Abstract
A japonica rice mutant, spotted-leaf 5 (sl5), was identified from YUN32 by EMS mutagenesis. The number of spots in leaves increased from maturity to late maturity in sl5, however, the leaves did not dry and withered. The sl5 mutant exhibited significantly lower height, spike length, primary branch number, second branch number, and 1,000-grain weight than YUN32. Genetic analysis shows that sl5 is controlled by a single recessive gene. OsSL5 was mapped into a 40-kb interval flanked by markers MX4 and MX5 on chromosome 7 by map-based cloning. Four ORFs, including one SPL5 gene, were identified in this region. Sequencing reveals that the G base at site 3,647 of the OsSL5 coding region was changed to A. The mutant OsSL5 site was different from that of the SPL5 mutant, with the background of indica rice. OsSL5 is thus a new SPL5 allele which encodes a putative splicing factor 3b subunit 3. QPCR shows that OsSL5 expression in the sl5 mutant is significantly lower than that in YUN32. The spotted leaf-related genes RLIN1, SPL28, and SPL18 expressions were significantly decreased, whereas the SPL7 and SL gene expressions significantly increased. The OsSL5 gene may be important for rice cell apoptosis regulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Spotted-leaf (spl) mutants are ubiquitous in plants and exhibit different phenotypes, colors, and spot sizes. These spots commonly appear to be brown. The spl mutants also exhibit other shades, including reddish-brown and white. Spots in this leaf mutant type are generated even without the presence of pathogens, mechanical or pesticide damages, and adverse conditions. Many spots are related to plant cell apoptosis and disease resistance. The spl mutants that exhibit natural, chemical, or physical mutagenesis are currently separated from maize (Johal et al. 1995), Arabidopsis thaliana (Dietrich et al. 1994), barley (Wolter et al. 1993) and rice (Takahashi et al. 2007).
Caused genes of spotted-leaf have been isolated and cloned in rice. SPL6 (Matin et al. 2010) and SPL28 (Qiao et al. 2010), for instance, are found in chromosome 1. The transport of compounds associated with SPL28-encoding clathrin is regulated between the vesicle and the cytoplasm during vesicle formation. Other genes, such as LRD1 (BL1) and SPL2 (Yoshimura et al. 1997; Kojo et al. 2006), are found in chromosome 2. spl2 mutant plants exhibited accumulated higher amounts of H2O2 and increased rates of cell death in response to wounding; BL4 (Yoshimura et al. 1997) and RLIN1 (Sun et al. 2011) are detected in chromosomes 3 and 4, respectively. The rlin1 mutant gene encodes coproporphyrin III oxidase. This gene is also involved in spot formation in leaves and stems. Such formations are mainly related to H2O2 metabolism and illumination. SPL7 (Yamanouchi et al. 2002; Kojo et al. 2006) and SPL8 (Liu et al. 2003) are found in chromosome 5. By contrast, no spots have been identified in the spl7 mutant during the seeding stage. However, small yellow spots have been found on leaves during the tillering stage to the heading stage. These spots disappear after exposing the leaves to high-temperature illumination or UV irradiation. SPL7 encodes heat stress transcription factor (HSF). HSF is activated by thermally stimulated factors when UV irradiation or illumination and temperature are decreased. HSF is then combined with a thermally stimulated protein gene promoter site in the nucleus to promote thermally stimulated protein expression and thermally stimulated response. Several genes have also been detected in other chromosomes. The lms1 mutant started to appear small brown spots on the leaves in elongation stage, and the spots gradually spread to whole leaves and sheaths (Ma et al. 2012). The lbsl1 displayed light brown spots during the whole growth period, and the LBSL1 gene was mapped between markers RM586 and RM588 (Feng et al. 2013). BL2 (Matin et al. 2010), small spots in the seedlings appeared at the four leaves stage and gradually grew into a large round and black area with a gray center on the leaf surface, the gene was not cloned. BL3 (Yoshimura et al. 1997), for example, are found in chromosome 6. SPL5 (Chen et al. 2012; 2013) and SPL9 (Liu et al. 2003) are detected in chromosome 7. The spl5 mutant plants exhibited small reddish brown spots scattered extensively on leaves, from the tillering stage to heading stage. SPL5 encodes for a splicing factor 3b subunit (SF3b3) and may regulate cell death by splicing mature RNA. OsLSD1 (Wang et al. 2005) and LMI1 (Liu et al. 2003) are found in chromosome 8. SPL10 (Babu et al. 2011) and SPL18 (OsAT1) (Mori et al. 2007) are located in chromosome 10. The spl10 showed lesions similar to spl5 mutant and have blast-resistant functions. Spots are mainly generated on the edge of the spl18 mutant leaves. The SPL18 gene encodes the OsAT1 protein, which is highly homologous to acyltransferase in tobacco. The OsAT1 protein is also important in the allergic reaction in tobacco. Studies verified that spot formation is related to the PR protein. SPL1 (SL) (Fujiwara et al. 2010), SPL11 (Zeng et al. 2004; Kojo et al. 2006) and SPL29 (t) (Li et al. 2010) are found in chromosome 12. SPL1 (SL) encodes a cytochrome P450 monooxygenase, which is important in serotonin formation and disease resistance in plants (Fujiwara et al. 2010). SPL11 regulates cell death and encodes a U-box-ARM-like protein, which induces E3 ubiquitin ligase catalytic activity. SPL11 can be potentially applied to breed rice blast fungus–resistant rice because this gene encodes the phenotype manifesting resistance to rice blast fungus (Zeng et al. 2004).
