Abstract
Seed yield is influenced by the combined effects of genes, including additive and non-additive interactions. Therefore, accurately predicting seed yield holds significant importance in rapeseed breeding. Nonetheless, limited information exists regarding yield estimation for canola using neural networks. This study employs multi-layer perceptron (MLP) neural network, radial basis function neural network and support vector machine, to forecast rapeseed yield. The models are trained using phenological, morphological, yield and yield-related data, as well as molecular marker information from 8 genotypes and 56 hybrids. Comparative analysis of the models reveals that the MLP model effectively forecasts hybrid yield with root mean square error (RMSE), mean absolute error (MAE) and coefficient of determination (R2) values of 226, 183, and 92%, respectively. Among the 40 primers examined, the ISJ10 primer demonstrates superior discriminatory power compared to others. The use of molecular and phenotypic data as inputs in the model highlights the MLP model’s superiority, presenting lower RMSE and MAE values, along with a higher R2, compared to direct crosses in predicting the performance of reciprocal crosses. The proposed neural network model enables performance estimation of hybrids prior to crossing parent studied, thereby enabling spring rapeseed breeders to focus on the most promising hybrids.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Background
Rapeseed (Brassica napus L.) stands as a pivotal oilseed plant cultivated across diverse regions like Europe, Canada, Australia and Iran, owing to its substantial genetic diversity (FAO 2020). The primary goal of rapeseed research involves expanding germplasm diversity to attain elevated yields. However, most critical agronomic traits of canola are limited. Quantitative traits are governed by numerous minor alleles, complicating the identification of chromosomal allele locations and their relative contributions to quantitative trait manifestation and phenotypic distribution (Sabouri et al. 2012; Liu et al. 2022). Genetic marker-based breeding has enabled the identification of quantitative trait alleles and the creation of genetic maps. Molecular marker-based breeding technology is a suitable and useful method due to the ease of improving the expansion of genetic diversity and the absence of time limits in rapeseed cultivation (Ton et al. 2020; Chugh et al. 2023; Singh et al. 2022). Genetic markers include morphological, cytogenetic, biochemical and molecular markers, with DNA-level polymorphic markers being especially important. Markers such as RAPD, SSR, AFLP, and ISSR are extensively employed for locating genes linked to polygenic and monogenic traits (Suping et al. 2021; Dolatabadian et al. 2022). Several investigations have explored genetic diversity within rapeseed germplasm utilizing identical and non-identical markers (Chai et al. 2019; Singh et al. 2017; Jesske et al. 2013). For instance, Motallebinia et al. (2019), assessed genetic diversity in 12 canola genotypes using 18 ISSR markers, identifying 60 polymorphic bands out of 106 amplified bands. Similarly, Safari and Mehrabi (2017), reported 100% polymorphism across 45 canola genotypes through 12 RAPD markers. Masoudi et al. (2017) appraised 60 wheat genotypes using three markers IPBS, ISSR and IRAP, observed 47 polymorphic bands out of 61 amplified bands, which is the highest and lowest percentage of polymorphism related to ISSR and IPBS primers.
Artificial neural networks (ANN) are modern computational methods for machine learning to predict responses to complex problems, partly inspired by the way the biological nervous system functions to process data and information. An ANN is a set of computational elements called neurons that function similarly to biological neurons. These networks are capable of learning and correcting their errors. Learning in these systems is done adaptively, ‘i.e.,’ using examples ‘the weight of synapses changes so that the system produces a correct response if new inputs are given. The characteristics of ANN include the ability to train the versatility of dispersion, capability information, generalization of parallel processing, robustness, and general modeling of physical processes, which deductive and inductive methods can do. The basis of the deductive method is based on mathematical theories and formulas’ in other words, modeling is done by relationships and constant coefficients of experiment (Kasabov 2019). However, ANN based modeling methods have to be more useful and flexible in dealing with possible non-linear relationships than linear regression (Jamshidi et al. 2016; Niazian and Niedbała 2020). The potential of molecular and phenotypic data in predicting crop yield has been harnessed through various ANN models (Wojciechowski et al. 2016; Sharma and Singh 2017; Torkashvand et al. 2017). Neural networks, offer great flexibility in precise access to pre-harvest yield prediction, gaining traction in genetic research (Ma et al. 2018; Singh et al. 2016; Gholipoor and Nadali 2019; Wang et al. 2019). The synergy of quantitative and qualitative data within network models yields enhanced predictions, that in the context of rapeseed yield, both types of data have been utilized (Zhang et al. 2020; Wawrzyniak et al. 2020).
