Abstract
Burbot (Lota lota) occur in the Wind River Basin in central Wyoming, USA, at the southwestern extreme of the species’ native range in North America. The most stable and successful of these populations occur in six glacially carved mountain lakes on three different tributary streams and one large main stem impoundment (Boysen Reservoir) downstream from the tributary populations. Burbot are rarely found in connecting streams and rivers, which are relatively small and high gradient, with a variety of potential barriers to upstream movement of fish. We used high-throughput genomic sequence data for 11,197 SNPs to characterize the genetic diversity, population structure, and connectivity among burbot populations on the Wind River system. Fish from Boysen Reservoir and lower basin tributary populations were genetically differentiated from those in the upper basin tributary populations. In addition, fish within the same tributary streams fell within the same genetic clusters, suggesting there is movement of fish between lakes on the same tributaries but that populations within each tributary system are isolated and genetically distinct from other populations. Observed genetic differentiation corresponded to natural and anthropogenic barriers, highlighting the importance of barriers to fish population connectivity and gene flow in human-altered linked lake-stream habitats.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Throughout their Holarctic range, burbot (Lota lota; Linnaeus) are an ecologically important species, sharing the position of apex predator with other large, piscivorous cold-water species. However, many populations are in decline due to anthropogenic disturbances such as overfishing (Paragamian, 2000; Hubert et al., 2008), dams and flow manipulation (Spence & Neufeld, 2002; Paragamian et al., 2005; Hubert et al., 2008), invasive species, food web alteration, decreased water quality, and habitat degradation (Stapanian et al., 2010). Dams and anthropogenic barriers can be especially harmful to burbot populations in river systems where burbot are known to undergo long migrations (Evenson, 1993; Paragamian, 2000; Paragamian & Wakkinen, 2008). Additionally, sampling difficulties, the complexity of burbot life history, their wide distribution, and the fact that they are often considered a “trash fish” has traditionally resulted in a lack of research on burbot biology, ecology, and population monitoring. Fortunately, recent decades have seen an expansion of burbot research, but more is needed to facilitate effective management and conservation throughout their varied habitats (Paragamian & Bennett, 2008; Paragamian & Stapanian, 2011).
The burbot of Wind River/Bighorn River drainage of Wyoming, USA, which is the southwestern-most watershed containing native burbot in North America, have not escaped the issues affecting global populations. Krueger (1996) concluded that without management of harvest and habitat quality, Wind River burbot populations could become threatened, and Hubert et al. (2008) identified overharvest and entrainment of burbot into canals as the main threats to this species. These issues persist within the region despite a relatively long history of population management. In 1949, the Wyoming Game and Fish Department closed all waters to winter burbot fishing, and since then, a variety of regulations have been implemented in an attempt to maintain sustainable fisheries (Hubert et al., 2008). In Wyoming, burbot is a popular sport fish, and is a traditional food source of the Eastern Shoshone and Northern Arapaho American Indian Tribes. As burbot conservation efforts continue within the region, more information on population trends and especially sub-population connectivity and gene flow is needed.
The Wind River Basin is also a unique area for studying burbot population connectivity because individual populations are isolated in lakes on Wind River tributary streams, which are separated by high-gradient stream segments, dams, and irrigation diversions. Burbot are very rarely found in the connecting stream and river segments. Therefore, fisheries managers in the region speculate that burbot are unable to move from lower elevation to higher elevation sites, and that movement between populations occurs solely in a downstream direction. If this is the case, then the entire basin may be acting as a metapopulation, with source populations towards the headwaters, and sink, or partial sink populations, in lower elevation waters. The dynamics of these populations, including dispersal, gene flow, connectivity, and population structure, may be highly influenced by natural and anthropogenic barriers.
The structure and connectivity of fish populations can be identified through a variety of approaches including presence–absence monitoring (Woodford & McIntosh, 2010), tagging and tracking of individuals (Dunnigan & Cameron, 2008), otolith microchemistry analyses (Thorrold et al., 2001), and genetics methods (Fontaine et al., 1997; Dunham & Reiman, 1999). Genetics methods may sometimes be preferred because sample collection is relatively cheap, non-lethal, and requires a relatively low time commitment. Genetic tools can be useful for studying burbot population connectivity as populations separated by natural and artificial barriers have been shown to diverge genetically (Paragamian et al., 1999). While burbot are relatively poor swimmers (Gardunio 2014), they have high fecundity and females can produce several million eggs apiece which result in a large number of larvae that undergo a passive drift phase (Roach & Evenson, 1993). This may result in the passive downstream movement of genetic material from headwater populations to lower elevation populations. Genetic differentiation or population structure can result from a reduction or cessation of homogenizing gene flow (i.e., larval drift and migration) between populations. Understanding the genetic structure of potentially connected fish populations can aid managers in determining the importance of population connectivity and the effects of both natural and artificial barriers.
Hawkins et al. (2011) investigated the genetic structure of Bull Lake, Dinwoody Lakes, and Boysen Reservoir burbot populations using microsatellites and mitochondrial DNA, and found evidence of genetic divergence between the Boysen Reservoir population and the Bull Lake and Dinwoody Lakes populations, suggesting that dispersal between Boysen reservoir population and the other populations may be limited. Additionally, this study found lower levels of genetic diversity in Bull Lake and the Dinwoody lakes than in Boysen Reservoir (Hawkins et al., 2011). To address management concerns about the importance of individual populations and the connections between them in the Wind River Basin, more detailed information is needed on the genetic structure of these populations.