The spl gene provides important genetic resources that can be used to investigate plant cell apoptosis. This gene is also important in breeding disease resistant rice. A stable spl mutant, designated as sl5, was obtained in this study by subjecting YUN32 to ethyl methyl sulfide (EMS)-induced mutagenesis. The agronomic, physiological, and biochemical characteristics of the mutant were analyzed. In the study, OsSL5 was found in the 40 kb range of chromosome 7 by using the map-based cloning method. The sequencing results show that OsSL5 from Japonica rice is a new allele of SPL5. QPCR (real-time quantitative PCR) was performed to analyze OsSL5 and multiple cloned spl-related gene expressions in sl5 and YUN32. The results also show that OsSL5 is important in apoptosis regulation. These results further provide a new genetic resource that could be used to analyze the rice spl formation mechanism.
Materials and methods
Plant materials and growth condition
The sl5 mutant was identified from the YUN32 mutant library which was genetated by EMS mutagenesis. The sl5 and YUN32 were planted in paddy field by normal management in Fuyang, Hangzhou, in 2013. Each of sl5 and YUN32 were prepared for fifteen rows in each plot, with six plants per row. This arrangement was repeated three times. Fifteen plants in the two middle rows were selected to investigate the agronomic characteristics at the mature stage. Plant height, effective panicles, spike length, primary and secondary branch numbers, seed setting rate, 1,000-grain weight, and other characteristics were surveyed. Mean values and significance for agricultural traits were performed through Student’s t test analysis.
Genetic analysis and allelic test
The sl5 mutant was used as the female parent to cross with the indica cultivar TN1, NJ06, and 93-11. The F1 plants self-fertilized to produce three F2 generations. The leaf phenotypes of the F1 and F2 plants were investigated. The segregation ratio of the normal and the spot leaf individuals from the F2 generation at the heading stage was studied. A Chi square test was conducted. For the allelic test, sl5 was crossed with the spl5 mutant and the leaf phenotype of F1 plants was observed.
Primary mapping of the OsSL5 gene
The F2 individuals with spotted leaves derived from a cross of the sl5 mutant and TN1 were used to map the OsSL5 gene. Genomic DNA was extracted using the cetyltrimethyl ammonium bromide method (Rogers and Bendich 1985). The DNA samples from 21 mutant plants were pooled equally. One hundred and sixty-three pairs of rice SSR primers were evenly distributed on 12 chromosomes and used for polymorphism screening. A total of 50 mutant individuals were used for linkage analysis of the sl5 gene and flanking markers. The volume of the PCR system was 10 μL and contains 1 μL of template DNA, 1 μL each of forward and reverse primers (10 μmol/L), 0.1 μL of dNTPs (10 μmol/L), 1 μL of 10× PCR buffer solution, and 0.05–10 μL of r-Taq enzyme with ddH2O. PCR was performed for 38 cycles as follows: pre-denaturation at 94 °C for 4 min, denaturation at 94 °C, annealing at 55 °C, and extension at 72 °C for 30 s. Temperature was held constant at 15 °C during PCR. The PCR products were subjected to 4–5 % agarose gel electrophoresis, observed, and photographed using a gel imager after gel-red staining.