There are different types of ANNs such as radial basis function (RBF) and multilayer perceptron (MLP) (Araghinejad et al. 2017), that have no dependency on any previous knowledge regarding the construction or inter-relationships between input and output signals. Therefore, the usage of these kinds of models such as ANN would be useful in modeling and optimizing in plant genetics such as, tissue culture (Jamshidi et al. 2016; Eren et al. 2023; Aasim et al. 2022) and molecular markers (Sandhu et al. 2021). An ANN model with MLP architecture predicted rapeseed yield based on meteorological (temperature and precipitation) and fertilization data, demonstrating lower MAPE errors values with the 15:15-18-11-1:1 structure (Niedbała 2019).
Support vector machine (SVM) is a learning system used both for classifying input data and estimating the data fit function so that the least error occurs in the data classification and regression. The data is divided into three categories: training, validation, and test so that training data causes SVM training, validation data is used to calibrate the parameters of the machine, and finally, this machine is used to classify or estimate test data. This method is based on constraint optimization theory that uses the principle of minimization of structural error and leads to a solution with overall optimum returns (Campbell and Ying 2011; Hesami and Jones 2020). Among these models, the SVM emerges as a widely adopted machine learning algorithm, adeptly addressing both classification and regression tasks (Noble 2006).
The integration of machine learning-based techniques into breeding introduces a novel avenue, promising accurate prediction of rapeseed hybrid performance. This paper aims to predict rapeseed yield using phenotypic and molecular data, employing diverse machine learning models. These trained models reduce the need for resource-intensive experiments, marking a significant advancement in rapeseed breeding research.
Materials and methods
During the 2017–2018 crop year, a Diallel genetic design was employed to cross eight parents (refer to Table 1) at the Gorgan City Natural and Agricultural Resources Research Station. Subsequently, in the autumn of 2018, a total of 8 parents and 56 hybrid offspring were cultivated in the research field using a randomized complete block design (RCBD) with three replicates. These plants were sourced from Dr. Payghamzadeh at the Gene Bank of Horticultural Products Research Institute, Golestan Agriculture and Natural Resources Research and Education Center, under the Agricultural Research, Education and Extension Organization (AREEO). The plant specimens, identified as voucher IDs (SPN-202, SPN-204, SPN-206, SPN-207, SPN-217, SPN-225, SPN-227, SPN-182) are accessible for study and verification at the Herbarium of the Research Institute (AREEO).
Molecular analysis
The researchers acquired seeds from eight distinct rapeseed genotypes originating from the Gorgan Agriculture and Natural Resources Research Station. Subsequently, the researchers planted 15–20 seeds from each genotype, including parents and hybrids within small pots in the greenhouse at Gonbad-Kavous University. To extract DNA from every genotype, plant samples were harvested at the three-leaf stage, pulverized with liquid nitrogen and then preserved in a − 20 °C freezer. For DNA extraction, the CTAB method as described by (Saghi Maroof et al. 1994) was employed on leaf samples, and the of the extracted DNA was assessed through 0.8% agarose gel electrophoresis. To explore the genomes of the studied genotypes, the researchers employed a set of 40 primers (refer to Table 2) capable of reproducing the genome of the genotypes studied. The PCR products were subsequently separated through electrophoresis, utilizing a 1.5% agarose gel, and the resulting gel was visualized under UV light.
Yield prediction utilizing MLP neural network
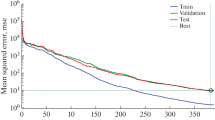
Neural networks exhibit capabilities encompassing classification, prediction and clustering. The training process involves increasing and decreasing the weight coefficients of input nodes. These networks generally comprise fundamental neural units forming an input layer, one or more hidden layers, and an output layer. The input signal propagates through the network in a direct layer-by-layer path, often referred to as the MLP architecture. The structure of a multilayer neural network is depicted in Fig. 1.
Yield prediction utilizing RBF neural network
The radial basis function (RBF) network is a type of ANN where each unit generates an output vector upon receiving input. Training this network employs the backpropagation training algorithm with a diminishing learning rate (BDLRF). This algorithm’s advantages encompass parameter adjustment the ease, reduced the learning time and enhanced network behavior depiction learning. The schematic depiction of a three-layer RBF network, comprising input, output and hidden layers, is presented in Fig. 2.
