V. marmonieri, genetic distances between individuals from below ground are with 0.00544 slightly higher than those from above ground (0.0046), but this difference is statistically not significant. Also for V. matildae, the distance between specimens from above ground is lower (0.0028) than below ground (0.0040) but the first estimate is based on a single comparison. When no habitat type is considered, intraspecific genetic diversities reach an average of 0.0052 and 0.0049, respectively, for V. marmonieri and V. matildae (Fig. 4). This difference is not significant. If the molecular clock of COI from the South African V. molopoensis is applied (Schön et al., 2003), then these distances would equal a separation time of 1.5–2.5 myr between Pilbara populations of the same species. As stated above, no intraspecific diversity could be observed within ITS of V. matildae or V. marmonieri. Average genetic distances between the three investigated Vestalenula species (V. matildae, V. marmonieri and V. molopoensis) are about five times higher for COI than for ITS (Fig. 5).
When comparing genetic distances between Vestalenula species, the highest divergence (0.285) is observed between V. marmonieri and V. matildae, whereas distances between V. molopoensis and the two Australian species are rather similar with means of 0.230 and 0.232 (Fig. 5). The average distance using ITS is also higher between V. marmonieri and V. matildae (0.053) than between other species. Vestalenula matildae and V. molopoensis differ on average by 0.043, while V. marmonieri and V. molopoensis differ by 0.018. If the molecular clock by Schön et al. (2003) is applied, then the distances between the two Australian species equal 88 myr for COI and 75 myr for ITS while the distance between V. molopoensis and the two species from Australia equals 61 myr (ITS) and up to 71 myr (COI), respectively. For the COI data of V. matildae and V. marmonieri, no obvious phylogeographic pattern could be observed when applying ANeCA. For V. matildae, the inference key of AneCA (Panchal, 2007) gave restricted dispersal by distance in non-sexual species as the result for the first clade level of nested clades. For the second level and the whole cladogram, however, no phylogeographic pattern was found. In V. marmonieri, ANeCA (Panchal, 2007) could not detect any phylogeographic pattern at any nested clade level.
Discussion
Variability of molecular markers
We extracted DNA from 23 individuals but could not generate sequences for both genomic regions from all the samples. This is because of three factors: the low density of both species in their habitats, the small size of darwinulid ostracods and, hence, the limited amount of DNA being available for PCR amplification. Also, ostracods in general, and darwinulids in particular, appear to be most reluctant to yield their DNA (Schön, 2001). It was not possible to increase the number of samples available for analysis because densities of both species in the sampled habitats were very low, and usually only one or two specimens were collected from the few sites with darwinulids. Repeated sampling at a site that had previously yielded darwinulids did not reliably yield more specimens.
The nuclear ITS region turned out to be not sufficiently variable to address our research questions. Genetic distances of ITS between the Vestalenula species are on average 2–5% (see Fig. 5) and thus much smaller than COI with an average of 23%. This is similar to the differences between ITS and mitochondrial COI observed in phylogenetic reconstructions of the family Darwinulidae at higher taxonomic levels (Schön et al., 2003; Martens et al., 2005).
In contrast, there was no intraspecific variability of ITS within the analysed V. marmonieri and V. matildae from Western Australia. The apparent discrepancy of molecular evolution between, as compared to within, species has been observed in other darwinulids (Schön et al., 2003). Whereas some variability of ITS could be identified in P. brasiliensis (Schön et al., in preparation), D. stevensoni appears similar to V. marmonieri and V. matildae in that no variability at all in ITS was detected by direct sequencing, not even when comparing individuals from Europe and Africa (Schön et al., 1998). The lack of genetic diversity in ITS has been attributed to highly efficient DNA repair (Schön & Martens, 1998) or homogenizing mechanisms such as gene conversion (Butlin, 2000; Schön & Martens, 2003). Some further statistical evidence for gene conversion in the ITS region of D. stevensoni (Schön & Martens, 2003), P. brasiliensis, P. aotearoa and Microdarwinula inexpectata and Microdarwinula n. sp. (Schön et al., in preparation) has meanwhile been obtained. It thus seems likely that homogenization events have also affected the nuclear genome of Australian Vestalenula although we cannot exclude the possibility that additional samples would show greater genetic differences in ITS.