Fine mapping of the OsSL5 gene
The published rice genome sequence (http://rpg.dna.affrc.go.jp/E//toppage.html) and BLAST (http://blast.ncbi.nlm.nih.gov/) program were used to compare the genome sequence of Oryza sativa L. ssp. japonica var. Nipponbare with that of O. sativa ssp. indica 93-11. Indel markers were developed using the Software Primer 5 to narrow down the target gene (Table 1).
Sequencing of candidate gene
The full-length genomic sequences of the candidate genes were downloaded from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). The OsSL5 gene was amplified by KOD FX fidelity enzyme (Toyobo) using wild type and sl5 genomic DNA as template. The amplified products were sequenced in Shanghai Sunny Biotechnology Co., Ltd. The sequencing primers are shown in Table 2. The wild type gene and its mutant site were confirmed after the sequencing results were compared.
Histological staining
The Thordal-Christensen et al. (1997) method was used to detect H2O2 reactant accumulation in the mutant leaf. The spotted sl5 mutant leaves and the wild-type YUN32 leaves of heading stage (approximately 10 cm) were immersed in a 1 mg/mL 3,3′-diaminobenzidine (DAB; pH 3.8) solution and incubated at 25 °C for 8 h. The leaves were boiled in 96 % ethanol for 10 min to induce decolorization. The decolorized leaves were immersed subsequently in fresh ethanol for 4 h at 25 °C to observe and record the resulting color.
QPCR for OsSL5 analysis and related spl gene expression
Real-time fluorogenic quantitative PCR was performed to determine the relative spot formation mechanism in OsSL5 and to analyze spl-related gene expressions in the wild type and the sl5 mutant. The total RNAs in the wild type and the mutant leaves at the four-leaf stage were extracted to synthesize the first-strand cDNA by reverse transcription. The volume of the PCR system was 10 μL and contained the following: 1 μL of cDNA template; 1 μL each of upstream primer and downstream primer (10 μmol/L); 5 μL of 2× SYBR green PCR master mix (Applied Biosystems); and 2 μL of ddH2O. Each sample has three replicates. PCR amplification was performed for 40 cycles under the following conditions: enzyme activation at 95 °C for 10 min, denaturation at 95 °C for 15 s, annealing at 60 °C for 5 s, extension at 72 °C for 15 s, and fluorescence detection at 76 °C for 3 s. 2−∆∆Ct was used to calculate relative gene expression. The spl-related gene expression primers are shown in Table 3 (Livak and Schmittgen 2001). Student’s t test analysis was performed to determine the significance of each gene’s expression.
Results
Phenotypic and agronomic characteristics of the mutant
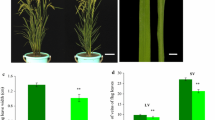
Sparse rust points were observed at the leaf tip when the second leaf of the sl5 mutant grew. Rust points were dispersed on the leaf surface during leaf growth. The agronomic characteristics of the sl5 mutant were investigated. The results show that plant height reduced from 85 to 71.7 cm, and spike length decreased from 19 to 14.8 cm. The internode also significantly reduced in mature leaves compared with the wild-type leaves. The secondary branch number of sl5 decreased from 31 to 5.3 and directly affected the sl5 grain number (Fig. 1; Table 4).
Phenotypes of wild-type and spotted-leaf mutant sl5. a Gross phenotypes of wild type and mutant: YUN32 (left) and sl5 (right); b leaf phenotypes of wild-type and mutant in mature leaves: YUN32 (left) and sl5 (right); c, d refer to the internodes and spikes of YUN32 and sl5, respectively. White bar 5 cm
Relationship between spot formation and reactive oxygen species accumulation
YUN32 (Fig. 2a) and sl5 mutant (Fig. 2b) were stained with DAB. No H2O2 accumulated in YUN32 (Fig. 2c). By contrast, numerous H2O2 deposition spots were observed in the mutant (Fig. 2d). This finding indicates that the emergence of spots on the sl5 leaf was caused by the excessive reactive oxygen species (ROS) in the rice cells.