Our research objective was to identify the genetic structure of all known Wind River Basin burbot populations. We used a genotyping-by-sequencing (GBS) method (Parchman et al., 2012) which is a relatively recent development in population genetics (Elshire et al., 2011). It involves using high-throughput DNA sequencing to generate millions of DNA sequences across an organism’s entire genome and sequences thousands of genetic markers quickly and at low cost for organisms with few previous genomic resources (e.g., Parchman et al., 2012; Hohenlohe et al., 2013; Wagner et al., 2013; Gompert et al., 2014). We used these data to identify genetically isolated populations, populations that share genetic material, and infer the movement of fish based on the distribution of specific genetic clusters.
Materials and methods
Study area
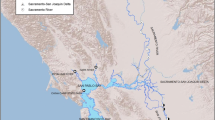
Burbot populations within the Wind River Basin are found primarily in tributaries that descend from the Wind River Mountains to the main stem of the Wind River and in Boysen Reservoir (Fig. 1). Heavy glaciation during the Bull Lake and Pinedale glaciations of the Quaternary ice-age produced lakes of varying size on three main Wind River tributaries (listed in order from upstream to downstream): Torrey Creek, Dinwoody Creek, and Bull Lake Creek. Missourian lineage burbot most likely colonized these lakes sometime after the recession of the Bull Lake and Pinedale glaciations (21,000 years ago), but the timing and mechanism of this colonization is unknown. Stable populations persist within Torrey Ring and Trail Lakes on Torrey Creek, Upper and Lower Dinwoody Lakes on Dinwoody Creek, and in Bull Lake on Bull Lake Creek (Hubert et al., 2008). Each of these tributary lakes or lake chains terminate in a glacial moraine, and the creeks then pass through steep whitewater sections before joining the Wind River. Additionally, burbot are abundant in Boysen Reservoir (Boysen Dam constructed in 1952) and in low numbers within the lower Wind River above Boysen Reservoir, and within lower portions of the Little Wind and Popo Agie Rivers (Bergstedt & Bergersen, 1992).
A variety of natural and anthropogenic barriers exist throughout the system. Burbot are likely to be able to pass upstream and downstream between Torrey, Ring, and Trail lakes, but a step series of cascades below Torrey Lake likely prevents upstream movement from the Wind River (Hubert et al., 2008). Similarly, there are no barriers between Upper and Lower Dinwoody Lakes, but a short steep river section may have historically prevented burbot from moving up into the system while a 2-m-high dam on the outlet of Lower Dinwoody Lake currently prevents upstream movement except during high flows (Hubert et al., 2008). Bull Lake Creek is lower gradient so upstream movement from the Wind River may have occurred historically, but Bull Lake Creek was dammed in 1938 to expand Bull Lake, and the dam now acts as a complete barrier to upstream movement into the lake (Hubert et al., 2008).
The Wind River between the confluences of Torrey Creek and Bull Lake Creek is braided and flows over mostly small to large gravel, and small cobble. The river is highly mobile and transportation rates of sediment and woody debris are high. As a result habitats suited to burbot appear to be rare. The main agricultural diversion on the Wind River is the Wind River Diversion Dam (constructed in 1923) located several miles downstream from the confluence with Bull Lake Creek (Fig. 1). This dam is a complete barrier to upstream fish movement. Several other small irrigation structures exist on the Wind River and its tributaries, but while all are entrainment hazards, none are thought to be barriers to movement.
The Wind River above Boysen Reservoir, the Little Wind River, and the Popo Agie and Little Popo Agie Rivers are meandering, low gradient rivers. Deeper water, finer sediments, low velocity, and more complex habitat provide refugia and foraging areas for burbot. These river sections have been relatively unaltered physically except for some channelization through the town of Riverton, WY. Agricultural practices contribute to an altered flow regime and elevated nutrient inputs, though agricultural pollution is not considered significant.
Burbot can spawn in both lakes and streams with some populations being adfluvial, migrating between lakes and streams to spawn. Bjorn (1940) and Williams (1959) hypothesized that spawning burbot in the Torrey Creek watershed moved between lentic and lotic environments, though this was never verified. There is no evidence of spawning migrations between the Torrey Creek, Dinwoody Creek, and Bull Lake Creek tributaries and the presence of a substantial number of natural and anthropogenic barriers makes this unlikely. However, spawning migrations between the Little Wind, Popo Agie, and Little Popo Agie rivers and Boysen Reservior may be possible, but are unverified.
Burbot collections
Our goal was to collect 30 individual burbot from each known population within the Wind River Drainage: Torrey, Ring, Trail, Upper and Lower Dinwoody, and Bull lakes, Boysen Reservoir, and the Little Wind, Popo Agie, and Little Popo Agie rivers. Scarcity of fish in several populations prevented us from sequencing a full set of 30 individuals. We also attempted to collect burbot from a suspected but unconfirmed population within the Wind River, but were unable to find any individuals despite multiple sampling events with techniques proven effective at other sites we sampled. Most genetic samples were collected by the Wyoming Game and Fish Department or the US Fish and Wildlife Service between 2011 and 2013 during routine fisheries management operations. We collected the remaining genetic samples from fish captured on our behalf by Wyoming Game and Fish Department during the fall 2013. No fish were sampled during the spawn, so captured fish were likely residents of the locations they were collected from, though some spawning migrants may have been captured in spring or fall.
All burbot were captured by either trammel net or boat electrofishing. Fish were collected with trammel nets from Torrey (n = 34), Ring (n = 13), Trail (n = 25), Upper Dinwoody (n = 36), Lower Dinwoody (n = 13), and Bull (n = 30) lakes, as well as Boysen Reservoir (n = 16) during September, October, and November of 2011, 2012, and 2013. Boat electrofishing was used to collect fish in the Wind (no fish found), Little Wind (n = 4), Popo Agie (n = 1), and Little Popo Agie (n = 19) rivers during spring and early summer in 2011, 2012, and 2013 (Fig. 1).