Yield prediction utilizing SVM model
Support vector machine stands as a supervised learning approach utilized for classification and regression tasks. Support vectors, a set of points in the data’s 1D space, establish category boundaries, effectively segmenting and categorizing data as displayed in Fig. 3. This algorithm aims to find a boundary between categories so that maximally distances itself from support vectors of each category. This method’s essence lies in processing data through kernel mathematical functions, mapping it into a new space, for analyzing complex, nonlinearly structured separated data. Various kernel functions are available, including linear, polynomial, cyclic, and radial, each producing distinct results upon function selection.
Input data for models
Trait assessment involved recording traits such as days to flower initiation from emergence date, days to flower termination from emergence date, flowering duration, physiological maturing, plant height (cm), no. lateral branches, branching height (cm), podding height (cm), main stem length (cm), pod length (cm), and stem diameter (mm). Additionally, no. pods per main stem, no. pod per lateral branches, no. pods per plant, no. grain per pod, 1000 grain weight (g), and yield (kg. ha−1) were noted for both direct and reciprocal crossings after the cultivation period. From each plot, five randomly chosen plants from the two central rows were tagged before flowering (BBCH: 32) and harvested at maturity (BBCH: 99) (Meier et al. 2009), to collect data on traits. Moreover, the entire plot was harvested for obtaining grain yield per plot. Detailed data collection procedures are outlined in Table 3. Genetic factors also played a role in this research, with genetic data from 40 markers in Table 2 being employed. Alleles were categorized as zero (absence of band) and one (presence of band).
Evaluation of model performance
When predicting the yield of rapeseed hybrids, a combination of phenological, morphological, and yield-related traits, along with seed yield components, and parental molecular data, were concurrently employed to train the models. The quantity and distribution of the training data are pivotal factors influencing prediction accuracy (Duan et al. 2015). The dataset was randomly divided into two segments: training and testing. Specifically, 80% of the data was allocated for training, while the remaining 20% was designated for model testing. Based on this method, a data is randomly selected from the data set so that each cultivar had an equal probability of being selected during the data sampling process (Yates et al. 2008). Additionally, validation involved comparing model-predicted values with actual values obtained from phenotypic and molecular data. To assess the models’ performance in predicting hybrid performance, the statistical criteria such as MAE, RMSE, and R2 were employed (Zhang et al. 2020). Correlation coefficient square (R2) is a measure that describes how closely the values of measurement and simulation are correlated (Eq. 1). In other words, when the measured values increase, the predicted values increase, or vice versa. The values of R2 are between zero and one, and the closer this value is to one, the more the values of measurement and prediction correlations are more than each other, and vice versa.
Mean square error (MSE) is a statistical scale of the difference between the objective values of the observational dataset and the predicted output values through the model (Eq. 2). It is the mean of all squares between the prediction and actual values. Error-values are squared to represent the effect of large error values better and, on the other hand, to remove the effect of the positive and negative values caused by subtraction. Root mean square error (RMSE) is the root of the MSE metric (Eq. 3).
In these equations, \({y}_{i} and \overline{{y }_{i}}\) are predicted value and actual value, \({y}_{ave}\). The average of data set values, and n is the number of observations.
The MATLAB 2018b software was employed to establish and train the neural network within the programming environment (Sajid et al. 2022).
Results
The analysis of the studied genotypes’ averages (Table 4) indicates that the estimated average yield for parents was 1975.17 (kg. ha−1), with the highest parent yield reaching 2853.49 (kg. ha−1) and the lowest at 1433.48 (kg. ha−1). Similarly, the hybrid yields revealed that the average yield of reciprocal crosses (2025.90 kg. ha−1) exceeded that of direct crosses (1974.12 kg. ha−1). The maximum seed yield observed among direct crosses and the reciprocal crosses was 3002.65 and 2969.07 (kg. ha−1), respectively. Conversely, the lowest yield recorded within the reciprocal crosses was 1237.67 (kg. ha−1).
