Abstract
Characterization and molecular mechanism dissection for lesion-mimic mutation can help to gain an in-depth insight into regulatory mechanisms of programmed cell death in plants. In present study, the barley lesion mimic mutant bspl (barley spotted 1eaf) was used to investigate the formation of the bspl lesion mimic phenotype in barley using proteomics and quantitative real-time (qRT)-PCR techniques. The experiment showed that the formation of the bspl phenotype was directly related to light. Cell necrosis spontaneously formed in multiple parts of the leaf, and H2O2 accumulated continuously along with the formation of lesions. A total of 31 differentially expressed protein spots were identified using two-dimensional electrophoresis and mass spectrometry analysis of the expressed proteins between the bspl and the wild-type. These differentially expressed proteins are involved in different biological processes, mainly related to defense, photosynthesis, nitrogen acid metabolism, and carbohydrate metabolism. The expression of 15 genes was analyzed by qRT-PCR, and the results showed that the transcription level was basically consistent with the change in the protein level. The decreased expression of ATPase β subunit, cytochrome B6-F complex, photosystem I reaction center subunit VI, and other proteins may lead to the blockage of electron transport during photosynthesis, leading to cell death. Moreover, the overexpression of chitinase, thaumatin-like protein, PR-1a and other proteins, and the increased expression of NADPH-thioredoxin reductase and dehydroascorbate reductase may increase the disease resistance and stress resistance of bspl. This study will provide a new insight into the molecular mechanism underlying bspl-induced cell death in plants.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
A lesion mimic mutant (LMM) is a type of mutant that can spontaneously form necrotic plaques on leaves, sheaths, and whole plants without obvious damage, stress, or infection by external pathogens. Many scholars have conducted in-depth studies on the mechanisms of lesion mimic mutants. At present, such mutants have been found in plants such as rice (Ge et al. 2015; Chen et al. 2018a), Arabidopsis thaliana (Dietrich et al. 1994; Badigannavar et al. 2002), maize (Hu et al. 1998), soybean (Kosslak et al. 1996), and barley (Buschges et al. 1997). Studies have shown that the causes of various types of lesion mimic mutants are as follows: (1) mutation or abnormal expression of plant disease resistance genes whereby HR (Hypersensitive Response) is abnormally stimulated, leading to the early onset or reduced efficiency of plant programmed cell death (PCD), resulting in necrotic symptoms similar to pathogen infection. For example, the SSI4 mutation of the TIR-NB-LRR protein in Arabidopsis causes constitutive expression of the mutant R protein and produces a lesion mimic phenotype (Shirano et al. 2002). (2) The loss of control of plant PCD triggers the production of the lesion mimic. For example, the Arabidopsis lsd1 gene encodes a similar transcription factor protein containing three zinc finger domains, which can activate plant PCD and defense responses, and also exhibit a lesion mimic phenotype (Dietrich et al. 1994). (3) Plant metabolism disorders directly or indirectly induce the occurrence of a lesion mimic. For example, the maize Les22 gene encodes uroporphyrinogen decarboxylase (UROD), which causes a significant decrease in UROD activity, thereby accumulating the reaction substrate urinary protoporphyrin III, which converts molecular oxygen into active oxygen under light conditions, resulting in a maize lesion mimic phenotype (Hu et al. 1998). Changes in the external environment can also induce the formation of a lesion mimic; for instance, the rice (Wang et al. 2015) and wheat lm3 (Wang et al. 2016) produce lesion mimics under light. These studies suggest that the causes of the induction of lesion mimics may be diverse and that the plant disease resistance signal transduction network is also very complex. Only by identifying and studying these lesion mimic mutants can the mechanisms of lesion mimic production be elucidated.
The proteome represents all the proteins expressed in cells, tissues, or organisms, and reflects the different combinations of proteins in different cells. Proteomics technology mainly includes two-dimensional gel electrophoresis (2DE) and mass spectrometry. 2DE is the core technology of proteomic research. Through 2DE technology, all protein components in cells or tissues can be displayed and separated. Mass spectrometry can be used to rapidly identify proteins to elucidate protein-related properties. Kang and Bae (2010) found 159 upregulated or downregulated protein spots in the protein 2DE analysis of the rice lesion mimic mutant spl6, among which ATP synthase, thioredoxin peroxidase (TPX), and protein disulfide isomerase (PDI) were not expressed in mutants but were abundantly expressed in the wild-type. The deletion of PDI and TPX may lead to the degradation of thylakoid membranes, which may form lesions. Han et al. (2014) studied the rice lesion mimic mutant chl1 using proteomics, indicating that differentially expressed proteins played important roles in inducing H2O2 accumulation and cell death, scavenging excess reactive oxygen species (ROS), and enhancing disease resistance and stress resistance.
