Abstract
Foxtail millet is small diploid (2n = 2x = 18), one of the oldest domesticated, self-compatible, C4 Panicoid cereal grains in Eurasia. Change in climatic conditions, ecological degradation, overexploitaion, and commercial cultivation has led to the genetic loss of landraces as well wild relatives of cultivated crops. Established genetic relations among the species are prerequisites for their future breeding programs to improve cultivars. Therefore, there is an urgent need to conserve plant genetic resources for sustainable agriculture. Keeping in view, we collected seven different species of Setaria that include S. italica, S. viridis, S. sphacelata, S. pumila, S. glauca, S. verticillata, and S. intermedia from Andhra Pradesh, India. The specimens were examined for species identification and taxonomically described. In the present study, the trait “awn” was used as a key taxonomic character for the differentiation of two species viz., S. verticillata and S. intermedia. Earlier S. intermedia was merged in S. verticillata. We observed tremendous variation in phenotypic traits among weedy, wild, landrace, and cultivars of Setaria germplasm, indicating potential genotypic variation. To confirm the genetic variation, the selected Setaria sps were genotyped through ISSR and SSR genetic markers. The average number of amplicons amplified for ISSR and SSR markers was 3.75 and 2.45 alleles per locus, respectively. The Polymorphic information content and Shannon information index (I) for ISSR and SSR markers had an average value of 0.34, 0.34, and 0.46, 0.58, respectively. Gene flow among seven Setaria species was relatively high (Nm > − 1.0). Two-way cluster analysis separated 12 accessions into two significant clusters using combined marker systems and agreed with Principal coordinate analysis. An Analysis of molecular variance confirms that a substantial genetic variation among seven Setaria species. Morphological cluster analysis was almost similar to molecular cluster analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Foxtail millet [Setaria italica (L.) P. Beauv.] commonly known as Italian millet or German millet or Russian millet or Chinese millet or Hungarian millet, is one of the oldest domesticated small diploid, C4 Panicoid cereal grains in Eurasia (Sakamoto 1987). It is a staple crop, requires minimal water for rapid maturation, and is used extensively for food, feed, and fodder. It has been adapted to arid and semi-arid zones, South and North America, North Africa, and Asia.
Previously, the cultivated foxtail millet was described as Panicum italicum and green foxtail as P. viridis (Linnaeus 1753). Different workers identified variations within the P. italicum to species levels, such as P. glomeratum Moench and P. germanicum Mill. (Beauvois 1812). Later on, all these species were elevated, classified, and transferred to the Setaria genus, and the variants were combined under S. italica (Beauvois 1812). The genus Setaria P. Beauv. (Poaceae) belongs to the subtribe Cenchrinae of the tribe Paniceae in the Poaceae sub-family Panicoideae. It comprises about 125 species and is cosmopolitan in distribution (Hubbard 1915; Li et al. 1998). Due to the presence of interconnected “setae” with the inflorescence, the subtribe was abruptly grouped under “bristle clade”. The presence of bristles as sterile branches in the inflorescence is one of the indicative characteristics of the genus Setaria. The number of bristles per spikelet is a trait often used to differentiate the species. Inflorescences are open or spiciform panicles relatively contracted with spikelets along branches, which can be reduced to racemes. The Setaria species identification is complicated and a challenging task for several reasons. The wild Setaria species were found to be very heterogeneous. Further, morphological variation among the species of the genus is significant. It may differ in several traits, such as days to flowering, days to maturity, color and shape of the seed, stigma color variation, color of bristle, and presence/absence of bristle etc.
Among the Setaria species understanding the phylogenetic relationships, including crop evolution, hybridization, organizing germplasm, identifying cultivars, and species relationships, is poorly understood, and no clear genetic hypotheses have been proposed. Thus, it is essential to have a clear understanding of the genetic diversity and relationship between wild-weed-landraces-crop complex for effective conservation, classification, and further utilization of foxtail millet germplasm resources. In this direction, globally, limited studies were carried out in foxtail millet as compared to maize (Zea mays L.), wheat (Triticum aestivum L.), and rice (Oryza sativa L.).
Foxtail millet improvement efforts should be directed towards reducing the crop duration, high yield with improved grain quality, resistance to biotic stress, and tolerance to abiotic stress. It is essential to search genetic resources for new traits among wild-weed-landraces to address these problems. The wild species are helpful to improve the gene pool of cultivated species (Hanson et al. 2007). Unlike high-yielding varieties (whose variability is limited due to homozygosity), wild relatives and landraces have tremendous genetic potential for crop improvement. Expansion of the genetic base is significant in species, in which inbreeding has resulted in a decline in genetic diversity. The genetic diversity of wild relatives and landraces has been widely used for crop origin studies (Spooner et al. 2005; Heerwaarden et al. 2011; Huang et al. 2012).