Molecular assessment
As evidenced by the data presented in Table 5, the employed primers yielded distinct and marked banding patterns. Among the 40 primers examined, the distribution consisted of one primer from the CAAT tag, twelve from the IPBS tag, eleven from the ISSR tag, seven from the ISJ tag, and nine from the SCoT tag. Evaluation of the genotypes resulted in the discovery of a total of 196 alleles, averaging 4.90 alleles per marker. Notably, 114 alleles were documented, with an average of 2.85 alleles per marker. The average proportion of polymorphism across all primers was computed at 58.16%. Significant percentages of polymorphism were observed among the IPBS, ISSR, ISJ, and SCoT markers. Specifically, the primers IPBS15 (80%), ISSR58 (100%), ISJ10 (100%), and SCoT1 and SCoT9 (both 80%) displayed notable polymorphism levels. The mean polymorphic information content (PIC) value attributed to the primers was calculated at 0.34. Among all markers, the SCoT9 primer stood out with the highest PIC value of 0.50, while the primers CAAT28, IPBS5, IPBS8, and ISSR47 registered the lowest values at 0. In evaluating the efficiency of primers in determining polymorphism, the Shannon index (I) serves as an important parameter. The primers ISJ10 and SCoT1 exhibited higher values of the Shannon index (I) compared to other markers. The overall mean value of the Shannon index (I) was calculated to be 0.29. Examination of the Nei genetic diversity index (H) indicated diversity values ranging from 0.39 to 0 across markers. Among the primers, ISJ10 (0.39), CAAT28, IPBS5, IPBS8, and ISSR47 (0) recorded the highest and lowest Nei genetic diversity values, respectively. The comprehensive average value of Nei genetic diversity was estimated at 0.19. Furthermore, the analysis of effective alleles (Ne) revealed that the ISJ10 primer had the highest effective allele value at 1.72, while primers CAAT28, IPBS5, IPBS8, and ISSR47 had the lowest number of effective alleles.
Assessment of MLP, RBF and SVM model performance
The construction and training of the MLP, RBF, and SVM models were carried out utilizing the fitrnet, newrb, and fitrsvm functions embedded within the MATLAB software. Outcomes of machine learning models employing distinct data partitioning approaches are outlined in Table 6. We explored various data splitting ratios, including 90–10, 80–20, 70–30, and 60–40, with the optimal performance observed under the 80–20 ratio. Examining the minimum, maximum, and average values across these data partitioning ratios revealed that no significant differences exist among the mentioned data partitioning ratios.
Assessment of MLP, RBF, and SVM model effectiveness based on 80–20 ratio for predicting hybrid performance using various criteria
The illustration of the effectiveness of the MLP, RBF, and SVM models in forecasting hybrid performance, as evaluated against the MAE criterion and utilizing an 80–20 ratio, is depicted in Fig. 4. Among models trained exclusively with genetic traits in direct crosses, the MLP model displayed a notably lower error compared to the other models. Conversely, in models trained with genetic traits in reciprocal crosses, the MAE errors were closely aligned. When utilizing phenotypic traits, both the MLP and RBF models trained for direct crosses demonstrated similar and improved MAE errors compared to the SVM model. Remarkably, for the SVM model trained with phenotypic traits, the lowest MAE error was observed in the context of reciprocal crosses as indicated in Fig. 4.
Comparison of multilayer perceptron (MLP), radial basis function (RBF), and support vector machine (SVM) model performance in predicting hybrid yield using MAE criterion. G1: Model trained using genetic traits in direct crosses; G2: Model trained using genetic traits in reciprocal crosses; P1: Model trained using phenotypic traits in direct crosses; P2: Model trained using phenotypic traits in reciprocal crosses; PG1: Model trained using phenotypic and genetic traits in direct crosses; PG2: Model trained using phenotypic and genetic traits in reciprocal crosses
Further examination of the MLP, RBF and SVM models’ effectiveness in predicting hybrid performance, evaluated through the RMSE criterion, is visualized in Fig. 5. Models trained with genetic traits in both direct and reciprocal crosses yielded RMSE errors closely clustered. Notably, the RMSE error was comparatively lower in reciprocal crosses than in direct crosses. The utilization of phenotypic traits led to a decreased RMSE error, particularly for the forward intersections of the RBF model and the reciprocal crosses of the SVM model. The incorporation of phenotypic and genetic traits, yielded models with decreased RMSE errors in both the direct crosses of the RBF model and the reciprocal crosses of the SVM model.