Genetic sample collection and DNA extraction
We collected genetic samples following the protocol suggested by Rogers (2007). We clipped greater than one cm2 of right pectoral fin from each individual using sterilized scissors and stored the samples in 90% ethanol at room temperature for up to 3 years. We extracted DNA from the preserved fin tissue using Qiagen DNeasy Blood and Tissue mini spin column kits (Qiagen Inc., Hilden, Germany, www.qiagen.com), according to the manufacturer’s instructions. We checked concentrations of extracted DNA with a NanoDrop ND-1000 spectrophotometer to ensure that DNA concentrations were appropriate for the preparation of reduced-complexity genomic libraries.
Reduced-complexity genomic library preparation and DNA sequencing
We prepared our extracted DNA for sequencing following the methodology described by Parchman et al. (2012). In summary, two restriction enzymes (EcoRI and MseI) were used to cut DNA at specific sites across the genomes of all individuals, resulting in millions of variable-length sequences from the same genomic regions among all fish. Individual 8–10 base-pair barcodes were then ligated to each fragment of each individual fish’s genome. This allowed us to later assign sequence reads back to individual fish. We then combined barcoded DNA fragments from all individuals, and used PCR to amplify barcoded DNA fragments. Barcoded DNA fragments from all samples were sent to the University of Texas Genomic Sequencing and Analysis Facility (GSAF; Austin, Texas, USA). The sequencing facility checked library quality using an Agilent Bioanalyzer, and then used a BluePippin system to select DNA fragments 250–300 base pairs in length, to reduce the number of unique fragments represented in the library. The completed reduced-complexity genomic library was then sequenced on the Illumina Hiseq 2500 platform using single-end reads, 100 basepairs in length.
Assembly and variant calling
DNA sequencing produced 113,727,470 short DNA sequences that passed contaminant filtering and matched to barcoded individuals. As a reference genome is not available for burbot, we created an artificial reference genome by doing a de novo assembly of 25 million sequences using smng (SeqMan NGen, DNAstar). This assembly resulted in an artificial reference genome composed of 53,789 contigs. We then attempted to assemble all 113,727,470 short sequence reads to this artificial reference genome using bwa (Li & Durbin, 2009). We excluded four individuals with fewer than 2,000 assembled reads from downstream analysis due to low sequence coverage (two individuals from Boysen Reservoir, one each from Torrey Lake and Trail Lake). For retained individuals, 88.67% of all DNA sequence data produced (100,836,745 sequenced DNA fragments) successfully assembled to the artificial reference genome.
From the assembled data, we identified 22,272 single nucleotide variants using samtools and bcftools (Li et al., 2009). For a variant site to be identified, we required that at least 60% of all individuals had at least one sequence read at that genetic site. After removing sites with a minor allele frequency of <1% and randomly selecting one site per contig to ensure independence of loci, we retained 11,197 SNPs for downstream analyses of population genetic structure. Mean sequence coverage at retained SNPs was approximately 13 reads per locus per individual.
Population genetic analyses
We used several complementary analyses to reveal genetic structure among the burbot populations in this study. Analyses were conducted on genetic data first for individual fish, and then also at the population level after we confirmed that geographic populations are composed of genetically similar individuals that differ from other populations. Our analyses also included both model-based and model-free analyses.
Genetic analyses of individuals
We used a hierarchical Bayesian model, entropy (Gompert et al., 2014), to estimate proportional ancestry of each individual fish in each of k genetic clusters. This model estimates ancestry independently for each individual, and incorporates no a priori information about membership of individuals in populations (much like the popular program STRUCTURE; Pritchard et al, 2000; Falush et al., 2003). Details of the entropy model are described in Gompert et al. (2014). For k = 1 to k = 12 genetic clusters, we ran three replicate chains for each model for 150,000 MCMC steps, and discarded the first 100,000 steps as burn-in. We accepted every 25th step, resulting in 2,000 samples from the posterior distribution for each of the three replicate chains (6,000 samples total) for each model for k genetic clusters. We assessed mixing and convergence of parameter estimates by plotting traces of parameter estimates across multiple chains. We used the posterior probability distributions generated by entropy to estimate mean, median, and 95% credible intervals for parameters of interest, including genotype at each locus for each individual, and the proportion of an individual’s ancestry that came from a genetic cluster (q). To determine what number of genetic clusters (k) is best statistically supported, we calculated DIC (deviance information criterion), a model choice criterion analogous to AIC. DIC is the model deviance penalized by the number of effective parameters, and identifies the model that best fits the data but is not overly complex. Both the deviance and effective number of parameters are estimated by entropy.
In addition to analyzing genetic data with entropy, we also conducted a model-free analysis to understand genetic variation in the individuals sampled without assuming any specific genetic structure. We used genotype probabilities for each individual at each genetic locus (11,197 SNPs, 187 individuals) to build a covariance matrix of genetic similarity among individuals, and then used this matrix in a principal component analysis (prcomp in R).
Population-level analyses
Principal component analysis and entropy show population genetic structure, but do not quantify genetic distance between genetically distinct clusters. Since entropy and PCA results confirm that individuals within a population are genetically more similar to one another than to individuals in other populations, we used population-level analyses to quantify genetic differentiation. We used genotype probabilities to calculate allele frequency at each locus in each population. From allele frequencies, we estimated genetic differentiation among populations by calculating Hudson’s F ST (Hudson et al., 1992) pairwise between all populations. We also used TreeMix software to infer relationships and possible gene flow (“migration events”) between populations (Pickrell & Pritchard, 2012). We ran TreeMix to build a maximum likelihood tree for the relationships among populations. TreeMix allows the addition of migration events to improve model fit. This is useful because it allows inference of contemporary and historical patterns of gene flow among populations. We explored trees with 1–5 migration events.