The genetic diversity of a species can be determined using molecular markers. Molecular markers are famed for detecting genetic diversity, genetic relationships between wild–weed–landrace–cultivar species because of their high efficiency, low sample number requirements, independence of tissue or environmental effects, and a low number of limitations on the growth stage (Bjorklund et al. 2009). Molecular markers are helpful for the management of crop genetic resources (Virk et al. 2000; Song et al. 2003). Among the molecular markers, ISSR markers are more reproducible than RAPD producing polymorphic bands (Semagn et al. 2006) Similarly, the SSRs have impeccable applications in molecular breeding due to their abundant genomic distribution, chromosome-specific, multi-allelic nature, and codominant inheritance. The SSRs are highly informative genetic markers (Cho et al. 2000) and have cross-transferability to closely related species (Yu et al. 2011; Gupta et al. 2012). The SSR markers have been used successfully for the evaluation of genetic diversity among several species, including finger millet (Babu et al. 2014), foxtail millet (Kim et al. 2012; Zhao et al. 2012; Gupta et al. 2012; Wang et al. 2011) and proso millet (Cho et al. 2010).
The study aimed at taxo-morphologically identifying and differentiating Setaria species and estimating morphological variability available in twelve lines of different Setaria species. Further utility of ISSR and SSR markers systems in revealing genetic relationships among the wild-weed-landraces-cultivated species at the molecular level.
Material and methods
Plant materials collection
A total of twelve Setaria lines were collected from different regions of Andhra Pradesh, India. These collections contain seven species of Setaria that include S. italica (2 landraces and 1 released cultivar), S. viridis (three accessions), S. pumila (two accessions), S. sphacelata (one accessions), S. glauca (one accession) S. verticillata (one accession) and S. intermedia (one accession) (Table A1).
Pot experiment
All seven Setaria species seeds were collected and sown in pots filled with red and clay soil. The pots were grown in a greenhouse at a day/night temperature of 30 ± 1 °C/37 ± 1 °C, and relative humidity varied from 50 to 80%. Two replications were maintained, and plants were grown by following standard farming practices. Observations were recorded from randomly selected three individual plants in triplicates for different qualitative traits like awn, type of awn (TA), seed color (SC), culm color (CC), and quantitative traits, namely plant height (PHT; cm), leaf length (LLT; cm), leaf width (LWT; cm), panicle length (PLT; cm) and panicle exertion (PE; cm).
DNA isolation and genotyping
Three grams of fresh young leaves were collected from all the species studied, frozen immediately in the liquid N2, and stored at − 80 °C. Total genomic DNA was isolated using the modified CTAB method (Murray and Thompson 1980). The quality and quantity of the isolated DNA was checked by spectrophotometric analysis and agarose gel and diluted to 7.5 ng/µl for marker analysis. Twelve Setaria lines were evaluated for polymorphism using 8 ISSR and 24 SSR markers (Table A2, 3). The PCR reaction was carried out for each marker in a total volume of 20 µl mixture containing 1 × PCR buffer (10 mM TRIS pH 9.0, 50 mM KCL, 1.5 mM MgCl2), dNTP's each 0.12 mM (Bio Basic, Canada), 0.4 mM of primers (forward and reverse), 40 ng of total genomic DNA and 1 U of taq DNA polymerase (Genei, India). PCR amplification was performed on an Eppendorf thermocycler (Mastercycler® X50) with the following temperature profile conditions: initial, preheating step 94 °C, 5 min was carried out to achieve a hot start. Subsequently, a touchdown (TD) procedure was carried out that consisted of denaturation at 94 °C followed by 35 cycles of 94 °C of denaturation for 1 min, the annealing temperature of 48 °C for ISSR, 49–55 °C for one minute for SSR with an extension time of one minute at 72 °C. The final extension step is carried out for 10 min at 72 °C. The amplification products were mixed with loading buffer (0.005% each of xylene cyanol and bromophenol blue, as tracking dyes) and resolved on agarose gel 1.2% for ISSR, 3% for SSR in a 0.5xTBE system with a voltage of 100v for 2 h. The gel was stained with ethidium bromide, and the products were visualized and documented (G Box Syngene, Synoptic, Ltd, UK). Clear and explicit bands were used for scoring. The reproducible amplified amplicons in ISSR and SSR markers were scored as binary data 1 and 0 for presence and absence, respectively.