Barley is a cereal crop that is widely cultivated across the world (Chen et al. 2018b). Its total annual yield is second only to wheat, maize, and rice. It has the advantages of short growth period, high stress resistance, early maturity, and wide adaptability (Lu 1996). Proteomics studies of barley have involved almost all tissues, organs, and developmental stages, and have focused on the biotic or abiotic stress, quality, and development. Finnie et al. studied the protein profiles of Barke and Morex barley grains and identified some specific protein differences during seed germination that could be used as markers to distinguish the two varieties (Finnie et al. 2006). Kristian et al. (2004) used 2DE to analyze salt-soluble proteins in barley grains and malt. In the range of pH 6–11, 380 and 500 protein spots were detected. These proteins were associated with antibacterial functions, glycolysis, and metabolism. In the study of barley lesion mutants, Buschges et al. cloned the gene for the barley lesion mutant mlo, which encodes a 60 kD protein and has sustained resistance to most powdery mildew pathogens (Buschges et al. 1997). Powdery mildew infection experiments showed that there were actin-dependent and -independent resistance pathways at the edge of the cell membrane that act synergistically and inhibit the expression of resistance signals by the transmembrane protein mlo (Miklis et al. 2007). Compared with rice, there are few studies on barley lesion mutants, and no studies have reported on the proteomics of barley lesions.
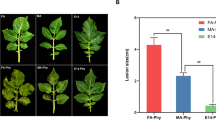
A barley lesion mutant bspl was screened from a barley mutant library induced by ethyl methane sulfonate (EMS) (Zhang et al. 2011). Compared with its wild-type—ZJU3, bspl exhibited red-brown lesion mimics at the top of the leaves at the seedling stage, which gradually spread across the entire leaves. Lesion mimics in the tillering stage do not increase, invoke leaf death, or affect the heading and fruiting of the plants (Fig. 1). Our previous study showed that the phenotype of the barley lesion mutant bspl1 was controlled by a recessive single gene, and it was speculated that the generation of lesions was related to the accumulation of ROS in the leaves (Sun et al. 2014). The present study used 2DE technology and mass spectrometry in the proteomic analysis of bspl and its wild-type to explore the formation of the bspl lesion mutant phenotype.
Materials and methods
Plant material and mutagenic treatment
The wild-type ZJU3 and the mutant bspl were planted in the experimental field of the Hangzhou Normal University (Xiasha Campus), and the leaves were stored at − 80 °C until the heading stage.
Leaf shading treatment
Before the lesion mimics appeared in the mutant, the upper leaf surfaces of the bspl and wild-type ZJU3 were covered with tin foil. When the leaf lesion mimics appeared in the light spot, the lesion mimics in the shading area were observed.
Detection of leaf cell death
Trypan blue staining was used to detect cell death in leaves of the lesion mimic mutant. Leaves of the mutant bspl and wild-type ZJU3 were collected at the same time and from the same position on the plant and were dipped in lactophenol–Trypan blue solution and stained in boiling water bath for 10 min. They were then placed at room temperature for 12 h and soaked in 2.5 g/mL trichloroacetaldehyde hydrate solution for decolorization of 4 days. A stereo microscope was used to observe the leaves and obtain images.
Detection of leaf H2O2 deposition
To detect the accumulation of H2O2 in the leaves of the lesion mimic mutant, we used diaminobenzidine (DAB) staining. The bspl and wild-type ZJU3 leaves in the same period were immersed in 1 mg/mL DAB solution (pH 3.8), irradiated at 25 °C for 8 h, and boiled in 96% ethanol and decolorized for 10 min, following which the leaves were immersed in fresh 96% ethanol solution (25 °C, 4 h). A stereo microscope was used to observe the leaves and obtain images.
Extraction of total protein from barley leaves
The total protein of the bspl and ZJU3 leaves stored at − 80 °C was extracted using the trichloroacetic acid (TCA)-acetone precipitation method, and the protein was quantified using the Bradford method (Kruger and Hammond 1994).
Two-dimensional electrophoresis
The total protein of the bspl and ZJU3 leaves was separated three times using 2DE. Gel staining was used the Coomassie Brilliant Blue method, according to the procedure of Candiano et al. (2004). The stained gel was scanned by ImageScanner III, and the image analysis was performed using the ImageMaster 2D Platinum 6.0 (GE Healthcare Life Sciences). The main operations included gel protein spot detection, image background subtraction, protein point gray value normalization, and matching of the protein spots between the different gels.
Mass spectrometry
The digested protein sample was dissolved in 0.1% formic acid and used for matrix-assisted laser desorption/ionization (MALDI)-time of flight (TOF) MS analysis. The mass spectrometer was an ABI 4700 MALDI-TOF-TOF mass spectrometer with a nitrogen: argon laser.
qRT-PCR of differential protein coding genes
RNA extraction was performed using TRIzol reagent (Invitrogen Shanghai, China) following the manufacturer’s instructions. Reverse-transcription was carried out using a M-MLV Reverse Transcriptase cDNA Synthesis Kit (Promega). qRT-PCR primers and probe design were performed using GenScript (https://www.genscript.Com/ssl-bin/app/primer) and fluorescent probes were synthesized by Invitrogen. qRT-PCR was performed using Takara’s Premix Ex TaqTM (Probe qPCR) and ABI7500 qRT-PCR instrument. The expression quantity was expressed using a relative expression amount, and the calculation was performed using 2− ΔΔCT method. Correlation analysis between gene expression and protein expression was performed using the IBM SPSS Statistics 20.0.