Comparison of multilayer perceptron (MLP), radial basis function (RBF), and support vector machine (SVM) model performance in predicting hybrid yield using root mean square error (RMSE) criterion. G1: Model trained using genetic traits in direct crosses; G2: Model trained using genetic traits in reciprocal crosses; P1: Model trained using phenotypic traits in direct crosses; P2: Model trained using phenotypic traits in reciprocal crosses; PG1: Model trained using phenotypic and genetic traits in direct crosses; PG2: Model trained using phenotypic and genetic traits in reciprocal crosses
The assessment of MLP, RBF, and SVM models’ efficacy in predicting hybrid performance based on the R2 criterion is portrayed in Fig. 6. Across diverse datasets and model training inputs, models trained with genetic traits in direct crosses exhibited the lowest R2 values. For models trained using genetic traits in reciprocal crosses, their R2 values closely aligned. In contrast, models utilizing phenotypic traits in both direct and reciprocal crosses showcased a superior R2 value for the MLP model compared to the other models. Incorporating both phenotypic and genetic traits resulted in models with comparable R2 values for both direct and reciprocal crosses.
Comparison of multilayer perceptron (MLP), radial basis function (RBF), and support vector machine (SVM) model performance in predicting hybrid yield using coefficient of determination (R2) criterion. G1: Model trained using genetic traits in direct crosses; G2: Model trained using genetic traits in reciprocal crosses; P1: Model trained using phenotypic traits in direct crosses; P2: Model trained using phenotypic traits in reciprocal crosses; PG1: Model trained using phenotypic and genetic traits in direct crosses; PG2: Model trained using phenotypic and genetic traits in reciprocal crosses
The ultimate configuration details for each machine learning model–MLP, RBF, and SVM–are elucidated in Table 7.
Discussion
Markers efficiency
The comparison of markers in terms of their discriminating power relies on crucial parameters such as polymorphic information content (PIC) and Nei index. Higher values of these parameters indicate heightened polymorphism, the presence of alleles or rare alleles within a marker band, and a marker’s proficiency in differentiation (Badirdast et al. 2018). Our exploration aimed to evaluate the performance and efficiency of markers to assess the extent of diversity among rapeseed parents. The outcomes unveiled that the mean percentage of total primer polymorphism and average PIC value of the primers were 58.16 and 0.34%, respectively, indicating the capacity to discern and characterize genetic diversity among the 8 canola parents. In a study by Motallebinia et al. (2019) involving canola and ISSR markers, polymorphic information content values ranged from 0.36 to 0.08. Furthermore, the highest effective allele (Ne) value was observed for ISJ10 primer with a value of 1.72, while the primers CAAT28, IPBS5, IPBS8, and ISSR47 displayed the lowest number of effective alleles. The discrepancy between the total alleles and effective alleles signifies the presence of rare alleles found in only a few genotypes, which can be exploited for identification purposes. A proper distribution of markers throughout the genome, achieved by selecting markers from different genome regions, enhances the accuracy of molecular diversity measurement due to a more comprehensive representation of the entire genome (Yeken et al. 2022; Tiwari et al. 2022; Pour-Aboughadareh et al. 2022; Heikal et al. 2022). Thus, our findings are consistent with previous research, indicating that the markers studied here exhibit a diversified distribution within the genome similar to SCoT and ISSR markers (Badirdast et al. 2021; Khodadadi et al. 2021; Shah-Ghobadi et al. 2018), underlining the genetic diversity across the parents.
Model performance
The assessment of model performance reveals that, in terms of RMSE, the MLP, RBF, and SVM models results yielded within the ranges of [207,405], [175,367], and [168,374], respectively (Fig. 4). Concerning MAE, the models exhibited values spanning [182, 322], [147,309], and [141,296], respectively (Fig. 5). In the context of R2, the model performance ranged from [0.64, 0.92], [0.63, 0.89], to [0.55, 0.89], respectively (Fig. 6). Evaluating models trained based on genetic traits in direct crosses unveiled that none of the MLP, RBF, or SVM models surpassed an accuracy of 65% (R2) in predicting hybrid performance. However, in reciprocal crosses, all three models exhibited an accuracy of 89% (R2). Turning to models trained using phenotypic traits, the MLP model demonstrated superior predictive capabilities in both direct and reciprocal crosses, with an accuracy of 89% and 92%, respectively. Furthermore, models trained with both phenotypic and genetic traits exhibited comparable accuracy across at the three models, with the highest values reaching 77% in direct crosses and 89% in reciprocal crosses. While the application of artificial intelligence-based methods for phenotypic and genetic prediction remains limited, the significance of neural networks in genetic enhancement has been underscored by previous research. Marini et al. (2004) successfully predicted corn and soybean yields based on environmental climatic conditions using neural networks, achieving explanatory coefficients of 0.77 for corn and 0.81 for soybean. Similarly, Rosado et al. (2020) demonstrated that employing ANN-MLP neural networks for bean genetic prediction, incorporating phenotypic and genetic traits, led to a 90% increase in model accuracy. This approach capitalizes on quantitative features to improve prediction accuracy. The results across various crop plants further endorse the efficiency of neural networks for crop performance prediction (Eren et al. 2023; Shamsabadi et al. 2022; Hara et al. 2023; Huang 2023).