The mitochondrial COI region, however, showed some variability. The observed intraspecific divergences of both Australian species were of the same order of magnitude as those between European populations of D. stevensoni (Schön et al., 1998; Schön, 2007) but lower than in P. brasiliensis (Schön et al., in preparation).
Genetic diversity above and below ground
In contrast to previous reports (Martens & Rossetti, 2002; S. Halse, pers. commun.), we found two (out of eight) V. matildae above ground. This allowed us to test for both species whether specimens from above ground indeed showed a higher accumulation of mutations, as compared to those from below ground. As with ITS, it is possible that additional samples would increase the estimates of intraspecific genetic variability in COI. For the more variable COI region, no clear separation between individuals from above and below ground could be confirmed in the current data set by phylogenetic trees or parsimonious networks (see Figs. 2, 3a, b). Three haplotypes of V. marmonieri (see Fig. 3a) and one of V. matildae (see Fig. 3b) were even shared between both habitat types, while the other haplotypes were only a few mutational steps apart, regardless of the habitat type from which they originated (Fig. 3a, b). The absence of any structuring is in line with comparisons of genetic distances between individuals from above ground to those from subterranean habitats—no significant differences were observed for any of the two species from the analysed samples. Therefore, our hypothesis that the investigated 13 populations of Western Australian Vestalenula from above ground might be more genetically diverse because UV or greater temperature fluctuations have introduced mutations is not supported with the current data set. Whether this is owing to active repair mechanisms against UV as has been suggested (Schön & Martens, 1998), or other factors, remains to be investigated. If the effects of UV are more moderate, more data might be required to detect any significant differences between animals from subterranean as compared to UV-exposed habitats. The same results would be obtained if regular genetic exchange between populations from below and above ground takes place.
Phylogeography
Analytical results of the phylogeography of both Western Australian species did not reveal any patterns, and genetic distance was not linked to geographic distance. In particular, we could not observe any clustering of haplotypes according to hydrological basins in the current data set. The pattern of Vestalenula ostracods is thus more similar to that of copepods than to that of amphipods and other ostracods from the same geographic region (see Karanovic, 2006, 2007; Finston et al., 2007). There are several possible explanations for the results obtained. Firstly, the data set may be too limited to detect any phylogeographic patterns although ‘insufficient sampling’ was not an outcome of the ANeCA analyses. Secondly, geographically widespread haplotypes have been identified in both species. They could represent superclones with a general purpose genotype as described for D. stevensoni (Van Doninck et al., 2002) and P. brasiliensis, but not for V. molopoensis (Van Doninck et al., 2003). Ecological characterization of the two Australian species would enable verification of this hypothesis, but might be difficult to achieve because of the low densities.
Thirdly, it is possible that the two Vestalenula species are no longer evolving at the molecular level or that evolution is extremely slow. We know from other darwinulids that molecular evolution in the group is indeed at least two orders of magnitude slower than in other invertebrates (Schön et al., 2003), and this slow rate fits with the geological age of the Pilbara region as well as with the estimated age of the two species. The Pilbara region is one of the oldest terrestrial regions in the world, having been above sea level for 3000 myr (Allwood et al., 2007) and having been geologically stable for at least 100 myr (Frakes et al., 1981).
The Pilbara (along with the rest of Australia) began breaking away from Gondwana around 170 myr and its fauna contains Gondwanan elements, although these require better study (Knott & Halse, 1999; Humphreys, 2008). Our absolute dating suggests that V. marmonieri and V. matildae have evolved from each other 75 myr (ITS clock; see above) to 88 myr (COI clock; see above) ago. The earlier populations of the two species may have been more divergent and might then have experienced a recent bottleneck. The intraspecific differences of COI indicate that the analysed haplotypes within each species separated from each other around 1.5 myr ago. This is also the time when some of the V. marmonieri and most of the V. matildae went underground. Whether this time estimate is linked to climatic changes or other obvious factors remains to be investigated but aridity in the Pilbara has been increasing since the mid-Miocene (Martin, 2006) with lakes perhaps becoming episodic, rather than permanent, from about 5–1 myr ago (see Martin, 2006).