Statistical analysis
The mean morphological traits data of three plants for each Setaria line (representing seven species) was subjected to multivariate statistical analysis. Genotypic coefficients of variation (GCV) and phenotypic coefficients of variation (PCV) were calculated by the method specified by Burton (1952), heritability in a broad sense (h2) calculated by the method provided by Burton and Vane (1953), and genetic advance by following Johnson et al. (1955). PCA and two-way cluster analysis were carried out by using JMP software (SAS 2012). In marker data analysis, each variant (bands) as an allele and every primer pair as a locus consideration of the heterozygous or homozygous state were counted. The band profiles were scored only distinct, reproducible, resolved bands were used in the genetic analysis. The number of alleles (Na) per locus, the effective number of alleles (Ae) per locus was calculated according to Kimura and Crow (1964), expected heterozygosity (He), and observed heterozygosity (Ho) was calculated for each SSR locus according to Nei (1973). Shannon’s information index (I) value was calculated using the genetic analysis package POPGENE Version 1.31 (Yeh et al. 1999). The diversity between the germplasm/selected lines was calculated by Euclidean Ward’s method by using SAS JMP statistical discovery software (SAS, 2012). Polymorphic information content (PIC) value was calculated using the formula 1–p2–q2, where p is the presence of band frequency, and q is the absence of band frequency (Mondal et al. 2009). Principal coordinate analysis (PCoA) and AMOVA were done using a distance matrix of combined marker data standardization provided by the GenAlEx package (Peakall and Smouse 2012).
Results
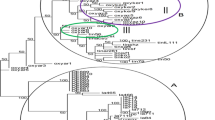
A total of twelve lines representing seven different Setaria species were used in the present study, the specimens were examined for identification and taxonomically described. From the present study, it is evident that bristle is used as a key taxonomic character for the separation of two species viz., S. verticillata and S. intermedia Fig. 1.
Taxonomic description
Characterization of Setaria italica (L.) P. Beauv
It is a cultivated tufted annual grass. Culms grow up to 1 m high, nodes glabrous. Leaf-sheaths 3–10 cm long, margins ciliate; ligule membranous; blades 10–35 × 0.5–2 cm, linear, flat, base cordate, apex acuminate, minutely scaberulous. Panicle length is up to 10 cm long, contracted; spikelets subtended by an involucre; involucral bristles persistent, 2–5 in a whorl, antrorsely barbed. Spikelets comprising one basal sterile florets and one fertile floret, 2 mm, ovoid or elliptic, dorsally compressed. Lower glume ovate, hyaline; upper glume broadly ovate or elliptic, and membranous. Basal florets barren; lower lemma ovate, membranous; lower palea scaly, empty; upper lemma fertile, ovate or elliptic, coriaceous, transversely rugose; upper palea elliptic, involute. Stamens 3. Stigmas are plumose, Caryopsis ellipsoid.
Fl. & Fr: Sep.-Nov.
Vern. Name: Korralu.
Voucher Specimen: Andhra Pradesh, Chittoor District, Punganur, 02–05-2012, P. Ramesh, A. Chandra Sekhar and P. Chandra Obul Reddy 4805 (YVU).
Characterization of Setaria viridis (L.) P. Beauv.
Annual. Culms erect or geniculately ascending, about 60 cm high, nodes glabrous. Leaf-sheaths outer margin hairy; ligule with a fringe of hairs; blades 3–26 × 0.5–1 cm, linear, flat, surface scaberulous. The inflorescence is a cylindric and dense panicle. Spikelets subtended by involucres; involucral bristles persistent, 4–14 in one whorl, 1–3 per spikelet; bristles antrorsely barbed. Spikelets elliptic, dorsally compressed, decidesous, false spikes not lobed or interrupted by cylindric; fertile one pedicelled. Lower glume oblate, membranous; upper glume elliptic; membranous. Basal florets barren; lower lemma similar to upper glume, oblong; lower palea membranous; upper lemma fertile, as lons as the upper glume, elliptic, surface rugulose; upper palea involute.
Fl. & Fr.: Sep.-Jan.
Voucher Specimen: Andhra Pradesh, Kurnool district, RARS Nandyal, 05–12-2013, P. Ramesh, A. Chandra Sekhar and P. Chandra Obul Reddy 4806 (YVU).
Characterization of Setaria sphacelata (Schumach.) Stapf & C.E.Hubb. ex Moss
Perennial. Culms 20–150 cm high, nodes glabrous. Leaf-sheaths hairy; ligule a fringe of hairs; blades 5–40 × 0.2–1.5 cm, linear, flat, or convolute. Inflorescence a spiciform panicle, 7.5–30 cm long; spikelets subtended by involucres; involucral bristles persistent, 6–14 in a whorl, antrorsely scaberulous, yellow or brown. Spikelets elliptic, dorsally compressed; fertile one sessile. Glumes as long as spikelets, ovate, membranous. Basal florets male; lower lemma ovate, membranous; upper lemma fertile, ovate, surface rugose; upper palea involute.