Genetic analysis of the OsSL5 gene
The mutant sl5 was crossed with indca cultivar TN1, NJ06, and 93-11 to determine the inheritance pattern of OsSL5. All F1 plants showed normal phenotype without spotted-leaf. This finding indicates that sl5 is controlled by a recessive gene. In three F2 populations investigated, the number of normal plants and spl plants fitted to a 3:1 ratio well (χ2 = 1.1228, 1.0756, 1.6260 < χ 20.05 = 3.84; Table 5). These results indicated that the spotted-leaf phenotype of sl5 is controlled by a single recessive gene. The F1 plants derived from the cross of sl5 and spl5 mutants exhibited leaf spots like spl5. Thus, OsSL5 is allelic to SPL5.
Mapping of OsSL5 gene
To isolate OsSL5 gene, map-based cloning approach was performed. A total of 163 SSR markers covering the whole rice genome were screened for linkage with OsSL5. The results showed that OsSL5 is located into a region flanked by two markers, namely, RM6574 and RM542 on chromosome 7. Ten additional indel markers within this region were also developed (Table 1). Among 2,500 F2 plants, 40 and 60 recombinants were identified by RM6574 and RM542, respectively. Further genetic analysis using 8 indel markers delimited OsLs5 into a 40-kb interval flanked by MX4 and MX5 (Fig. 3). Only four open reading frames (ORFs) were found in this region (Fig. 3b). Of the four ORFs, two were coded for hypothetically unknown function protein. The other two were coded for proteins with known functions (i.e., S1 RNA-binding domain-containing protein and splicing factor 3B subunit 3; Table 6). This result suggested that the SPL5 gene is one of the candidate genes.
Mutant OsSL5 gene positioning, and the confirmation of the selected gene. a Fine positioning of the OsSL5 gene; b ORF prediction in the target interval; c structure and mutant sites of the OsSL5 candidate gene; d difference between YUN32 and sl5 amino acids. Black arrows represent base mutant site; thick black arrows show the difference in amino acids
Candidate gene sequencing analysis
The OsSL5 and SPL5 genes were sequenced to determine whether the spots on sl5 leaves are caused by the SPL5 gene. The SPL5 gene is 12,900 bp in length, including 15 exons and 14 introns (Fig. 3c). The sl5 and YUN32 were sequenced for SPL5 gene using eleven sequencing primers (Table 2) and compared. The CDS of SPL5 is 4,068 bp in length. The sequencing results showed that the G in the 3,647 position of the SPL5 coding region in sl5 was changed to A (Fig. 3c), thereby altering the amino acid sequence (Fig. 3d). Several japonica and indica rice varieties were sequenced to further ascertain whether the base substitution leads to leaf spots. No mutation sites were found in these rice varieties. The other three candidate genes in this region were further sequenced. The results did not reveal any difference in SPL5 between the wild-type (YUN32) and the sl5 mutant. The cDNA sequence of wild type and sl5 amplified by primer X12 was also compared (Table 2) and showed no difference.
Analysis of spl-related gene expression
The sl5 mutant seedling leaf showed a spot-like characteristic with a dry tip. To determine the relationship between the sl5 mutant gene and other spl-related genes, we conducted real-time fluorescence QPCR and compared the expression levels of spl-related genes in the sl5 mutant and the wild-type. The following genes were analyzed: SPL7 (heat stress transcription factor), RLIN1 (coproporphyrin original III oxidase), SPL28 (clathrin-associated protein compound), SPL18 (acyltransferase), SPL11 (E3 ubiquitin ligase), and SL (cytochrome P450 monooxygenase). The result showed that the OsSL5 expression in the mutant was significantly lower than that in the wild-type. The SPL7 and SL gene expressions were significantly increased. By contrast, the SPL28, RLIN1 and SPL18 gene expressions significantly decreased, but the SPL11 expression remained unchanged (Fig. 4). The change in the expression of OsSL5 in the mutant may affect the expression of other related genes. A significant difference was observed between OsSL5 and other spl-related gene expressions. This difference may affect spl related gene regulation in rice.