A fourth explanation for the pattern of genetic variation is that the two species are widespread and disperse easily with V. marmonieri dispersing more often between above and below ground habitats. However, since darwinulid species cannot produce drought-resistant eggs, as do most other non-marine ostracods, it seems rather unlikely that the observed patterns can be explained by efficient dispersal unless this occurs through groundwater acquifers.
In conclusion, we could not observe any obvious phylogeographic patterns in V. marmonieri, nor in V. matildae from Western Australia. Also, we found no clear indication from our analysed samples that V. marmonieri or V. matildae from above ground may be negatively affected by UV radiation. We calculated that Western Australian V. marmonieri and V. matildae may have split from each other >70 myr ago, but that recent genetic diversities within each species are low. The ecological or historical factors leading to this low genetic diversity remain to be investigated.
References
Allwood, A. C., M. R. Walter, I. W. Burch & B. S. Kamber, 2007. 3.43 billion-year-old stromatolite reef from the Pilbara Craton of Western Australia: ecosystem-scale insights to early life on Earth. Precambrian Research 158: 198–227.
Altschul, S. F., W. Gish, W. Miller, E. W. Myers & D. J. Lipman, 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410.
Anonymous, 1984. Streamflow Records of Western Australia to 1982, 3 Vols. Public Works Department, Perth.
Artheau, M., 2007. Geographical review of the ostracod genus Vestalenula (Darwinulidae) and a new subterranean species from southern France. Invertebrate Systematics 21: 471–486.
Bell, G., 1982. The Masterpiece of Nature. Croom Helm, London.
Birky, C. W. Jr., 1996. Heterozygosity, heteromorphy, and phylogenetic trees in asexual eukaryotes. Genetics 144: 427–437.
Butlin, R. K., 2000. Virgin rotifers. Trends in Ecology & Evolution 15: 389–390.
Clement, M., D. Posada & K. A. Crandall, 2000. TCS: a computer program to estimate gene genealogies. Molecular Ecology 9: 1657–1660.
Eberhard, S. M., S. A. Halse, M. R. Williams, M. D. Scanlon, J. S. Cocking & H. J. Barron, 2007. Exploring the relationship between sampling efficiency and short range endemicity for stygofauna in the Pilbara region, Western Australia. Freshwater Biology 54: 885–901.
Finston, T. L., M. S. Johnson, W. F. Humphreys, S. M. Eberhard & S. A. Halse, 2007. Cryptic speciation in two widespread subterranean amphipod genera reflects historical drainage patterns in an ancient landscape. Molecular Ecology 16: 355–365.
Fisher, R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford.
Folmer, O., M. Black, W. Hoeh, R. Lutz & R. Vrijenhoek, 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3: 294–299.
Frakes, L. A., B. McGowran & J. M. Bowler, 1981. Evolution of Australian environments. In George, A. S. (ed.), Flora of Australia Volume 1 – Introduction, 1st ed. Flora of Australia Series, CSIRO Publishing, Canberra: 1–14.
Guindon, S. & O. Gascuel, 2003. PhyML – a simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52: 696–704.
Hamilton, W. D., 1980. Sex versus non-sex versus parasite. Oikos 35: 282–290.
Hammer, Ø., D. A. T. Harper & P. D. Ryan, 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4: 9 pp.
Humphreys, W. F., 2008. Rising from Down Under: developments in subterranean biodiversity in Australia from a groundwater fauna perspective. Invertebrate Systematics 22: 85–101.
Karanovic, T., 2006. Subterranean copepods (Crustacea, Copepoda) from the Pilbara region in Western Australia. Records of the Western Australian Museum Supplement 70: 1–239.