Functional classification of proteins
Differentially expressed proteins identified by MALDI-TOF-TOF MS/MS were obtained from http://www.uniprot.org/, http://www.ebi.ac.uk/interpro/, http://www.gramene.org/, http://www.ncbi.nlm.nih.gov, and other database search results that obtained comprehensive functional classifications. The classified proteins were combined with their corresponding metabolic or regulatory pathways and the physiological and biochemical changes in barley for functional analysis.
Gene ontology (GO) enrichment analysis
The GO classification of the corresponding protein genes was performed using an online tool (http://www.ricechip.org/) that identifies enriched terms based on a P value.
Results
Leaf shading treatment
No lesion mimic was observed in the shaded bspl leaves, and thus it was concluded that lesion mimic formation in barley occurs only under light conditions (Fig. 2a).
The results of leaf shading treatment, Trypan blue and DAB staining. a Before the lesion mimics appeared in the mutant, the upper leaf surfaces of the bspl and wild-type ZJU3 were covered with tin foil. It can be seen from (ii) that the mutant has no lesion mimics in the shading area. Bar 2 cm. (i) Wild type ZJU3; (ii) mutant bspl. b Necrotic cells and tissues can be stained dark blue by trypan blue. The results showed that the wild-type leaves were evenly stained without deep blue spots; multiple locations of bspl leaves were stained dark blue spots. (i) Wild type ZJU3, Bar 1 cm; (ii) mutant bspl, Bar 1 cm. c H2O2 reacts with DAB to form a red-brown precipitate for detecting the accumulation of H2O2 in cells. It was found by staining that the red–brown site on the mutant leaves coincided with the site of the lesion mimic. (i), (iii) wild type ZJU3, Bar: 1 cm; (ii), (iv) mutant bspl, Bar: 1 cm; (i), (ii) before dyeing; (iii), (iv) after dyeing. (Color figure online)
Detection of leaf cell death
Trypan blue staining is a common method for detecting plant cell death, and necrotic cells and tissues are stained dark blue. We found that the wild-type leaves were uniform in color and had no dark blue spots, whereas the bspl leaves developed dark blue spots on multiple positions, indicating spontaneous cell necrosis (Fig. 2b).
Detection of leaf H2O2 deposition
H2O2 can react with DAB to form a red-brown precipitate and localize to the site of peroxidase activity, thereby identifying the accumulation of H2O2 in cells. It was found that the red-brown spots on the bspl leaves were consistent with the sites of the lesion mimics (Fig. 2c).
Mapped protein spots
2DE images were obtained using ImageScanner, and 2DE images of the total protein of bspl and ZJU3 were analyzed using ImageMaster 2D Platinum software. More than 1300 protein spots were detected in the three biological replicates, and protein spots with upregulated and downregulated expression abundance of ≥ 1.5-fold (bspl/ZJU3) were screened. The molecular weight of these protein spots ranged from 19 to 49 KD, and the isoelectric point was around 4–9 (Fig. 3a).
Representative 2-DE gel pattern and qRT-PCR analysis. a Representative 2-DE gel pattern of leaf blade proteins of ZJU3 and bspl mutant. The horizontal axis represents the isoelectric point of the protein; the longitudinal axis represents the molecular weight of the protein. IEF was performed with IPG dry strips (pH 3–10 NL, 24 cm length). SDS–PAGE was run in a 10–16% gradient SDS gel. (i): ZJU3; (ii) mutant bspl. Arrows indicate differentially expressed protein points. b qRT-PCR analysis of differentially expressed proteins in the barley lesion mimic mutant, bspl. The X axis represents the name of the differential protein; the Y axis represents the relative expression of the differentially expressed protein at the transcriptional level
Mass spectrometric analysis of bspl differential protein spots in the barley lesion mimic mutant
A total of 31 differentially expressed proteins were identified by mass spectrometry. These proteins were involved in different biological processes, including defense, redox, photosynthesis, carbohydrate, amino acid metabolism, and other related processes (Table 1).
Changes in differential proteins in the bspl mutant at the transcriptional level
To further analyze the necrosis and resistance mechanisms of bspl, the expression of bspl proteins and changes in transcription levels were compared. The qRT-PCR analysis indicated that the expression levels of chitinase, ribulose bisphosphate carboxylase small subunit, ATPase β subunit, cytochrome B6-F complex, and other proteins were basically consistent with the protein levels (Fig. 3b). Correlation analysis showed that there was a good correlation between gene expression and protein level expression (R2 = 0.7549, P < 0.001).