Discussion
Spl mutants are important in plant apoptosis and disease resistance. This study shows that the death of the spl mutant is related to ROS, as observed in various mutants, including spl2 and spl7 in rice (Yoshimura et al. 1997; Yamanouchi et al. 2002); lsd1 (Wang et al. 2005) and acd2 (Mach et al. 2001) in Arabidopsis; and les22 (Hu et al. 1998) and lls1 (Gray et al. 1997) in maize. Plants develop a radical scavenging system through evolution. This system maintains ROS levels under relatively stable conditions. Metabolic disorders occur if this balance is disrupted. High-content ROS highly oxidizes the cell membrane, thereby affecting cell permeability. Spot-like characteristic was observed on the leaves. Extremely high-content ROS eventually causes cell death. Nevertheless, the disease resistance of spl plants is enhanced to a certain degree; for example, spl11 enables rice to exhibit significant resistance to rice blast and bacterial blight (Yin et al. 2000). Sl (spl1) and spl28 affect rice blast and bacterial blight in rice (Mizobuchi et al. 2002). Wu et al. (2008) mutated rice IR64 by using butane, γ-ray, and fast neutrons to obtain 11 new spl mutants with different phenotypes. Spots appeared on the leaf and sheath in the two-leaf stage of the spl1-2 mutant. These spots increased in size afterward. Spots were also observed on the glume and decreased the 1,000-grain weight. Dark brown spots appeared on the tip of the leaf of Spl3-2 3 weeks after seeding, and yellow rust spots were found on other parts of the leaf. Yellow strip spots were detected on the leaf vein of spl6-2 6 weeks after seeding. After 2 months, the spots became longer with distinct phenotype. Small and discontinuous white spots appeared on the spl20 leaf 2 weeks after seeding until the tillering stage. Eleven mutants were inoculated with filamentous fungi. Among these mutants, five exhibited resistance. The spl17, spl26, spl20, and G862 mutants are resistant to rice blast fungus, particularly Ca89 and P06-6. The spl17 and spl26 mutants are also resistant to bacterial blight, which affects rice disease-resistant. Therefore, the spl gene is important in the theoretical research and disease resistance breeding.
In this study, the OsSL5 gene encoding for SF3b3 belongs to the SF3b3 splicing family. However, homologous genes in plants have yet to be determined. Other homologous genes have been found in fungi and animals. For example, the homologous gene prp12-1 in yeast and animals is involved in splicing and cell differentiation (Habara et al. 2001). Yamasaki et al. (2008) found that SF3b3 can form an acceptor by combining with a C-type lectin taken from macrophages to induce non-steady apoptosis. Inflammatory cells, such as neutrophils, are then generated and enter damaged tissues to mediate cell death. Menon et al. (2008) reported that SF3b3 is related to cullin-RING E3 ubiquitin ligase and stabilizes genomes during the cell cycle. Kerins et al. (2010) isolated the SF3b3 homologous gene TEG-4 in nematodes, which positively regulates the GLP-1 signal transmission downstream pathway and controls embryonic stem cell proliferation and meiosis. The SF3b3 gene in mice significantly down-regulates the gene that may induce apoptosis of early sac cells in the embryonic stem cell sac (Jincho et al. 2008). In this study, the sl5 mutant initially generated spots on the leaf tip in the seeding stage. These spots spread to the whole leaf during growth. The spots further increased in size and number at the late stage of growth. This finding indicates that the OsSL5 gene could control cell apoptosis. The plant height, spike length, primary branch number, and secondary branch number of the sl5 mutant were significantly lower than those of the wild type. The most significant observation in this study is the decrease in the secondary branch number. Spotted leaf formation and changes in several parameters were complicated because of various factors. These factors include the interactions and relationships between genes and the single or multiple pathways involved in signal transduction. The OsSL5 gene is an allelic SPL5 gene (Chen et al. 2012). However, the severity of the spots in the sl5 mutant was less than that in spl5. This condition may be attributed to the A-to-G single base substitution in the OsSL5 gene encoding region, which slightly affected the amino acid sequence. The SPL5 encoding region lacks the base G, and this insufficiency terminates protein translation at early stages. Therefore, SPL5 may participate in cell apoptosis regulation. QPCR results show that the PR1 gene (Yin et al. 2000) is highly expressed in spl5 and spl1 (sl) mutants. The PR1 gene induces and activates PBZ1 protein expression in the nucleus after rice is infected with rice blast. This report indicates that the OsSL5 gene induces rice blast resistance. Studies have also shown that SPL5 regulates precursor mRNA splicing, amino acid metabolism, photosynthesis, glycolysis, ROS metabolism, and stress resistance (Chen et al. 2013). However, the specific regulatory mechanism of SPL5 splicing remains unclear. The sl5 is a new weak allelic mutant in the background Japonica rice of the SPL5 gene from indica rice (Chen et al. 2012). Further studies on mutants in different subspecies backgrounds will elucidate the molecular mechanism of rice spl formation.