For each population, we also estimated two genetic diversity statistics using samtools and bcftools (Li et al., 2009). We estimated π, the expected heterozygosity, and Watterson’s theta (θ W). To obtain these estimates, we used 20 iterations of the expectation–maximization (EM) algorithm (described in Li, 2011) to achieve convergence of both statistics. This method includes invariant loci in addition to loci containing polymorphisms.
Results
After filtering sequence data and removing four individuals with low coverage, we were able to identify 11,197 SNPs from 187 individuals. Mean sequence coverage for retained SNPs was approximately 13 reads per locus per individual, and this set of SNPs was used in all downstream analyses.
The entropy analysis indicated that k = 2 is the best supported number of genetic clusters (DIC = 5231065.01) in the Wind River system. The primary genetic structure was a divergence between lower basin river and reservoir populations, and upper basin tributary populations. Boysen Reservoir, the Little Wind River, and the Little Popo Agie River populations exhibited distinct ancestry from all other populations, except for several Little Popo Agie River fish that displayed ancestry similar to the other populations (Fig. 2; Table 1). Although the k = 2 model was best supported by DIC, higher values of k also provide useful information about population genetic structure. At k = 3, an additional ancestral cluster emerged in Torrey, Ring, and Trail lakes, while the Dinwoody drainage and Bull Lake share ancestry under this model. At k = 4, no new population-specific ancestry clusters appeared, but the fourth cluster made up a greater proportion of ancestry in the Bull Lake population than the Upper and Lower Dinwoody populations, suggesting weak divergence between Bull Lake and the Dinwoody Lakes. Estimates of an individual fish’s genetic ancestry (q) had very little associated uncertainty (mean 95% credible intervals were <0.05 for k = 2 to k = 4), confirming that the model precisely estimates ancestry.
Proportion of ancestry (y axis) of individual fish (x axis) estimated by a hierarchical Bayesian model, entropy, from all sampled populations within the Wind River drainage. The model was run with assigned numbers of genetic clusters (k) 1–12. Shown are k = 2–4 due to decreased relevance at k > 4. The Bull Lake grouping includes fish from Bull Lake only. The Torrey Creek grouping includes fish from Torrey, Ring, and Trail lakes. The Dinwoody grouping includes fish from Upper and Lower Dinwoody lakes. The Boysen/rivers grouping includes fish from Boysen Reservoir, the Little Wind River, the Popo Agie River, and the Little Popo Agie River
Principal component analysis of the genotype probabilities for each individual at each genetic locus also revealed major axes of genetic variation (Fig. 3). PC1 explains most genetic variation (78.82%), and separates Boysen Reservoir and the river populations (Little Wind, Popo Agie, and Little Popo Agie rivers) from all tributary populations (Bull Lake, Dinwoody lakes, and Torrey Creek lakes) (Fig. 3a). PC2 (15.50% of genetic variation) shows differences among tributary populations, and separates Torrey Creek populations (Torrey, Ring, and Trail lakes) from the Bull Lake and Dinwoody populations. Finally, PC3 (0.84% of variation) shows weak genetic differentiation between Bull Lake and Dinwoody burbot (Fig. 3b).
Principal component analysis plots of a principal components one and two, and b principal components two and three for the genetic covariance of individual burbot from Upper Dinwoody Lake (ud), Lower Dinwoody Lake (ld), Torrey Lake (tol), Ring Lake (rl), Trail Lake (trl), Bull Lake (bl), Popo Agie River (par), Little Popo Agie River (lpa), Little Wind River (lwr), and Boysen Reservoir (br)
To quantify genetic differentiation among populations, we first calculated FST pairwise between all populations (Table 2; Hudson et al., 1992). Populations in the Upper Wind River were strongly differentiated from populations in the Lower Wind River populations, as previously indicated by PCA and entropy analysis. To augment our understanding of genetic differentiation among populations, we also used TreeMix to infer historical relationships and ongoing migration (i.e., gene flow) among populations (Pickrell & Pritchard, 2012). TreeMix results concur with previous analyses that indicate that Boysen Reservoir and the river populations were strongly differentiated from other populations (Fig. 4). Torrey, Ring and Trail lakes, which are located along a single tributary, were genetically distinct from the other populations, but had greater similarity to Bull Lake and the Dinwoody lakes than to the Boysen/river populations. Bull Lake burbot are weakly differentiated from Dinwoody burbot. The addition of migration events to our maximum likelihood trees improved model fit to the data (increased the model log-likelihood and decreased model residuals), but model log-likelihood increases were minimal after three migration events (Table 3). The first migration event (Fig. 4) indicates migration from Torrey, Dinwoody, and Bull Lakes to the Little Popo Agie River, indicating occasional contributions from tributary populations to downstream riverine populations. The second migration event suggests historical gene flow between Bull Lake and the Dinwoody lakes and the Torrey Creek lakes. The third migration event suggests occasional, infrequent gene flow from Bull Lake downstream to Boysen reservoir (Fig. 4).