Basic functional classification analysis of differential proteins in the bspl mutant
The identified differentially expressed proteins were classified according to their functions. It was found that the upregulated proteins were mainly associated with redox, defense, sugar metabolism and transport, protein modification, and other functions, while the downregulated proteins were mainly concentrated in transportation, electron transfer, and other functions (Fig. 4a). These results indicate that the functions of bspl transportation and electron transfer were seriously affected and that other functions such as redox and defense had been activated.
Basic functional classification, proposed subcellular localization and GO analysis of differentially expressed proteins in the barley lesion mimic mutant, bspl. a Basic functional classification of differentially expressed proteins in the barley lesion mimic mutant, bspl. (i) Upregulated protein spots; (ii) downregulated protein spots. b Proposed subcellular localization of differentially expressed proteins in the barley lesion mimic mutant, bspl. (i) Up-regulated protein spots; (ii) down-regulated protein spots. c GO analysis of differentially expressed proteins in the barley lesion mimic mutant, bspl. The horizontal axis represents the number of protein spots; the longitudinal axis represents the GO database was used to obtain relevant biological information regarding the cellular components, molecular functions and biological processes of the differential proteins. (Red, blue and green indicates up-regulation protein spots; Purple indicates down-regulation protein spots). (Color figure online)
Subcellular localization analysis of differential proteins in the bspl mutant
Through subcellular localization analysis, it was found that the upregulated proteins were mainly localized in the cytoplasm and chloroplast, while the downregulated proteins were mainly located in the chloroplast thylakoid membrane and chloroplast thylakoid lumen (Fig. 4b).
GO functional classification of the differential proteins in the bspl mutant
The GO database was used to obtain relevant biological information regarding the biological processes, cellular components, and molecular functions of the differential proteins (Fig. 4c). According to the statistics of the GO secondary function classification, in the biological process classification, downregulated proteins were mainly concentrated in photosynthesis process, whereas upregulated proteins were mainly concentrated in defense reaction, redox, metabolism, carbohydrate metabolism, and other processes. The downregulated proteins involved in cell components were mainly localized in the thylakoids and chloroplast thylakoid membranes, while the upregulated proteins were mainly localized in the chloroplast, cytoplasm, and extracellular regions. In terms of molecular function, the downregulated proteins mainly performed binding functions, while the upregulated proteins mainly performed catalytical functions.
Discussion
During leaf senescence, the chlorophyll content, photosynthetic efficiency, and membrane composition of the cell membrane system change (Lim et al. 2007). The mutant bspl in this study spontaneously formed reddish-brown spots on the leaves from the seedling stage, which were evenly distributed on the leaves and were accompanied by brown spots throughout the remainder of the treatment. Tyrosin staining is an effective method for testing cell membrane integrity and cell death. Some cells of the mutant bspl leaves were stained dark blue (Fig. 2b), indicating that the membrane system of the cells was incomplete or that the cells had died. DAB reacts with H2O2 to form a brown–red precipitate. Therefore, DAB staining can detect the accumulation of H2O2. Compared with the wild-type ZJU3, some cells of the mutant bspl leaves were stained brown–red (Fig. 2c), indicating the accumulation of H2O2. Light is an important external factor for lesion induction. The wheat lesion mimic mutant lm3 produces lesions under light conditions (Wang et al. 2016). Mutant bspl was found to have no lesion mimic in the shaded bspl leaves (Fig. 2a), possibly because light promotes the production of ROS in plants.
Pathogenesis-related (PRs) proteins are encoded by plant PR genes. Based on the biochemistry and amino acid composition, PRs are divided into 17 protein families (PR-1–PR-17) (Loon et al. 1999). The PR-1 protein is encoded by the Ypr1 gene, which is a representative member of the PR-1a protein. The PR-1a protein is relatively evolutionarily conserved, and the PR-1 proteins from different plants of the same type are similar in physical and chemical properties and in molecular structure, amino acid composition, and serum response (Loon et al. 2006). Huijsduijnen et al. (1985) found that the expression level of PR-1 in plants was extremely low, but that the expression level could be increased by up to a thousand times following TMV infection. PR-1 protein was also found that it exhibits the highest level of accumulation of the PR family following pathogen infection in plants (Cornelissen et al. 1986; Lcvan et al. 1999). Muradov et al. (1993) found that when powdery mildew-resistant barley was inoculated with powdery mildew for 12 h, the PRb-1 gene transcript was significantly accumulated. Temporary silencing of PR-1 expression in barley by double-stranded RNA interference technology makes the barley powdery mildew enzymatic fungus more likely to penetrate the cell wall (Schultheiss et al. 2003). The protein expression of PR-1a in the barley lesion mimic mutant bspl increased more than 70-fold, suggesting that the PR-1a protein may have a certain anti-disease effect in bspl.