Conclusion
The recent years utilation of artificial intelligence in analysis, modeling, and forecasting has gained prominence. This study harnessed diverse algorithms with distinct structures to predict rapeseed hybrid performance. Utilizing both molecular and phenotypic data inputs in the model revealed that the MLP model exhibited reduced RMSE and MAE values and a heightened R2 in predicting reciprocal crosses, outperforming direct crosses. Training the MLP model based on molecular and phenotypic data yielded a R2 of up to 89%, highlighting its capability to approximate real data more accurately. The proposed neural network model empowers breeders to predict hybrid performance of parent combinations prior to crossing, streamlining efforts towards optimal hybrid outcomes. The remarkable versatility of neural networks has spurred advancements in learning and predictive models using both phenotypic and molecular data enabling comprehensive exploration of various plant traits. Further research is warranted to explore the potential of other machine learning models.
Availability of data and material
The data set used or analyzed during the current study is available to the corresponding author and will be made available upon reasonable request.
Abbreviations
- MLP:
-
Multi-layer perceptron
- MAPE:
-
Mean absolute percentage error
- CTAB:
-
Cetyltrimethylammonium bromide
- PCR:
-
Polymerase chain reaction
- RBF:
-
Radial basis function
- MAE:
-
Mean absolute error
- RMSE:
-
Root mean square error
- ANN:
-
Artificial neural network
References
Aasim A, Katırcı R, Akgur O, Yildirim B, Mustafa Z, Azhar Nadeem M, Shahzad Baloch F, Karakoy T, Yılmaz G (2022) Machine learning (ML) algorithms and artificial neural network for optimizing in vitro germination and growth indices of industrial hemp (Cannabis sativa L.). Ind Crop Prods 181:114801
Araghinejad S, Hosseini-Moghari SM, Eslamian S (2017) Application of data-driven models in drought forecasting. In: Eslamian S (ed) Handbook of drought and water scarcity. CRC Press, New York, pp 423–440
Badirdast H, Salehi-Lisar SY, Sabouri H, Movafeghi A, Gholamalalipour Alamdari E (2018) Identification of informative alleles controlling rice traits under flooding and drought stress conditions. Plant Genet Res 5:39–54
Badirdast H, Salehi-Lisar SY, Movafeghi A, Gholamalalipour Alamdari E (2021) Identification of ISSR, IRAP and iPBS markers containing information on rice characteristics under two conditions of flooding and drought stress. Cell Mol Res 34:34–44
Campbell C, Ying Y (2011) Learning with support vector machines. Springer Nature Switzerland AG 2011, Springer Cham. https://doi.org/10.1007/978-3-031-01552-6
Chai L, Li H, Zhang J, Wu L, Zheng B, Cui C, Jiang L (2019) Rapid identification of a genomic region conferring dwarfism in rapeseed (Brassica napus L.) YA2016-12. J Agron 9:129–143
Chugh V, Kaur D, Purwar S, Kaushik P, Sharma V, Kumar H, Rai A, Singh CM, Kamaluddin Dubey RB (2023) Applications of molecular markers for developing abiotic-stress-resilient oilseed crops. Life 13:88
Dolatabadian A, Cornelsen J, Huang Sh, Zou Z, Fernando WD (2022) Sustainability on the farm: breeding for resistance and management of major canola diseases in Canada contributing towards an IPM approach. Can J Plant Pathol 44:157–190
Duan H, Tan F, Yi X, Zhang H, Hou M, Dan Moghan JEM (2015) A predictive model of different growth of escherichia coli in freshcut lettuce based on MATLAB 7.0. In: 2nd International Conference on Civil, Materials and Environmental Sciences, Atlantis Press, pp 114–118
Eren B, Türkoğlu A, Haliloğlu K, Demirel F, Nowosad K, Özkan G, Niedbała G, Pour-Aboughadareh A, Bujak H, Bocianowski J (2023) Investigation of the influence of polyamines on mature embryo culture and DNA methylation of wheat (Triticum aestivum L.) using the machine learning algorithm method. Plants 12(18):3261
FAO. STAT (2020) Food and Agriculture Organization of the United Nations. Database crops production. https://www.fao.org/faostat/en/#data/QC
Gholipoor M, Nadali F (2019) Fruit yield prediction of pepper using artificial neural network. Sci Hortic 250:249–253