Conclusion
A spotted leaf mutant sl5, which exhibits more spots at maturity, was successfully isolated in this study. Genetic analysis shows that the mutant is controlled by a single recessive gene, and the allelic test shows that the OsSL5 gene is allelic to SPL5. The OsSL5 gene was determined to be anchored on chromosome 7 through map-based cloning, and the mapping interval was narrowed to 40 kb by using the most recently designed markers. The candidate genes were amplified, and the sequences were aligned with that of the wild type. The OsSL5 gene was finally cloned at a mutation site different from that of the SPL5 mutant gene. The QPCR analysis of the related spotted-leaf genes in the OsSL5 mutant plant reveals complex connection between these genes.
References
Babu R, Jiang CJ, Xu X, Kottapalli KR, Takatsuji H, Miyao A, Hirochika H, Kawasaki S (2011) Isolation, fine mapping and expression profiling of a lesion mimic genotype, spl (NF4050-8) that confers blast resistance in rice. Theor Appl Genet 122(4):831–854. doi:10.1007/s00122-010-1490-7
Chen X, Hao L, Pan J, Zheng X, Jiang G, Jin Y, Gu Z, Qian Q, Zhai W, Ma B (2012) SPL5, a cell death and defense-related gene, encodes a putative splicing factor 3b subunit 3 (SF3b3) in rice. Mol Breed 30(2):939–949
Chen X, Fu S, Zhang P, Gu Z, Liu J, Qian Q, Ma B (2013) Proteomic analysis of a disease-resistance-enhanced lesion mimic mutant spotted leaf 5 in rice. Rice 6(1):1. doi:10.1186/1939-8433-6-1
Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL (1994) Arabidopsis mutants simulating disease resistance response. Cell 77(4):565–577
Feng B, Yang Y, Shi Y, Shen H, Wang H, Huang Q, Xu X, Lv X, Wu J (2013) Genetic analysis and gene mapping of light brown spotted leaf mutant in rice. Rice Sci 20(1):13–18
Fujiwara T, Maisonneuve S, Isshiki M, Mizutani M, Chen L, Wong HL, Kawasaki T, Shimamoto K (2010) Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J Biol Chem 285(15):11308–11313. doi:10.1074/jbc.M109.091371
Gray J, Close PS, Briggs SP, Johal GS (1997) A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 89(1):25–31
Habara Y, Urushiyama S, Shibuya T, Ohshima Y, Tani T (2001) Mutation in the prp12+ gene encoding a homolog of SAP130/SF3b130 causes differential inhibition of pre-mRNA splicing and arrest of cell-cycle progression in Schizosaccharomyces pombe. RNA 7(5):671–681
Hu G, Yalpani N, Briggs SP, Johal GS (1998) A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 10(7):1095–1105. doi:10.1105/tpc.10.7.1095
Jincho Y, Sotomaru Y, Kawahara M, Ono Y, Ogawa H, Obata Y, Kono T (2008) Identification of genes aberrantly expressed in mouse embryonic stem cell-cloned blastocysts. Biol Reprod 78(4):568–576. doi:10.1095/biolreprod.107.064634
Johal GS, Hulbert SH, Briggs SP (1995) Disease lesion mimics of maize: a model for cell death in plants. BioEssays 17(8):685–692
Kerins JA, Hanazawa M, Dorsett M, Schedl T (2010) PRP-17 and the pre-mRNA splicing pathway are preferentially required for the proliferation versus meiotic development decision and germline sex determination in Caenorhabditis elegans. Dev Dyn 239(5):1555–1572
Kojo K, Yaeno T, Kusumi K, Matsumura H, Fujisawa S, Terauchi R, Iba K (2006) Regulatory mechanisms of ROI generation are affected by rice spl mutations. Plant Cell Physiol 47(8):1035–1044. doi:10.1093/pcp/pcj074
Li X, Wang P, Qu Z, Sun X, Wang B, Deng X (2010) Genetic analysis and fine mapping of a lesion mimic mutant C23 in rice. Sci Agric Sin 43:3691–3697
Liu D, Cheng Z, Liu G, Liu G, Wang B, Zhao X, Zhu L (2003) Identification and gene mapping of a rice lesion mimic mutant (lmi). Chin Sci Bull 48:831–835
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Ma JY, Chen SL, Zhang JH, Dong YJ, Teng S (2012) Identification and genetic mapping of a lesion mimic mutant in rice. Rice Sci 19(1):1–7