Thaumatin-like proteins (TLPs) belong to the PR-5 protein family and are encoded by the Ypr5 gene. Since the amino acid sequence of the PR-5 protein family is highly homologous to the amino acid sequence of the thaumatin protein, it is called a thaumatin protein. TLPs are generally expressed at low levels in plant-specific tissues and organs, and their expression activities differ. For example, the expression of TLPs was lowest in immature strawberry fruits and highest in the leaves and corolla, and the expression of TLPs was rapidly upregulated when the plants were subjected to external stress (Min et al. 2004). TLPs in barley, apple, and cherry all possess β-1,3-glucanase activity, which binds and hydrolyzes β-1,3-glucan, and the activity of β-1,3-glucanase in barley was found to be remarkable (Osmond et al. 2001). Menubouaouiche et al. (2003) believed that TLPs themselves could not directly destroy the cell wall of fungi in plants, but could induce the defense system in plants to resist fungal infection through the products of the β-1,3-glucanase reaction. The increased expression of TLPs in the barley lesion mutant bspl may contribute to the enhanced disease resistance of bspl. At present, chitinase activity has been found in four protein families, namely PR-3, PR-4, PR-8, and PR-11. Chitinase mainly hydrolyzes the β-1,4 glycoside bond in the chitin polymer, which is one of the cell wall components of fungi, to produce N-acylglucosamine oligomers. Chitinase activity is low in plants, and the activity of chitinase is rapidly increased after being induced by pathogenic and abiotic factors, which have a protective effect on plants to a certain extent. Robert et al. (2002) showed that the induction of chitinase genes in grapefruit depends on the pathogen of infection. In the present study, the expression of chitinase was increased by 85-fold in bspl. The expression of PR-1a, TLPs, and chitinase in bspl may be increased under the induction of disease resistance signals, which may help to enhance the disease resistance of bspl.
The protein expression levels of the ATPase β subunit and cytochrome B6-F complex decreased in bspl. Chloroplast ATPase is an enzyme necessary for photosynthetic electron transport and photosynthetic phosphorylation during plant photosynthesis. The enzyme consists of two parts, CF1 and CF0. CF1 is located outside the thylakoid membrane and is an enzyme complex composed of nine subunits of α3, β3, γ, δ, and ε. The β subunit is a catalytic subunit with a function of binding to ADP and can convert ADP into ATP in the presence of a transmembrane proton gradient (Hinkle and Mccarty 1978). The cytochrome BF complex is a multi-subunit membrane protein composed of four peptides of cytochrome b6 (b563), cytochrome f, subunit IV, and iron-sulfur protein, and is an important electron transporter that acts synergistically between photosystem (PS) I and PSII. Both the ATPase β subunit and the cytochrome B6-F complex are involved in the photosynthetic electron transport chain process. The decrease in the expression of these proteins also leads to a decrease in photosynthetic capacity and accumulation of H2O2 in the chloroplast, thereby inducing cell death.
Among the identified differential proteins, the protein expression of chitinase and pathogenesis-related protein PR-1a was increased by several tens of times, and these two proteins may increase bspl to enhance disease resistance. PR-1a is mainly located in the cytoplasm, and thus it can be inferred that pathogen resistance may occur in the cytoplasm. The protein expression of the ATPase β subunit, the cytochrome B6-F complex, and the PSI reaction center subunit VI decreased, and was mainly localized in the chloroplast, which may be the main factor affecting bspl photosynthesis. In the basic functional classification, mainly focusing on transportation and electron transfer, it was concluded that the electron transfer pathway was affected during bspl photosynthesis, which reduced the photosynthetic efficiency of bspl and might account for the formation of the bspl phenotype.
Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) is a key enzyme for immobilizing CO2 during plant photosynthesis and is one of the key enzymes in the dark reaction of photosynthesis. Rubisco consists of large and small subunits. The substrate activation center and catalytic center of the whole enzyme are located on the large subunit, which is non-specific and inefficient compared with the general enzyme. It can both catalyze carboxylation and catalyze oxygenation and has low catalytic efficiency (Mei et al. 2007). Lorimer et al. (1976) showed that the reaction rate of the Rubisco enzyme is not simply related to the total amount of enzyme protein, but is more greatly affected by the degree of Rubisco activity. It was found that the quantity of O2 in rice leaves is consistent with the content of Rubisco activase and is exponentially related to the ratio of Rubisco activase/Rubisco content in rice leaves (Fukayama et al. 1996). The expression of Rubisco activase was found to decrease in barley bspl, which resulted in a decrease in the photosynthetic efficiency of bspl. Excessive absorption of light energy by the photosystem for a long period of time produces photoinhibition and causes the accumulation of reducing substances in the chloroplast, resulting in the excessive reduction in photosynthetic electron transport chain activity and ROS production (Tang 2014), thereby inducing cell death in bspl.