Fl. & Fr.: Sep.- Mar.
Voucher Specimen: New Delhi, NIPGER, 14–03-2013, P. Ramesh, A. Chandra Sekhar and P. Chandra Obul Reddy 4807 (YVU).
Characterization of Setaria glauca (L.) P. Beauv.
Annual. Culms robust, about 3 m high, nodes bearded. Ligule with a fringe of hairs; Leaf-blades ca 80 × 6 cm, linear, flat inflorescence a cylindric spiciform panicle; spikelets subtended by an involucre; involucral bristles persistent, numerous, in whorls, base bluntly stipitate, inner bristles longer than the outer one, glabrous or ciliate. Spikelets 3 mm long, obovate, dorsally compressed, persistent, fertile, one pedicelled. Lower glume absent; upper glume oblong, membranous. Basal florets male or barren; lower lemma oblong, margins ciliolate; upper lemma fertile, ovate, coriaceous, coarsely rugose, boat-shaped and slightly reduced upwards, broadly elliptic, pubescent; upper palea coriaceous. Stamens 3; anther tip penicillate. Caryopsis obovoid.
Fl. & Fr.: Aug.-Jan.
Voucher Specimen: Andhra Pradesh, YSR Kadapa district, Yogi Vemana University, 10–09-2014, P. Ramesh, A. Chandra Sekhar, and P. Chandra Obul Reddy 4802 (YVU).
Characterization of Setaria pumila (Poir.) Roem. & Schult
Annual. Culms erect or decumbent, tufted, to 130 cm high, nodes glabrous. Leaf-sheaths 2–5 cm long, keeled, glabrous, ciliate at the mouth; ligules hairy; blades 5–15 × 0.2–0.6 cm, linear-lanceolate, flat, base cordate or rounded, apex acuminate, sparsely hairy. Panicles spiciform, 5–10 cm long, cylindric, brownish-yellow; spikelets subtended by an involucre; involucral bristles persistent, 6–8 in one whorl, antrorsely scaberulous. Spikelets ovate, gibbous, 1–3 × 2 mm; fertile one pedicelled. Lower glume ovate, 1–1.7 mm long, chartaceous, 3-nerved. Upper glume ovate, 1.5–2 × 1.5 mm, chartaceous, 5-nerved. Lower lemma elliptic-ovate, 1–2.5 × 1.5 mm, chartaceous, 7-nerved. Lower palea elliptic 1.5 × 1 mm, hyaline, 2-keeled, 2-nerved. Stamens 3, anthers 1 mm long. Upper lemma boat-shaped 2.5–3 × 2 mm, keeled, rugose. Stamens 3, anthers 1 mm long. Ovary oblong. Styles 1 mm, stigmas 1 mm. Caryopsis ovaid.
Fl. & Fr.: April- Nov.
Voucher Specimen: Andhra Pradesh, YSR Kadapa district, Utkur, 31–07-2014, P. Ramesh, A. Chandra Sekhar and P. Chandra Obul Reddy 4804 (YVU).
Characterization of Setaria intermedia Roem. & Schult
Annual. Culms erect or decumbent, tufted, to 80 cm high; nodes glabrous. Leaf-sheaths 5–8 cm long, keeled, hairy, margin ciliate; ligules hairy; blades 8–20 × 0.5–0.8 cm, linear-lanceolate, base rounded, apex acuminate, hairy. Panicles spiciform; spikelets subtended by involucres; involucral bristles persistent, 1–4 in a whorl, antrorsely scaberulous. Spikelets elliptic; dorsally compressed; fertile one sessile. Lower glume orbicular, 1 × 0.5 mm, chartaceous, 3-nerved; upper glume broadly ovate, 1–1.6 × 1 mm, chartaceous, 5-nerved. Lower lemma ovate-lanceolate, 1–2.5 × 1.5 mm, chartaceous, 5-nerved; lower palea orbicular, 1.5 × 1 mm, h.yaline, keeled, 2-nerved; upper lemma broadly ovate, 1–2 × 1 mm, crustaceous, rugose, keeled; upper palea involute, rugose. Stamens 3, anthers 1 mm long. Ovary oblong 0.5 mm long. Caryopsis ellipsoid.
Fl. & Fr.: Aug.-Oct.
Voucher Specimen: Andhra Pradesh, YSR Kadapa district, Krishanapuram, 28–11-2012, P. Ramesh, A. Chandra Sekhar and P. Chandra Obul Reddy 4801 (YVU).