Karanovic, I., 2007. Candoninae (Ostracoda) from the Pilbara region in Western Australia. Crustaceana Monographs 7: 1–432.
Knott, B. & S. A. Halse, 1999. Pilbarophreatoicus platyarthricus n.gen., n.sp. (Isopoda: Phreatoicidae: Amphisopodidae) from the Pilbara region of Western Australia. Records of the Australian Museum 51: 33–42.
Kondrashov, A. S., 1988. Deleterious mutations and the evolution of sexual reproduction. Nature 336: 435–440.
Kondrashov, A. S., 1993. Classification of hypotheses on the advantage of amphimixis. Journal of Heredity 84: 372–387.
Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson & D. G. Higgins, 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948.
Maddison, D. R. & W. P. Maddison, 2001. MacClade 4: Analysis of Phylogeny and Character Evolution. Version 4.02. Sinauer Associates, Sunderland, MA.
Mark Welch, D. & M. Meselson, 2000. Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science 288: 1211–1214.
Martens, K. (ed.), 1998. Sex and Parthenogenesis: Evolutionary Ecology of Reproductive Modes in Non-Marine Ostracods. Backhuys Publications, Leiden.
Martens, K. & G. Rossetti, 2002. On the Darwinulidae (Crustacea, Ostracoda) from Oceania, with the description of Vestalenula matildae n. sp. Invertebrate Systematics 16: 195–208.
Martens, K. & I. Schön, 2008. Opinion: ancient asexuals: darwinulids not exposed. Nature 453: 587.
Martens, K., G. Rossetti & D. J. Horne, 2003. How ancient are ancient asexuals? Proceedings of the Royal Society of London Series B 270: 723–729.
Martens, K., G. Rossetti, R. K. Butlin & I. Schön, 2005. Molecular and morphological phylogeny of the ancient asexual Darwinulidae (Crustacea, Ostracoda). Hydrobiologia 538: 153–165.
Martens, K., I. Schön, C. Meisch & D. J. Horne, 2008. Global diversity of ostracods (Ostracoda, Crustacea) in freshwater. Hydrobiologia 595: 185–193.
Martin, H. A., 2006. Cenozoic climatic change and the development of the arid vegetation in Australia. Journal of Arid Environments 66: 533–563.
Maynard Smith, J., 1980. Selection for recombination in a polygenic model. Genetic Research 35: 269–277.
Muller, H. J., 1932. Some genetic aspects of sex. American Naturalist 66: 118–138.
Muller, H. J., 1964. The relation of recombination to mutational advance. Mutational Research 1: 2–9.
Panchal, M., 2007. The automation of nested clade phylogeographic analysis. Bioinformatics 23: 509–510.
Pinto, R. L., C. E. F. Rocha & K. Martens, 2003. On two new species of the genus Vestalenula Rossetti & Martens, 1998 (Crustacea, Ostracoda, Darwinulidae) from semi-terrestrial habitats in Sao Paulo State (Brazil). Zoological Journal of the Linnean Society 139: 305–313.
Pinto, R. L., C. E. F. Rocha & K. Martens, 2004. On the genus Penthesilenula Rossetti & Martens, 1998 (Crustacea, Ostracoda, Darwinulidae) from (semi-) terrestrial habitats in São Paulo State (Brazil), with the description of a new species. Journal of Natural History 38: 2567–2589.
Posada, D., 2008. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256.
Posada, D., K. A. Crandall & A. R. Templeton, 2000. GeoDis: a program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Molecular Ecology 9: 487–488.
Rossetti, G. & K. Martens, 1998. Taxonomic revision of the Recent and Holocene representatives of the family Darwinulidae (Crustacea, Ostracoda), with a description of three new genera. Bulletin van het Koninklijk Belgisch Instituut voor Natuurwetenschappen Biologie 68: 55–110.
Roughgarden, J., 1991. The evolution of sex. American Naturalist 138: 934–953.
Schmidt, H. A., K. Strimmer, M. Vingron & A. von Haeseler, 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18: 502–504.