NADP-malic enzyme (ME) is one of the key enzymes in malic acid metabolism in living organisms. In the presence of divalent metal ions, ME catalyzes the oxidative decarboxylation of malic acid to produce pyruvic acid, CO2, and NAD(P)H. Currently, genes encoding NADP-ME in higher plants are not unique and are encoded by a family of multiple small genes. Studies have shown that C3 plant NADP-ME is associated with plant defense responses or stress responses, but the specific mechanism by which NADP-ME is involved in plant defense responses or stress responses is unclear. Liu et al. showed that the activity of NADP-ME in rice seedling leaves and roots increased by more than 50% under salt and drought conditions, and the resistance of NADP-ME overexpressing transgenic Arabidopsis plants was significantly enhanced (Liu et al. 2007). The expression of NADP-ME was also increased in bspl, which may increase the resistance of bspl.
NADPH-thioredoxin reductase (TrxR) is an NADPH-dependent dimeric selenoenzyme containing a flavin adenine dinucleotide (FAD) domain, the primary function of which is to catalyze the reduction of –S2 on thioredoxin (Trx) to (–SH)2 to maintain the reduced form of Trx by NADPH. Trx plays an important role in redox regulation and oxidation defense. In the presence of NADPH, the reduced form of Trx acts as an electron donor for thioredoxin peroxidase, reducing H2O2 to H2O (Zhang and Xiong 2005). The increased expression of NADPH-thioredoxin reductase in bspl may be to clear excess H2O2 in the leaves. Ascorbic acid (ASA) exists mainly in the reduced form (ASC) in living cells and reacts directly with ROS, such as H2O2, O2−, and HO−, to protect chloroplasts from oxidative damage (Noctor and Foyer 1998). Dehydroascorbate reductase (DHAR) reduces the ASC/DHA ratio by reducing the oxidized ascorbic acid (DHA) to the reduced ascorbic acid (ASC) via the ascorbate–glutathione cycle and plays an important role in maintaining the normal metabolic level of plant ASA and the redox balance of the plant (Wheeler et al. 1998; Boussiba et al. 1975). DHAR expression was upregulated in bspl, thereby promoting the redox balance of the bspl mutant in vivo to remove excess ROS.
The expression of TrxR and DHAR in bspl was more prominent than other similar proteins. They were mainly localized in the cytoplasm, and their basic functional classification was mainly related to redox, defense, glucose metabolism and transport, protein modification, and other functions. It is concluded that the metabolism of TrxR and DHAR in bspl is more vigorous in order to maintain the balance of oxidation and reduction in vivo, so as to eliminate excessive ROS. Additionally, excessive ROS may be one of the explanations for the formation of the bspl phenotype.
Conclusion
The phenotypic formation of bspl is directly related to light. Cell necrosis spontaneously occurred in multiple parts of the leaf, and H2O2 accumulated continuously with the formation of lesions. Proteomic analysis of bspl and ZJU3 showed that the decreased expression of Rubisco activator, ATPase beta subunit, cytochrome B6-F complex, and PSI reaction center subunit VI affected the electron transport pathways during photosynthesis, resulting in the accumulation of ROS in the chloroplasts and the formation of lesion mimic phenotypes. Increased expression levels of chitinase, pathogenesis-related proteins PR-1a, thaumatin-like proteins, NADPH-thioredoxin reductase, and dehydroascorbate reductase may increase the disease resistance of bspl. It is speculated that these differentially expressed proteins are involved in the formation of the bspl phenotype of barley lesion mimic mutants (Fig. 5).