Characterization of Setaria verticillata (L.) P. Beauv.
Annual. Culms erect, tufted, to 90 cm high, nodes glabrous. Leaf-sheaths 5–8 cm long, glabrous; ligules hairy; blades 5–20 × 0.5–1 cm, linear-lanceolate, base rounded, apex acuminate, sparsely pubescent. Panicles spiciform, 5–8 cm long, yellow; spikelets subtended by involucres; involucral bristles persistent, 1–4 in a whorl, retrorsely scaberulous. Spikelet oblong-ovate, 1.5–3 × 2 mm, dorsally compressed. Lower glume orbicular, 1–1.5 mm long, chartaceous, 3-nerved; upper glume ovate, 1.5–2 × 1.5 mm, chartaceous, 5-nerved. Lower lemma oblong-ovate, 1.5–2 × 1.5 mm, chartaceous, 5-nerved; lower palea elliptic, hyaline, keeled, 2-nerved; upper lemma oblong, boat-shaped, 2–2.5 × 2 mm, crustaceous, transversely rugose; upper palea oblong, 1.5 × 1 mm, crustaceous, rugose. Stamens 3, anthers 1 mm long. Ovary ovate. Styles 1 mm long; stigma 1 mm long. Caryopsis ellipsoid.
Fl. & Fr.: June-Dec.
Voucher Specimen: Andhra Pradesh, YSR Kadapa district, Krishanapuram, 05-12-2013, P. Ramesh, A. Chandra Sekhar and P. Chandra Obul Reddy 4803 (YVU).
Phenotypic performance
A total of seven qualitative and five quantitative traits mean data were presented in Tables A4 and 5. The present study showed significant variation for the trait PHT (33 to 140 cm), LLT (19–38 cm), LWT (0.5–2.5 cm) PE (0.5–16.2 cm) and PLT (2.5–14 cm) varied respectively (Table A6). As expected, a high degree of variation was observed for all the quantitative traits among seven Setaria species. In quantitative traits studies in these accessions, the genotypic coefficient of variation (GCV %) was almost equal to or lower than the phenotypic coefficient of variation (PCV %) Table 1. The mean data were subjected to correlation, PLT showed significant positive associations (p < 0.001) with PHT. Similarly, LLT showed a significant positive association with LS. PE showed significant negative associations (p < 0.001) with LLT and LWT (Table A6).
Principal components analysis (PCA)
Two-dimensional scaling for relationships among Setaria accessions and morphological traits accounts for the larger proportion of the total variance in PC1, PC2 and PC3 revealed by PCA (Fig. 2a). A total of seven qualitative and five quantitative traits were subjected to PCA. The PCA's first two PCs (PC1 and PC2) for morphological traits explained 63.8% of the total variation. PC1 was contributed by PLH, LL, LW, PAL, TOA, and SEC accounts for 39.09% of the total variation. Similarly, awn, AC, TOP, and LSC were the most important contributors to PC2, accounting for 24.7% of the total variation. The significant characters contributing to PC3, was PAE, and APS, which account for 11.19% of the total variation. Eigenvectors for the first four components of PCs were presented in Tables A7, 8.
Morphological diversity in seven Setaria species. a Principal component analysis in the seven Setaria species based on morphological traits. b Two-way cluster analysis based on Euclidean Ward’s method for morphological traits. Colored bars indicated in the two-way cluster corresponding to the trait mean data
Genetic similarity/distances based on the morphological traits
Assessment of genetic diversity based on morpho-agronomical traits enhances the identification of accessions with a similar genetic pool. This is apparent from the current findings that twelve Setaria accessions representing seven different species in the present study collected from different locations in AP state displayed a distinct variation. The grouping pattern of these accessions is based on morpho-agronomical diversity using Euclidean Ward’s method cluster analysis; grouped into two major clusters (Fig. 2b). Cluster size varied between groups. Among the clusters, cluster II has the most accessions (three accessions of S. italica, two accessions of S. viridis, two accessions of S. pumila, one accession each of S. sphacelata and S. glauca). These accessions in cluster II were from S. italica, a cultivated form of Setaria species and its wild relative S. viridis known to have traits for high yield, with more panicle length, width, and biomass. On the other hand, two Setaria species, S. intermedia and S. verticillata, were grouped in cluster I with high in plant height, tillering and early maturity. The results were strengthened by PCA analysis with morpho-agronomic traits, in which the 12 accessions were grouped into two clusters (cluster I with two accessions and cluster II with ten accessions). Overall, results yielded from both Euclidean Ward’s method, and PCA agrees with each other.