TreeMix graph of relationships among sampled burbot populations. This is a maximum likelihood tree constructed from the data, with three migration events (instances of gene flow) added to improve model fit (increase model log-likelihood and decrease model residuals). Burbot were sampled from Upper Dinwoody Lake (ud), Lower Dinwoody Lake (ld), Torrey Lake (tol), Ring Lake (rl), Trail Lake (trl), Bull Lake (bl), Little Popo Agie River (lpa), Little Wind River (lwr), and Boysen Reservoir (br) in the Wind River Basin. The drift parameter is an estimate of genetic differentiation between the sampled populations, and is analogous to F ST. Migration events corresponding to directional gene flow are indicated by arrows. Arrow color indicates migration weight, with darker orange indicating a stronger genetic effect on the destination population
Genetic diversity was similar across populations (Fig. 5), and was uniformly low. The highest diversity was found in the Little Popo Agie River (heterozygosity = 0.003254), consistent with occasional gene flow (identified by TreeMix) from upstream populations. The lowest genetic diversity was in Lower Dinwoody lake (heterozygosity = 0.000979) and Bull Lake (heterozygosity = 0.001231). Overall, differences in genetic diversity among populations were quite small.
Genetic diversity estimates for burbot populations in Upper Dinwoody Lake (ud), Lower Dinwoody Lake (ld), Torrey Lake (tol), Ring Lake (rl), Trail Lake (trl), Bull Lake (bl), Little Popo Agie River (lpa), Little Wind River (lwr), and Boysen Reservoir (br) in the Wind River Basin. Bars represent expected heterozygosity (π), while points represent Watterson’s θ w. Values of these two genetic diversity statistics are similar, and genetic diversity is generally low in all sampled populations
Discussion
We found evidence of strong genetic divergence among Wind River Basin burbot populations, with both natural and anthropogenic barriers likely contributing to limited gene flow between genetic clusters. There was a clear genetic divergence between the upper and lower Wind River populations, which are separated by a large diversion dam. Each tributary lake population in the Upper Wind River also appeared to be genetically distinct. Populations in the Torrey cluster (Torrey, Ring, and Trail lakes) were relatively strongly differentiated from populations in the Dinwoody Lakes and Bull Lake. Bull Lake burbot were also weakly genetically differentiated from Dinwoody fish. Each of these patterns was confirmed by multiple analyses, including clustering analysis with entropy, principal components analysis, estimates of genetic differentiation (F ST), and analysis of population relationships with TreeMix. The results suggest that efforts to restore and maintain connectivity should focus on the lower Wind River populations that appear to be less impacted by barriers, while upper Wind River tributary populations should be considered independently of one another.
We found the strongest statistical support for two primary genetic groups with a division between the lower Wind River cluster found in Boysen Reservoir and the Little Wind, Popo Agie, and Little Popo Agie rivers, and all of the upper Wind River tributary populations. This split between upper and lower Wind River populations corresponds geographically to a large irrigation diversion, the Wind River Diversion Dam, which is likely restricting movement of individuals and inhibiting homogenizing gene flow. The dam was built in 1923 and has a fish ladder, but burbot are unlikely to have ever used it due to its steepness. More recently, shifting sediments have made the fish ladder inaccessible to all fish. Bergstedt and Bergersen (1992) conducted an extensive survey of the Wind River, and of the 17,805 fish they collected only 60 were burbot, and only one of those was captured above the Wind River Diversion dam, which suggests that the diversion is limiting the movement of fish into the upper Wind River. While the dam is probably maintaining genetic differentiation, it remains unclear whether the dam caused genetic differentiation between Upper and Lower Wind River populations, or whether that divergence predated the construction of the dam.
The genetic similarity among the Little Wind, Popo Agie, Little Popo Agie, and Boysen Reservoir fish was expected. These rivers are deep, low gradient, meandering, silt- and sand-bottomed streams without any presumed barriers to dispersal. In other similarly structured systems, such as the Kootenai River system in Western North America, or in rivers of the Meuse Basin in France, it has been shown that burbot move freely within these systems, often motivated by spawning (Paragamian, 2000; Dillen et al., 2008; Paragamian & Wakkinen, 2008). Therefore, it is likely that migratory fish, especially spawning migrants, are maintaining gene flow between the Little Wind, Popo Agie, and Little Popo Agie Rivers and Boysen Reservoir.
Genetic differentiation between tributary populations within the Upper Wind River genetic cluster (e.g., the clear genetic division resolved between the Torrey Creek lakes, the Dinwoody lakes, and Bull Lake) is probably best explained by natural and anthropogenic migration barriers present in the Wind River and the tributaries themselves. Natural and anthropogenic migration barriers are well-documented drivers of genetic differentiation between populations of fish. For example, brown trout (Salmo trutta) populations within 3 km of each other in Sweden were found to have significantly different microsatellite allele frequencies, and these differences were linked to the presence of various natural barriers (Carlsson et al., 1999). In addition, habitat fragmentation due to anthropogenic barriers affected lake sturgeon population genetics (McDermid et al., 2014). Similar barriers are abundant in the Wind River Basin. The upper Wind River is steeper in gradient, shallower, and has more of a riffle run, small cobble-bottomed structure than the lower Wind River. The tributaries between the lakes and the Wind River descend glacial moraines with higher gradients still. These rivers and streams flow through large boulders and a plunge-pool structure with very high velocity segments. Gardunio (2014) found that burbot are poor jumpers and swimmers so these habitat conditions may pose as natural migratory barrier to fish that are attempting to move upstream. In addition, dams built to expand Bull Lake and Lower Dinwoody Lake during the 1930’s and 1940’s prevent all movement of fish into the lakes from the outlet rivers.