Hara P, Piekutowska M, Niedbała G (2023) Prediction of pea (Pisum sativum L.) seeds yield using artificial neural networks. Agric 13:661
Heikal YM, El-Esawi MA, Galilah DA (2022) Morpho-anatomical, biochemical and molecular genetic responses of canola (Brassica napus L.) to sulphur application. Environ Exp Bot 194:104739
Hesami M, Jones AMP (2020) Application of artificial intelligence models and optimization algorithms in plant cell and tissue culture. Appl Microbiol Biotechnol 104:9449–9485
Huang Y (2023) Improved SVM-based soil-moisture-content prediction model for tea plantation. Plants 12:2309
Jamshidi S, Yadollahi A, Ahmadi H, Arab M, Eftekhari M (2016) Prediction in vitro culture medium macro-nutrients composition for pear rootstocks using regression analysis and neural network models. Front Plant Sci 7:274
Jesske T, Olberg B, Schierholt A, Becker HC (2013) Resynthesized lines from domesticated and wild Brassica taxa and their hybrids with (B. napus L). genetic diversity and hybrid yield. Theor Appl Genet 126:1053–1065
Kasabov NK (2019) Time-space, spiking neural networks and brain-inspired artificial intelligence. Springer, Berlin
Khodadadi S, Dashti H, Saberi R, Malekzadeh K, Tajabadi pour A (2021) Genetic diversity of pistachio cultivars and genotypes in terms of resistance to crown and root rot (Phytophthora drechsleri) and its relationship with SCoT molecular markers. J Mod Genet 16:235–248
Liu S, Raman H, Xiang Y, Zhao C, Huang J, Zhang Y (2022) De novo design of future rapeseed crops: challenges and opportunities. Crop J 10:587–596
Ma W, Qiu Z, Song J, Li J, Cheng Q, Zhai J, Ma C (2018) A deep convolutional neural network approach for predicting phenotypes from genotypes. Planta 248:1307–1318
Marini F, Zupan J, Magrì AL (2004) On the use of counter propagation artificial neural networks to characterize Italian rice varieties. Anal Chim Acta 510:231–240
Masoudi H, Sabouri H, Taliey F, Jafarby J (2017) Genetic diversity and association analysis for morphophenolgic traits and resistance to powdery mildew using ISSR, IRAP and IPBS markers. Crop Biotech 7:41–56
Meier U, Bleiholder H, Buhr L, Feller C, Hack H, Heß MD, Lancashire P, Schnock U, StauB R, Boom Th, Weber E, Zwerger P (2009) The BBCH system to coding the phenological growth stages of plants–history and publications. J Kulturpflanzen 61:41–52
Motallebinia S, Sofalian O, Asghari A, Rasoulzadeh A, Fathi B (2019) Study of drought tolerance indices and their relationship with ISSR markers in some canola (Brassica napus L.) cultivars. Plant Genet Res 6:99–114
Niazian M, Niedbała G (2020) Machine learning for plant breeding and biotechnology. Agriculture 10:436
Niedbała G (2019) Application of artificial neural networks for multi-criteria yield prediction of winter rapeseed. Sustainability 11:533
Noble WS (2006) What is a support vector machine? Nat Biotechnol 24:1565–1567
Pour-Aboughadareh A, Poczai P, Etminan A, Jadidi O, Kianersi F, Shooshtari L (2022) An analysis of genetic variability and population structure in wheat germplasm using microsatellite and gene-based markers. Plants 11:1205
Rosado RDS, Cruz CD, Barili LD, Souza Carneiro JE, Carneiro PCS, Carneiro VQ, Silva JT, Nascimento M (2020) Artificial neural networks in the prediction of genetic merit to flowering traits in bean cultivars. Agric 10:638
Sabouri H, Navabpour M, Mohammad E (2012) Determination of genetic structure of agronomic rice traits using classical and molecular approach. J Plant Product 18:45–72
Safari S, Mehrabi A (2017) Genetic relationships of rapeseed cultivars revealed by RAPD markers. J Crop Breed 8:170–177
Saghi Maroof MA, Biyaoshev RM, Yang GP, Zhang Q, Allard RW (1994) Extra ordinarily polymorphic microsatellites DNA in barly species diversity, chromosomal location, and population dynamics. Proc Acad Sci USA 91:4566–5570
Sajid SS, Shahhosseini M, Huber I, Hu G, Archontoulis SV (2022) County-scale crop yield prediction by integrating crop simulation with machine learning models. Front Plant Sci 13:1000224