Mach JM, Castillo AR, Hoogstraten R, Greenberg JT (2001) The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci USA 98(2):771–776. doi:10.1073/pnas.021465298
Matin MN, Saief SA, Rahman MM, Lee DH, Kang H, Lee DS, Kang SG (2010) Comparative phenotypic and physiological characteristics of spotted leaf 6 (spl6) and brown leaf spot2 (bl2) lesion mimic mutants (LMM) in rice. Mol Cells 30(6):533–543. doi:10.1007/s10059-010-0151-7
Menon S, Tsuge T, Dohmae N, Takio K, Wei N (2008) Association of SAP130/SF3b-3 with Cullin-RING ubiquitin ligase complexes and its regulation by the COP9 signalosome. BMC Biochem 9:1. doi:10.1186/1471-2091-9-1
Mizobuchi R, Hirabayashi H, Kaji R, Nishizawa Y, Yoshimura A, Satoh H, Ogawa T, Okamoto M (2002) Isolation and characterization of rice lesion-mimic mutants with enhanced resistance to rice blast and bacterial blight. Plant Sci 163(2):345–353
Mori M, Tomita C, Sugimoto K, Hasegawa M, Hayashi N, Dubouzet JG, Ochiai H, Sekimoto H, Hirochika H, Kikuchi S (2007) Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol Biol 63(6):847–860. doi:10.1007/s11103-006-9130-y
Qiao Y, Jiang W, Lee J, Park B, Choi MS, Piao R, Woo MO, Roh JH, Han L, Paek NC, Seo HS, Koh HJ (2010) SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol 185(1):258–274. doi:10.1111/j.1469-8137.2009.03047.x
Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 5(2):69–76
Sun C, Liu L, Tang J, Lin A, Zhang F, Fang J, Zhang G, Chu C (2011) RLIN1, encoding a putative coproporphyrinogen III oxidase, is involved in lesion initiation in rice. J Genet Genomics 38(1):29–37. doi:10.1016/j.jcg.2010.12.001
Takahashi A, Agrawal GK, Yamazaki M, Onosato K, Miyao A, Kawasaki T, Shimamoto K, Hirochika H (2007) Rice Pti1a negatively regulates RAR1-dependent defense responses. Plant Cell 19(9):2940–2951. doi:10.1105/tpc.106.047142
Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11(6):1187–1194
Wang L, Pei Z, Tian Y, He C (2005) OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Mol Plant Microbe Interact 18(5):375–384. doi:10.1094/MPMI-18-0375
Wolter M, Hollricher K, Salamini F, Schulze-Lefert P (1993) The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol Gen Genet 239(1–2):122–128
Wu C, Bordeos A, Madamba MR, Baraoidan M, Ramos M, Wang GL, Leach JE, Leung H (2008) Rice lesion mimic mutants with enhanced resistance to diseases. Mol Genet Genomics 279(6):605–619. doi:10.1007/s00438-008-0337-2
Yamanouchi U, Yano M, Lin H, Ashikari M, Yamada K (2002) A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc Natl Acad Sci USA 99(11):7530–7535. doi:10.1073/pnas.112209199
Yamasaki S, Ishikawa E, Sakuma M, Hara H, Ogata K, Saito T (2008) Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat Immunol 9(10):1179–1188. doi:10.1038/ni.1651
Yin ZC, Chen J, Zeng LR, Goh ML, Leung H, Khush GS, Wang GL (2000) Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol Plant Microbe Interact 13(8):869–876. doi:10.1094/MPMI.2000.13.8.869
Yoshimura A, Ideta O, Iwata N (1997) Linkage map of phenotype and RFLP markers in rice. Plant Mol Biol 35(1–2):49–60
Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16(10):2795–2808. doi:10.1105/tpc.104.025171
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31171535; 31101135), Zhejiang Provincial Science and Technology Bureau (2012C22039), and Hangzhou Scientific and Technological Program (20130432B04). The authors are grateful to the editors and the anonymous reviewers for their valuable comments and help.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Chang-wei Ge and Zhi-guo E have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Ge, Cw., E, Zg., Pan, Jj. et al. Map-based cloning of a spotted-leaf mutant gene OsSL5 in Japonica rice. Plant Growth Regul 75, 595–603 (2015). https://doi.org/10.1007/s10725-014-9962-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-014-9962-4