Among the tributary lakes (Bull, Dinwoody, Torrey), the lakes in the Torrey tributary are most strongly differentiated genetically, with weaker genetic divergence between the Bull Lake and the Dinwoody lakes populations. This could suggest that the Torrey Creek populations were colonized earlier and have thus had more time to diverge from the Bull and Dinwoody lakes populations, or alternatively that gene flow between Torrey Creek and the other lakes is more strongly limited by distance and barriers. Distance and habitat factors could also explain the limited movement of burbot between the Torrey Creek populations and the lower tributaries compared between Bull Lake and the Dinwoody Lakes. Torrey Lake is further from Bull Lake (over 65 km) than Lower Dinwoody is from Bull Lake (over 40 km), the Wind River gradient increases noticeably between the Dinwoody Creek and Torrey Creek confluences, and Torrey Creek between Torrey Lake and the Wind River is longer and much steeper than Dinwoody or Bull Lake creeks. It is unknown how burbot were able to first colonize the upper tributary lakes, so it is difficult to speculate on the probability of these potential scenarios.
While fish movement is likely limited between the tributary clusters, there are fewer factors impeding the movement between lakes on the same tributary. The greatest distance between any two adjacent lakes on the tributaries is under 1 km and gradients are less steep, shorter, and fewer in number, especially in low winter flows when burbot are most likely to attempt spawning migrations (Paragamian & Wakkinen, 2008; McPhail & Paragamian, 2000). In fact, Bjorn (1940) noted spawning aggregations of burbot in Torrey Creek and Hagen (1952) captured burbot staging to spawn at the inlet to Ring Lake. Therefore, fish movement, and as a result gene flow, has maintained homogenous genetic structure between shared tributary populations.
While upstream movement into the tributaries is impaired by physical conditions and dams, these conditions do not explain why we did not identify extensive downstream gene flow that would result in a lack of differentiation between Boysen Reservoir and the upstream tributary populations. In other fish systems with large barriers to dispersal, such as waterfalls separating populations of Trinidad guppies, extensive downstream gene flow has still been documented (e.g., Fitzpatrick et al., 2015). The migration events identified by TreeMix suggest that in our study system there was historically gene flow both down river and upriver, but that migration is not continuing at a rate sufficient to homogenize genetic structure. This is especially surprising given that burbot have high fecundity and passive larval drift. Given these characteristics, it seems that larvae should drift from upstream populations and colonize lower populations with greater frequency than our data support. One potential explanation is that conditions are too adverse between higher and lower populations for delicate larval burbot to survive the journey. The high-gradient stream segments, and dam outlets and spillways between the tributary lakes and the Wind River may be turbulent enough to kill many larvae. Also, all of the streams and the Wind River have very high densities of drift feeding salmonids and cyprinids, which may result in high predation on drifting larvae. However, this would not explain the lack of downstream movement by larger fish. It may be because they do not emigrate from tributaries in large numbers, do not survive downstream passage, or they do not breed once reaching the suitable habitat in Boysen Reservoir and the lower rivers. Even if a small number of individuals do successfully move downstream, our limited sample size in Boysen Reservoir and the lower rivers might have failed to capture individuals with immigrant ancestry. A movement study found limited to no downstream outmigration of any size class of burbot from Bull Lake or Torrey Lake (personal observation, Z. Underwood).
Expanding the sample sizes from some populations in the lower Wind River basin may provide additional information on the amount and direction of gene flow. Larger sample sizes might have allowed us to identify individual migrants. Additionally, there are population gaps that we were unable to sample. The Wind River above and below the Wind River Diversion dam may be an important conduit for fish moving from the tributary populations to the lower basin populations. These reaches of the Wind River may also hold small, resident burbot populations with potentially intermediate genetic structure. We also considered the need for greater spatial coverage of samples within each of the individual populations as burbot and other fishes sometimes have population boundaries not delineated by simple stream or lake boundaries (Burger et al., 1997). For example, in Koocanusa Reservoir, Montana, Dunnigan and Cameron (2008) found that burbot rarely or never crossed the historic river thalweg, suggesting that spatially distinct subpopulations existed within one reservoir.
Interestingly, genetic diversity was similarly low across most populations, regardless of connectivity or location. McDermid et al. (2014) found similar results wherein genetic diversity of lake sturgeon (Acipenser fulvescens) populations did not differ greatly despite altered population genetic structure, reduced gene flow, and reduced population size due to anthropogenic barriers. If gene flow or extremely large population size was needed to maintain genetic diversity in tributary populations in the upper Wind River, we might have expected diversity to consistently be markedly reduced in Bull Lake, the Torrey Creek lakes, and the Dinwoody lakes relative to the highly connected lower Wind River populations. Genetic diversity is indeed slightly lower in Lower Dinwoody and Bull Lake, possibly because these are smaller, more isolated populations. Genetic diversity was elevated in one lower basin tributary, the Little Popo Agie River, likely due to the occasional gene flow from upper basin tributaries previously identified with TreeMix. In comparison with other river fish (i.e., Catostomus species; Mandeville et al., 2015), diversity in all populations is quite low. In general, gene flow between burbot populations appears to be highly sensitive to migration barriers.
We identified substantial population structure within the Wind River region, but did not identify any obvious source–sink interactions occurring in the region. There appears to be very little movement of fish out of the upper basin tributaries that influences the genetic structure of the lower basin populations. Movement of burbot between Boysen Reservoir, the Little Wind River, the Popo Agie, and the Little Popo Agie River also does not appear to be directional, with any one population acting as a source or sink. Given our findings, fisheries managers and conservationists in the region may want to focus attention on conserving individual populations in the upper basin, and maintaining connectivity between populations in the lower basin.
Our results suggest that natural barriers have shaped gene flow in the upper portion of the basin, while anthropogenic barriers could affect population dynamics in the lower portion of the basin. In general, the presence and scale of natural and anthropogenic barriers is an important factor to consider in burbot conservation efforts. Maintaining population connectivity is especially important for adfluvial burbot populations in areas with low geographic relief, but may be a less important goal when working with burbot and other weak swimming fish species in landscapes with high geographic relief that historically has naturally limited gene flow.