ISSR marker system
Assessing the genetic diversity by molecular markers will be more reliable and consistent. In the present investigation, sixteen ISSR markers were used for genetic diversity analysis in twelve accessions. Among the sixteen ISSRs studied, eight produced reproducible amplicon patterns consisting of thirty DNA band positions, which are 100% polymorphic among the studied accessions (Table 2). The number of amplification products produced by each primer ranged from two (UBC 866) to five (UBC 851), with an average of 3.75 amplicons and amplicon size ranging from 900 bp to 2.0 kb. The PIC value ranged from 0.30 (UBC 866) to 0.43 (UBC 851) with an average value of 0.34 (Table 2). The effective number of alleles ranged from 1.28 (UBC 824) to 1.75 (UBC 862), with an average of 1.52. The I ranged from 0.35 (UBC 824) to 0.58 (UBC 862), with an average of 0.46 (Table 2).The amplified markers in this present study can be used for the unique fingerprinting of Setaria sps.
SSR marker system
A set of 24 SSRs distributed across the nine chromosomes of the foxtail millet genome was used to assess the genetic diversity in Setaria sp. The amplicons thus generated were ranged from 140 to 350 bp respectively. Diversity analysis revealed that the number of alleles per locus varied from 1 to 6, with a mean of 2.45 alleles per locus (Table 3). The observed number of alleles ranged from one to five, with an average of 2.33. The effective number of alleles ranged from 1 (b196 and sigms1250) to 3.78 (b165), with an average of 1.72 across all loci. Genetic variation of Setaria species as a self-pollinating plant is caused mainly by allele frequencies. The various loci of the allele frequencies were differently distributed among the Setaria species (Table A9). Allelic frequencies showed wide variations, ranging from 0.04 to 0.95, and the polymorphism level ranged from 50 to 100%. The PIC value for twenty-four SSR markers varied from 0.14 to 0.5 with a mean of 0.34. The I ranged from 0 to 1.46 with an average of 0.58. In the present study among the Setaria species, the expected heterozygosity ranged from 0 to 0.77 with an average value of 0.37. At the same time, the observed heterozygosity varied between 0 and 1, with a mean of 0.33 (Table 3). Observed heterozygosity was one at two loci (P100, P6), indicating high diversity of the Setaria wild species. Two loci (Sigms1467, b166) showed the heterozygote deficit indicating significant positive Fis values with an average of − 0.75 (Table 3). Of the 24 loci, five loci have the highest index of genetic variation (Fst) 1, only one locus P6 has the lowest index of genetic variation 0, and the average value is 0.67. The locus with the lowest gene flow was at locus P6 (Nm, 0), and the highest gene flow is at locus B163 (Nm, 0.80) with an average of 0.12 (Table 3). Three loci (B234, Sigms3204, Sigms1222) had three rare alleles (with a frequency < 0.05) and nine loci had nine abundant alleles (frequency > 0.50) remaining loci had 45 intermediate alleles (0.05 < frequency < 0.50).
Genetic relationships based on ISSR and SSR markers
With pooled ISSR and SSRs markers, grouped the 12 accessions into two significant clusters I and II (Fig. 3a). There was no significant difference in the grouping of genotypes with ISSR or SSR alone with pooled ISSR and SSRs data. Even though the pooled analysis also showed two major clusters with two accessions S. verticillata and S. intermedia, in cluster I and ten accessions corresponding to five Setaria sps. viz., S. italica1, S. italica2, S. italica3, S. viridis2, S. viridis1, S. viridis3, S. sphacelata, pumila1 S. pumila2, and S. glauca in cluster II, and the pooled results were in agreement with diversity analysis based on alone ISSR, SSR data and PCoA analysis (Fig. 3b). The genetic distances among 12 accessions were represented in (Table A10). Further, Analysis of molecular variance (AMOVA) revealed the geographical location of the 12 accessions showed a maximum diversity with 84% within the cultivars, and wild species and minimum diversity with 16% within the groups of accessions (Table A11 and Fig. 3c).
Molecular diversity revealed by using combined ISSR and SSR markers systems. a Phylogenetic relationships seven Setaria species based on Euclidean Ward’s method of similarity coefficients. Colored bars indicated in the two-way cluster corresponding to the primer amplification profile b Principal coordinate analysis depicting relationships among seven Setaria species based on the genetic similarity matrix derived from SSR and ISSR based markers. c Analysis of molecular variance (AMOVA) among the seven Setaria species based on combined marker systems
Discussion
Wild species and landraces contribute sufficiently to make up the genetic variability in crop species and provide food security. In the last decade, wild relatives of crops and landraces have been lost at an alarming rate due to several reasons, such as the introduction of high-yielding varieties, degradation of the ecosystem, and climate change. Therefore, an urgent need to conserve the wild genetic resources for their sustainable utilization in breeding programs for crop improvement.