Sandhu KS, Lozada DS, Zhang Zh, Pumphery MO, Carter AH (2021) Deep learning for predicting complex traits in spring wheat breeding program. Plant Sci 11:13325
Shah-Ghobadi H, Shabanian N, Khadivi A, Rahmani MS (2018) Analysis of genetic diversity of Pistacia atlantica Desf. populations from Zagros forests using ISSR IRAP and SCoT Molecular Markers. IJRFPBGR 26:177–195
Shamsabadi EE, Sabouri H, Soughi H, Sajadi SJ (2022) Using of molecular markers in prediction of wheat (Triticum aestivum L.) hybrid grain yield based on artificial intelligence methods and multivariate statistics. Russ J Genet 58:603–611
Sharma LK, Singh TN (2017) Regression-based models for the prediction of unconfined compressive strength of artificially structured soil. Eng Comput 34:1–12
Singh A, Ganapathysubramanian B, Singh AK, Sarkar S (2016) Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci 21:110–124
Singh S, Singh VV, Ambawat S, Dubey M, Singh D (2017) Screening and estimation of allelic differentiation in Indian mustard using SSR markers for background selection. Int J Curr Microbiol Appl Sci 6:2506–2516
Singh VK, Bhoyar PI, Sharma V (2022) Application of genomics and breeding technologies to increase yield and nutritional qualities of rapeseed-mustard and sunflower. In: Technologies in plant biotechnology and breeding of field crops, Springer, Singapore, pp 103–131
Suping GUO, Yuan YAN, Ba DAN (2021) Application of molecular marker technologies in stress resistance breeding of rapeseed. Asian J Agric Res 12:36–40
Tiwari S, Singh Y, Upadhyay P, Koutu G (2022) Principal component analysis and genetic divergence studies for yield and quality-related attributes of rice restorer lines. Indian J Genet Plant Breed 82:94–98
Ton LB, Neik TX, Batley J (2020) The use of genetic and gene technologies in shaping modern rapeseed cultivars (Brassica napus L.). Genes 11:1161
Torkashvand AM, Ahmadi A, Nikravesh NL (2017) Prediction of kiwifruit firmness using fruit mineral nutrient concentration by artificial neural network (ANN) and multiple linear regressions (MLR). J Integ Agric 16:1634–1644
Wang L, Wang P, Liang S, Qi X, Li L, Xu L (2019) Monitoring maize growth conditions by training a BP neural network with remotely sensed vegetation temperature condition index and leaf area index. Comput Electron Agric 160:82–90
Wawrzyniak J (2020) Application of artificial neural networks to assess the mycological state of bulk stored rapeseeds. Agric 10:567
Wojciechowski T, Niedbała G, Czechlowski M, Nawrocka JR, Piechnik L, Niemann J (2016) Rapeseed seeds quality classification with usage of VIS-NIR fiber optic probe and artificial neural networks. In: 2016 International Conference on Optoelectronics and Image Processing (ICOIP), IEEE, pp 44–48
Yates DS, David SM, Daren SS (2008) The practice of statistics, 3rd edn. Freeman, New York
Yeken MZ, Emiralioğlu O, Çiftçi V, Bayraktar H, Palacioğlu G, Özer G (2022) Analysis of genetic diversity among common bean germplasm by start codon targeted (SCoT) markers. Mol Biol Rep 49:3839–3847
Zhang J, Zhao B, Yang C, Shi Y, Liao Q, Zhou G, Xie J (2020) Rapeseed stand count estimation at leaf development stages with UAV imagery and convolutional neural networks. Front Plant Sci 11:617
Acknowledgements
The authors would like to express their gratitude to all contributors who have played a role in the development of this research.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors MAN, LA, KP, HS, SJS participated in the sampling, laboratory and bioinformatics department. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Ethical approval and consent to participate
Research, collecting and measurement of plant material are in accordance with national (AREEO) and international guidelines and the IUCN policy statement for plant experiments.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Norouzi, M.A., Ahangar, L., Payghamzadeh, K. et al. Investigation of genetic diversity of different spring rapeseed (Brassica napus L.) genotypes and yield prediction using machine learning models. Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-024-01915-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-024-01915-6