A model of the regulatory networks of bspl differentially expressed proteins. Rubisco ribulose-1,5-bisphosphate carboxylase/oxygenase; DHAR dehydroascorbate reductase, TrxR NADPH-thioredoxin reductase, down downregulated proteins, up upregulated proteins, ROS reactive oxygen species—H2O2, O2−, HO2−, –OH
References
Badigannavar AM, Kale DM, Eapen S, Murty GS (2002) Inheritance of disease lesion mimic leaf trait in groundnut. Hered 93:50–52. https://doi.org/10.1093/jhered/93.1.50
Boussiba S, Rikin A, Richmond AE (1975) The role of abscisic acid in cross-adaptation of tobacco plants. Plant Physiol 56:337–339. https://doi.org/10.1104/pp.56.2.337
Buschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A (1997) The barley mlo gene: a novel control element of plant pathogen resistance. Cell 88:695–705. https://doi.org/10.1016/S0092-8674(00)81912-1
Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B (2004) Blue silver: a very sensitive colloidal coomassie g-250 staining for proteome analysis. Electrophoresis 25:1327–1333. https://doi.org/10.1002/elps.200305844
Chen P, Hu H, Zhang Y, Wang Z, Dong G, Cui Y, Qian Q, Ren D, Guo L (2018a) Genetic analysis and fine-mapping of a new rice mutant, white and lesion mimic leaf1. Plant Growth Regul 85:425–435. https://doi.org/10.1007/s10725-018-0403-7
Chen G, Wang Y, Wang X, Yang Q, Quan X, Zeng J, Dai F, Zeng F, Wu F, Zhang G, Chen Z (2018b) Leaf epidermis transcriptome reveals drought-Induced hormonal signaling for stomatal regulation in wild barley. Plant Growth Regul. https://doi.org/10.1007/s10725-018-0450-0
Cornelissen BJ, Hooft van Huijsduijnen RA, Van Loon LC, Bol JF (1986) Molecular characterization of messenger rnas for ‘pathogenesis related’ proteins la, lb and lc, induced by tmv infection of tobacco. EMBO J 5:37–40. https://doi.org/10.1002/j.1460-2075.1986.tb04174.x
Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL (1994) Arabidopsis mutants simulating disease resistance response. Cell 77:565. https://doi.org/10.1016/0092-8674(94)90218-6
Finnie C, Bakjensen KS, Laugesen S, Roepstorff P, Svensson B (2006) Differential appearance of isoforms and cultivar variation in protein temporal profiles revealed in the maturing barley grain proteome. Plant Sci 170:808–821. https://doi.org/10.1016/j.plantsci.2005.11.012
Fukayama H, Uchida N, Azuma T, Yasuda T (1996) Relationships between photosynthetic activity and the amounts of rubisco activase and rubisco in rice leaves from emergence through senescence. Jpn J Crop Sci 65:296–302
Ge C, Pan EZ, Jiang J, Zhang H, Zeng X, Dong D, Hu G, Xue J D (2015) Map-based cloning of a spotted-leaf mutant gene OsSL5 in Japonica rice. Plant Growth Regul 75:595–603. https://doi.org/10.1007/s10725-014-9962-4
Han XY, Yang Y, Chu-Lang YU, Zhang WH, Sheng-Hai YE, Chen B (2014) A proteomic study on a disease-resistance-enhanced rice lesion mimic mutant. Chin J of Rice Sci 28:559–569. https://doi.org/10.3969/j.issn.1001-7216.2014.06.001
Hinkle PC, Mccarty RE (1978) How cells make ATP. Sci Am 238:104. https://doi.org/10.1038/scientificamerican0378-104
Hooft van Huijsduijnen RA, Cornelissen BJ, van Loon LC, van Boom JH, Tromp M, Bol JF (1985) Virus-induced synthesis of messenger rnas for precursors of pathogenesis-related proteins in tobacco. Embo J 4:2167. https://doi.org/10.1002/j.1460-2075.1985.tb03911.x
Hu G, Yalpani N, Briggs SP, Johal GS (1998) A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell. https://doi.org/10.1105/tpc.10.7.1095
Kang SG, Bae H (2010) Proteome analysis and characterization of phenotypes of lesion mimic mutant spotted leaf 6 in rice. Proteomics 7:2447–2458. https://doi.org/10.1002/pmic.200600961
Kosslak RM, Dieter JR, Ruff RL, Chamberlin MA, Bowen BA, Palmer RG (1996) Partial resistance to root-borne infection by phytophthora sojae in three allelic necrotic root mutants in soybean. J Hered 87:415–422. https://doi.org/10.1093/oxfordjournals.jhered.a023030
Kristian Sass BJ, Sabrina L, Peter R, Birte S (2004) Two-dimensional gel electrophoresis pattern (ph6–11) and identification of water-soluble barley seed and malt proteins by mass spectrometry. Proteomics 4:728–742. https://doi.org/10.1002/pmic.200300615
Kruger NJ, Hammond JBW (1994) The bradford method for protein quantitation. Methods Mol Biol 32:9–15. https://doi.org/10.1385/0-89603-268-X:9
Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58:115–136. https://doi.org/10.1146/annurev.arplant.57.032905.105316
Liu S, Cheng Y, Zhang X, Guan Q, Nishiuchi S, Hase K (2007) Expression of an nadp-malic enzyme gene in rice (Oryza sativa L.) is induced by environmental stresses; over-expression of the gene in arabidopsis confers salt and osmotic stress tolerance. Plant Mol Biol 64:49–58. https://doi.org/10.1007/s11103-007-9133-3
Loon LC, Strien EA (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of pr-1 type proteins. Physiol Mol Plant Pathol 55:85–97. https://doi.org/10.1006/pmpp.1999.0213