The genus Setaria P. Beauv. is represented by about 125 species and about 10 species are widespread in Eurasia (Dekker, 2003; Austin, 2006; Wang et al. 2007). Of which, seven species (S. italica, S. viridis, S. pumila, S. sphacelate, S. glauca, S. verticillata and S. intermedia) were included in the present study. The taxonomy of the Setaria species is very complex, and morphological variation is large. The accurate classification has been confounded by the high degree of overlapping morphological characters both within and between species of the genus, and the diverse polyploidy levels. The morphology of the roots, clums, and leaf of Setaria is unusual among genera of Paniceae in the level of variation. The number and position of bristles in the inflorescence vary considerably among Setaria species (Morrone et al. 2014). One of the diagnostic characteristics of the genus is the presence of bristles as sterile branches in the inflorescence, and the number of bristles per spikelet is an attribute often used to identify species. The species of Setaria differ widely in inflorescence architecture and leaf form (Morrone et al. 2014).
The diploid genome of S. italica was designated as the A genome by Li et al. (1945), diploid S. viridis shares the A genome with S. italica, and verified by different studies like hybrid fertility, cytogenomic, enzymatic, and molecular markers (Li et al. 1945; Benabdelmouna et al. 2001a, b). The morphological differences in S. pumila were found in the spikelet length and lower floret, palea, and the presence of short rhizomes (Hitchcock 1971). Both S. viridis and S. pumila have been domesticated and cultivated as crops (De Wet et al. 1979). Foxtail millet (S. viridis subsp. italica) and S. viridis (S. viridis subsp. viridis) are subspecies of S. viridis, and not separate species, and are interfertile, have continuous and overlapping genetic variation, evidence of the weedy origins of the crop (Prasada Rao et al. 1986; Wang et al. 1995; Darmency et al. 1987; Willweber-Kishimoto 1962). Both taxa are considered as subspecies of S. italica (S. italica subsp. viridis; foxtail millet, S. italica subsp. italica) (Prasada Rao et al. 1986). Tetraploids classified as S. verticillata and S. verticilliformis each have one genome from the diploid S. adhaerens and one from S. viridis (Benabdelmouna et al. 2001b; Layton and Kellogg 2014). The genus is of agricultural importance as it included some food crops Yellow foxtail (S. pumila (Poir.) Roem. & Schult. or S. glauca (L.) P. Beauv.) is also occasionally cultivated as a cereal crop across Southern India. Some wild foxtail species S. verticillata has been harvested as wild cereal by local people in Australia, South America, Africa, and Asia (De Wet et al. 1979; Austin 2006).
Local populations of landraces and crop wild species provide a valuable resource for plant breeding as well as for the preservation of genetic diversity (Kölliker et al. 2003). Genetic diversity studies are important in a crop species in the selection of parents for hybridization and assist in the evaluation of germplasm because any crop improvement depends upon the magnitude of its genetic diversity (Chaudhary and Singh 1982). We have also observed tremendous variation in phenotypic traits among collected wild, weed, landrace, and cultivars of Setaria germplasm. These species give an indication of potential genotypic variation. However, many phenotypic traits (quantitative traits) are influenced by environmental factors (Simioniuc et al. 2002; Smýkal et al. 2008). Therefore, the selection of the important traits for a breeding program should be based on the degree of variability along with genetic advance and heritability.
In the present study, we applied ISSR and SSR markers to estimate the extent of genetic diversity. A total of 30 and 57 alleles were detected using ISSR and SSR markers systems. The total number of amplicons amplified by ISSR markers ranged from 2 to 5 an average of 3.75 and 24 SSR markers ranged from 1 to 6, with a mean of 2.45 alleles per locus. Previous studies on foxtail millets had reported in alleles per locus varied from 2.1 to 16.69 (Gupta et al. 2013, 2014; Pandey et al. 2013; Chander et al. 2017). The PIC value ranged from ISSR and SSR markers were 0.30 to 0.43 with an average value of 0.34 and 0.14–0.5 with a mean of 0.34. The average PIC value in the present study was comparable to earlier reports (Reddy et al. 2002; Gupta et al. 2012; Kim et al. 2012). The mean observed number of alleles for ISSR markers 2, for SSR markers ranged from 1 to 5. The Effective number of alleles for ISSR and SSR primers ranged from 1.28 to 1.75 with an average of 1.52 and 1to 3.78 with an average of 1.72. The Shannon index for ISSR and SSR primers from 0.35 to 0.58 with an average of 0.46 and 0–1.46 with an average of 0.58, respectively. The Fis value estimates the genetic diversity and correlates the allelic variation among the individuals of the same population. The genetic variation (Fis) value noticed in the present study ranged from 0 to 1, with a mean value of 0.67. In general, allele frequencies in a population are denoted as identical when fis value is 0, and it is highly diverse when a fis value is 1. In the present study, the mean of Fis 0.67 indicated a significant difference at the molecular level in the species under study. The locus with the highest gene flow is B163 (Nm, 0.80), the lowest is (Nm, 0.23), and the average gene flow of the 24 loci is 0.12. In a previous study, the gene flow between S. italica and S. viridis was identified (Wang et al. 2010; Jia et al. 2013). In the present study, the gene flow detected among the species indicates that they do not face any genetic drift and hence high genetic differentiation was observed. If no gene flow, inbreeding is dominant in the population which show effect on population structure. This results in loss of genetic variation, finally population prone to extinction (Barrett and Kohn 1991).