References
Bergstedt, L. & E. P. Bergersen, 1992. Impacts of Water Management on the Fishery Resources of the Wind River on the Wind River Indian Reservation. Annual Report by CSU Cooperative Fish & Wildlife Research Unit, Fort Collins, CO.
Burger, C. V., W. J. Spearman & M. A. Cronin, 1997. Genetic differentiation of sockeye salmon subpopulations from a geologically young Alaskan lake system. Transactions of the American Fisheries Society 126: 926–938.
Bjorn, E. E., 1940. Preliminary observations and experimental study of the ling, Lota maculosa (LeSueur), in Wyoming. Transactions of the American Fisheries Society 69: 192–196.
Carlsson, J., K. H. Olsen, J. Nilsson, O. Overli & O. B. Stabell, 1999. Microsatellites reveal fine-scale genetic structure in stream-living brown trout. Journal of Fish Biology 55: 1290–1303.
Dillen, A., J. Coeck, & D. Monnier, 2008. Habitat Use and Seasonal Migrations of Burbot in Lowland Rivers in North France. In Paragamian, V. L. & D. H. Bennett (eds), Burbot: Biology, Management, and Culture. American Fisheries Society, Symposium 59, Bethesda, MD: 29–42.
Dunham, J. B. & B. E. Rieman, 1999. Metapopulation structure of bull trout: influences of physical, biotic, and geometrical landscape characteristics. Ecological Applications 92: 642–655.
Dunnigan, J. L., & L. S. Cameron, 2008. Home Range and Movement Patterns of Burbot in Koocanusa Reservoir, Montana. In Paragamian, V. L. & D. H. Bennett (eds), Burbot: Biology, Management, and Culture. American Fisheries Society, Symposium 59, Bethesda, MD: 43–54.
Elshire, R. J., J. C. Glaubitz, Q. Sun, J. A. Poland, K. Kawamoto, E. S. Buckler & S. E. Mitchell, 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: e19379.
Evenson, M. J., 1993. Seasonal movements of radio-implanted burbot in the Tanana River drainage. Alaska Department of Fish and Game, Division of Sport Fish 93–97: 1–25.
Falush, D., M. Stephens & J. K. Pritchard, 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Fitzpatrick, S. W., J. C. Gerberich, J. A. Kronenberger, L. M. Angeloni & W. C. Funk, 2015. Locally adapted traits maintained in the face of high gene flow. Ecology Letters 18: 37–47.
Fontaine, P. M., J. J. Dodson, L. Bernatchez & A. Slettan, 1997. A genetic test of metapopulation structure in Atlantic salmon (Salmo salar) using microsatellites. Canadian Journal of Fisheries and Aquatic Sciences 54: 2434–2442.
Gardunio, E. I., 2014. Jumping and Swimming Performance of Burbot and White Suckers: Implications for Barrier Design. Master’s thesis, Colorado State University, Fort Collins, CO.
Gompert, Z., L. K. Lucas, C. A. Buerkle, M. L. Forister, J. A. Fordyce & C. C. Nice, 2014. Admixture and the organization of genetic diversity in a butterfly species complex revealed through common and rare genetic variants. Molecular Ecology 23: 455–4573.
Hagen, G. O., 1952. Ling Hatching Experiment, Cokeville, Wyoming. Cheyenne, WY: Report to the Wyoming Game and Fish Commission
Hawkins, D., B. Adams, S. Roth & D. Skates, 2011. Genetic characterization of Burbot (Lota lota maculosa) from three lakes in the Wind River watershed, WY. Project Report, U.S. Fish and Wildlife Service, Lander, WY.
Hohenlohe, P. A., M. D. Day, S. J. Amish, M. R. Miller, N. Kamps-Hughes, M. C. Boyer, C. C. Muhlfeld, F. W. Allendorf, E. A. Johnson & G. Luikart, 2013. Genomic patterns of introgression in rainbow and westslope cutthroat trout illuminated by overlapping paired-end rad sequencing. Molecular Ecology 22: 3002–3013.
Hubert, W., D. Dufek, J. Deromedi, K. Johnson, S. Roth, & D. Skates, 2008. Burbot in the Wind River Drainage of Wyoming: Knowledge of Stocks and Management Issues. In Paragamian V. L. & D. H. Bennett (eds), Burbot: Biology, Management, and Culture. American Fisheries Society, Symposium 59, Bethesda, MD: 187–200.
Hudson, R. R., M. Slatkin & W. P. Maddison, 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132: 583–589.
Krueger, K. L., 1996. Assessment of Lentic Sport Fisheries for Burbot and Sauger in the Bighorn/Wind River Drainage, Wyoming. Master’s thesis, Universtiy of Wyoming, Laramie, WY.
Li, H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993.
Li, H. & R. Durbin, 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760.
Li, H., B. Handsaker, A. Wysoker, T. Fennell, J. Ruan, N. Homer, G. Marth, G. Abecasis & R. Durbin, 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079.
Mandeville, E. G., T. L. Parchman, D. B. McDonald & C. A. Buerkle, 2015. Highly variable reproductive isolation among pairs of Catostomus species. Molecular Ecology 24: 1856–1872.
McDermid, J. L., S. Nienhuis, M. Al-Shamlih, T. J. Haxton & C. C. Wilson, 2014. Evaluating the genetic consequences of river fragmentation in lake sturgeon (Acipenser fulvescens Rafinesque, 1817) populations. Applied Icthyology 30: 1514–1523.