Loon LC, Rep M, Pieterse CM (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162. https://doi.org/10.1146/annurev.phyto.44.070505.143425
Lorimer GH, Badger MR, Andrews TJ (1976) The activation of ribulose-1,5-bisphosphate carboxylase by carbon dioxide and magnesium ions. equilibria, kinetics, a suggested mechanism, and physiological implications. Biochemistry 15: 529 https://doi.org/10.1021/bi00648a012
Lu L (1996) Chinese barley science. China Agriculture Press, Beijing
Mei Y, Hai-Lan LI, Xie J (2007) Ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco). Acta Phytophysiol Sin 78:155–162. https://doi.org/10.13592/j.cnki.ppj.2007.02.054
Menubouaouiche L, Vriet C, Peumans WJ, Barre A, Van Damme EJ, Rougé P (2003) A molecular basis for the endo-β-1,3-glucanase activity of the thaumatin-like proteins from edible fruits. Biochimie 85:123–131. https://doi.org/10.1016/S0300-9084(03)00058-0
Miklis M, Consonni C, Bhat RA, Lipka V, Schulze-Lefert P, Panstruga R (2007) Barley mlo modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiol 144:1132–1143. https://doi.org/10.1104/pp.107.098897
Min K, Ha SC, Hasegawa PM, Bressan RA, Yun DJ, Kim KK (2004) Crystal structure of osmotin, a plant antifungal protein. Proteins 54:170. https://doi.org/10.1002/prot.10571
Muradov A, Petrasovits L, Davidson A, Scott KJ (1993) A cdna clone for a pathogenesis-related protein 1 from barley. Plant Mol Biol 23:439–442. https://doi.org/10.1007/BF00029021
Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Biol 49:249–279. https://doi.org/10.1146/annurev.arplant.49.1.249
Osmond RI, Hrmova M, Fontaine F, Fincher GB (2001) Binding interactions between barley thaumatin-like proteins and (1,3)-β-D-glucans. Kinetics, specificity, structural analysis and biological implications. Eur J Biochem 268:4190–4199
Robert N, Roche K, Lebeau Y, Breda C, Boulay M, Esnault R (2002) Expression of grapevine chitinase genes in berries and leaves infected by fungal or bacterial pathogens. Plant Sci 162:389–400. https://doi.org/10.1016/S0168-9452(01)00576-3
Schultheiss H, Dechert C, Király L, Fodor J, Michel K, Kogel KH (2003) Functional assessment of the pathogenesis-related protein pr-1b in barley. Plant Sci 165:1275–1280. https://doi.org/10.1016/S0168-9452(03)00336-4
Shirano Y, Kachroo P, Shah J, Klessig DF (2002) A gain-of-function mutation in an arabidopsis toll interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14:3149–3162. https://doi.org/10.1105/tpc.005348
Sun Y, Wenyi LU, Zhang Y, Wang G, Sun J, Xue D (2014) Genetic and physiological analyses of barley lesion mimic mutant bspl. J Hangzhou Norm Univ 13:602–605. https://doi.org/10.3969/j.issn.1674-232X.2014.06.008
Tang S (2014) Study on the light adaptation of leaves in alternate breathing pathways. Dissertation/master’s thesis. Northwest Normal University, Lan Zhou
Van LL, Rep M, Pieterse CM (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162. https://doi.org/10.1146/annurev.phyto.44.070505.143425
Wang J, Ye B, Yin J, Yuan C, Zhou X, Li W (2015) Characterization and fine mapping of a light-dependent leaf lesion mimic mutant 1 in rice. Plant Physiol Biochem 97:44–51. https://doi.org/10.1016/j.plaphy.2015.09.001
Wang F, Wu W, Wang D, Yang W, Sun J, Liu D (2016) Characterization and genetic analysis of a novel light-dependent lesion mimic mutant, lm3, showing adult-plant resistance to powdery mildew in common wheat. PLoS ONE 11:e0155358. https://doi.org/10.1371/journal.pone.0155358
Wheeler GL, Jones MA, Smirnoff N (1998) The biosynthetic pathway of vitamin c in higher plants. Nature 393:365–369. https://doi.org/10.1038/30728
Zhang XT, Xiong YM (2005) The progress in studies of thioredoxin reductase. Foreign Med Sci-Sect Medgeogr 26:148–151
Zhang XQ, Xue DW, Zhou WH, Fei-Bo WU, Zhang GP (2011) Screening and identification of the mutants from two-row barley cultivar zju3 induced by ethyl methane sulfonate (EMS). J Zhejiang Univ 37:169–174. https://doi.org/10.3785/j.issn.1008-9209.2011.02.008
Acknowledgements
This work is funded by National Key Technology Research and Development Program (2015BAD01B02) to DX and the National Natural Science Foundation of China (31401316) to XZ. We would like to thank LetPub (http://www.letpub.com) for providing linguistic assistance during the preparation of this manuscript. The authors would also like to thank Professor Longbiao Guo for improving English language.
Author information
Authors and Affiliations
Contributions
DX and XZ conceived the research work. XZ, BT, YF, TT and JZ conducted experiments, and prepared the figures and tables. DX, XZ and BT wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Rights and permissions
About this article
Cite this article
Zhang, X., Tian, B., Fang, Y. et al. Proteome analysis and phenotypic characterization of the lesion mimic mutant bspl in barley. Plant Growth Regul 87, 329–339 (2019). https://doi.org/10.1007/s10725-018-00474-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-018-00474-y