Schön, I., 2001. Primers and PCR conditions for non-marine ostracods. BioTechniques 31: 1012–1016.
Schön, I., 2007. Did Pleistocene glaciations shape genetic patterns of European ostracods? A phylogeographic analysis of two species with asexual reproduction. Hydrobiologia 575: 33–50.
Schön, I. & K. Martens, 1998. Opinion: DNA-repair in an ancient asexual: a new solution to an old problem? Journal of Natural History 32: 943–948.
Schön, I. & K. Martens, 2003. No slave to sex. Proceedings of the Royal Society London Series B 270: 827–833.
Schön, I., R. K. Butlin, H. I. Griffiths & K. Martens, 1998. Slow molecular evolution in an ancient asexual ostracod. Proceedings of the Royal Society London Series B 265: 235–242.
Schön, I., K. Martens, K. Van Doninck & R. K. Butlin, 2003. Evolution in the slow lane: molecular rates of evolution in sexual and asexual ostracods (Crustacea: Ostracoda). Biological Journal of the Linnean Society 79: 93–100.
Schön, I., D. Lamatsch & K. Martens, 2008. Lessons to learn from ancient asexuals. In Egel, R. & D.-H. Lankenau (eds), Genomic Dynamics and Stability, Vol. 2. Meiosis and Recombination. Crossing Over and Disjunction. Springer Publishers, Berlin: 341–376.
Schön, I., G. Rossetti & K. Martens, 2009. Ancient asexual darwinulids: ancient scandals or scandalous gossip? In Schön, I., K. Martens & P. Van Dijk (eds), Lost Sex. The Evolutionary Biology of Parthenogenesis. Springer Academic Publishers, Dordrecht: 217–240.
Schurko, A. M., M. Neiman & J. M. Logsdon, 2009. Signs of sex; what we know and how we know it. Trends in Ecology & Evolution 24: 208–217.
Smith, R. J., T. Kamiya & D. J. Horne, 2006. Living males of the ‘ancient’ asexual Darwinulidae (Ostracoda, Crustacea). Proceedings of the Royal Society London Series B 273: 1569–1578.
Swofford, D. L., 1998. PAUP. Phylogenetic Analysis Using Parsimony (and Other Methods), Version 4.0. Sunderland Associates, Sunderland, MA.
Templeton, A. R., 1998. Nested clade analyses of phylogeographic data: testing hypotheses about gene flow and population history. Molecular Ecology 7: 381–397.
Van Doninck, K., I. Schön, L. De Bruyn & K. Martens, 2002. A general purpose genotype in an ancient asexual. Oecologia 132: 205–212.
Van Doninck, K., I. Schön, F. Maes, L. De Bruyn & K. Martens, 2003. Ecological strategies in the ancient asexual animal group Darwinulidae. Freshwater Biology 48: 1285–1294.
Van Valen, L. M., 1973. A new evolutionary law. Evolutionary Theory 1: 1–30.
White, T. J., T. Bruns, S. Lee & J. Taylor, 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In Innis, M. A., D. G. Gelfand, J. J. Sninsky & T. J. White (eds), PCR Protocols: A Guide to Methods and Applications. Academic Press, London: 315–322.
Acknowledgements
IS thanks the Edith Cowan University, Perth, Western Australia, for two fellowships as visiting fellow in 2006 and 2008/2009. KM and IS are grateful to the Department of Environment and Conservation and Bennelongia for financial support during their two scientific stays in Perth (2006 and 2008/2009). KM and IS also acknowledge the Belgian Belspo (project MO/36/015) for funding and financial contribution of the project 1.5.172.09 (krediet aan navorsers) of the FWO Vlaanderen (Fund for Scientific Research, Flanders).
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Luigi Naselli-Flores
Rights and permissions
About this article
Cite this article
Schön, I., Martens, K. & Halse, S. Genetic diversity in Australian ancient asexual Vestalenula (Ostracoda, Darwinulidae): little variability down under. Hydrobiologia 641, 59–70 (2010). https://doi.org/10.1007/s10750-009-0057-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-009-0057-6