McPhail, J. D., & V. L. Paragamian, 2000. Burbot Biology and Life History. In Paragamian, V. L. & D. W. Willis (eds), Burbot: Biology, Ecology, and Management American Fisheries Society, Fisheries Management Section, Publication Number 1, Bethesda, MD: 11–23
Paragamian, V. L., 2000. The Effects of Variable Flows on Burbot Spawning Migrations in the Kootenai River, Idaho, USA, and British Columbia, Canada. In Paragamian, V. L. & D. W. Willis (eds), Burbot: Biology, Ecology, and Management American Fisheries Society, Fisheries Management Section, Publication Number 1, Bethesda, MD: 111–123
Paragamian V. L. & D. H. Bennett (eds), 2008. Burbot: Biology, Management, and Culture. American Fisheries Society, Symposium 59, Bethesda, MD.
Paragamian, V. L. & M. A. Stapanian, 2011. Preface. Journal of Applied Ichthyology 27: 3.
Paragamian, V. L., & V. D. Wakkinen, 2008. Seasonal Movement of Burbot in Relation to Temperature and Discharge in the Kootenai River, Idaho, USA and British Columbia, Canada. In Paragamian V. L. & D. H. Bennett (eds), Burbot: Biology, Management, and Culture. American Fisheries Society, Symposium 59, Bethesda, MD: 55–77.
Paragamian, V. L., M. Powell & J. Foler, 1999. Mitochondrial DNA analysis of burbot in the Kootenai River basin of British Columbia, Montana, and Idaho. Transactions of the American Fisheries Society 128: 854–886.
Paragamian, V. L., R. Hardy & B. Gunderman, 2005. Effects of regulated discharge on burbot migration. Journal of Fish Biology 66: 1199–1213.
Parchman, T. L., Z. Gompert, J. Mudge, F. Schilkey, C. W. Benkman & C. A. Buerkle, 2012. Genome-wide association genetics of an adaptive trait in lodgepole pine. Molecular ecology 21: 2991–3005.
Pickrell, J. K. & J. K. Pritchard, 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS genetics 8: e1002967.
Pritchard, J. K., M. Stephens & P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Roach, S. M., & M. J. Evenson, 1993. A Geometric Approach to Estimating and Predicting the Fecundity of Tanana River Burbot. Alaska Department of Fish and Game. Juneau, AK. Fisheries Data Series 93–98.
Rogers, K. B., 2007. A Suggested Protocol for Collecting Cutthroat Trout Tissues for Subsequent Genetic Analysis. Colorado Divison of Wildlife, Fort Collins, CO.
Spence, C., & M. Neufeld, 2002. Radio Telemetry Studies of Duncan Reservoir Burbot. Report prepared by the Ministry of Water, Land and Air Protection, for the BC Habitat Conservation Trust Fund and the Bonneville Power Administration, Nelson BC.
Stapanian, M. A., V. L. Paragamian, C. P. Madenjian, J. R. Jackson, J. Lappalainen, M. J. Evenson & M. D. Neufeld, 2010. Worldwide status of burbot and conservation measures. Fish and Fisheries 11: 34–56.
Thorrold, S. R., C. Latkoczy, P. K. Swart & C. M. Jones, 2001. Natal homing in a marine fish metapopulation. Science 291: 297–299.
Wagner, C. E., I. Keller, S. Wittwer, O. M. Selz, S. Mwaiko, L. Greuter, A. Sivasundar & O. Seehausen, 2013. Genome-wide rad sequence data provide unprecedented resolution of species boundaries and relationships in the Lake Victoria cichlid adaptive radiation. Molecular Ecology 22: 787–798.
Williams, F., 1959. Progress Report on Life History Investigations of the Burbot. Wyoming Game and Fish Commission, Daniel.
Woodford, D. J. & A. R. McIntosh, 2010. Evidence of source-sink metapopulations in a vulnerable native galaxid fish driven by introduced trout. Ecological Applications 20: 967–977.
Acknowledgements
We could not have accomplished this research without extensive help in the field and lab. Joe Deromedi, Paul Gerrity, and Kevin Johnson with the Wyoming Game and Fish Department, and Mike Mazur with US Fish and Wildlife collected the majority of our fish for genetic sampling, and were invaluable resources during the planning stages of this project. Sean Lewandoski also collected a number of burbot for us. Alex Buerkle provided direction and advice relating to the genetics sample collection and analyses, and provided extensive computing and lab resources. David Underwood provided figure design support. Carlin Girard, Eric Gardunio, and Mary Kathryn Hooley offered intellectual contributions to data interpretation and discussed initial drafts of this manuscript. Dave McDonald and two anonymous reviewers provided feedback that further improved the manuscript. All fish were treated humanely and anesthetized before all surgical procedures in accordance with University of Wyoming Animal Care and Use Committee protocol #A-3216-01. Funding was provided by the Wyoming Game and Fish Department and the U.S. Geological Survey. Use of trade, product, or firm names does not imply endorsement by the U.S. Government.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: M. Power
This article was originally intended to appear in ‘Ecology, Culture, and Management of Burbot’ guest edited by Martin A. Stapanian & Christopher A. Myrick (published in volume 757 of Hydrobiologia).
Data accessibility Raw sequence data (.fastq files) are available at the National Center for Biotechnology Information (NCBI) SRA: SRX1078958. SNP files and necessary scripts are available at http://datadryad.org; doi:10.5061/dryad.7842r.
Rights and permissions
About this article
Cite this article
Underwood, Z.E., Mandeville, E.G. & Walters, A.W. Population connectivity and genetic structure of burbot (Lota lota) populations in the Wind River Basin, Wyoming. Hydrobiologia 765, 329–342 (2016). https://doi.org/10.1007/s10750-015-2422-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-015-2422-y