Combined marker system applied in two-way cluster analysis, cluster I included two species S. veticillata and S. intermedia. It was reported that the S. veticillata was an allopolyploid with an AABB genome with a chromosome number 36 or 54, while S. intermedia share an unknown genome (Benabdelmouna et al. 2001a, b; Lata et al. 2013). Previous classical taxonomical studies failed to differentiate these two species and are considered single species. However, a few taxonomists classified these two species as separate entities based on variation in the awn morphology. Interestingly grouping based on molecular markers in the present study has separated these two species into two groups. Cluster II in sub cluster a contain S. italica1, S.italica2, S.italica3, S.viridis1, S.viridis2, S.viridis3 and S. sphacelata, which implies that the S. viridis genotypes are close to S. italica. These accessions had the least diversity amongst the five identified species groups. S. italica and S. viridis have the haploid chromosomal number of 09, with an annual life cycle and a similar AA genome. Grouping of S. sphacelata with that of the S. italica and S. viridis indicates the probable genome nature of S. sphacelata, which is known for its earlier unknown genome with a perennial life cycle (Lata et al. 2013). The subcluster b consists of two species, namely S.glauca, and S.pumila. The genomic nature of these two species is unknown, which are known for their high inbreeding nature with an annual life cycle. S. glauca shows a close morphological and molecular relation with S. pumila (Lata et al. 2013). In addition to cluster analysis, PCoA of 12 Setaria accessions gave a grouping pattern resembling two-way cluster analysis. AMOVA from combined (ISSR and SSR) marker systems indicate a significant genetic variation among the seven species studied in the present investigation and provided a clue for their unknown genome nature for some species.
Conclutions
This study provided genetic relationships among seven Setaria species based on taxo-morphological and molecular tools. A key taxonomical character “awn” was used for the differentiation of two species viz., S. verticillata and S. intermedia. Which was earlier S. intermedia was merged in S. verticillata. Further morphological and molecular studies have strongly supported these results. These two species were clearly separated. This phylogenetic study also proposed the possible genome nature of unknown Setaria species. We have also identified three rare alleles, 45 intermediate alleles, and nine abundant alleles. These wild and landraces contain adaptive traits in specific accessions that could be useful in identifying trait associations. These wild relatives and landraces of Setaria contain many expedient traits which can be utilized in crop improvement. In the present study we have identified polymorphic markers, that are useful in crop improvement.
References
Austin DF (2006) Foxtail millets (Setaria: Poaceae): abandoned food in two hemispheres. Econ Bot 60:143–158
Babu BK, Agrawal PK, Pandey D et al (2014) Comparative genomics and association mapping approaches for opaque2 modifier genes in finger millet accessions using genic, genomic and candidate gene-based simple sequence repeat markers. Mol Breed 34:1261–1279
Barrett SCH, Kohn JR (1991) Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Fack DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York
Beauvois P (1812) Essai d’une nouvelle agrostographie; ou nouveaux genres des Gramiùnees. Paris
Benabdelmouna A, Darmency MA, Darmency H (2001a) Phylogenetic and genomic relationships in Setaria italica and its close relatives based on the molecular diversity and chromosomal organization of 5S and 18S–5.8S-25S rDNA genes. Theor Appl Genet 103:668–677
Benabdelmouna A, Shi Y, Abirached-Darmency M et al (2001b) Genomic in situ hybridization (GISH) discriminates between the A and the B genomes in diploid and tetraploid Setaria species. Genome 44(4):685–690
Bjorklund M, Ranta E, Kaitala V et al (2009) Quantitative trait evolution and environmental Cchange. PLoS ONE 4:e4521
Burton GW (1952) Quantitative inheritance in grass. Proc 6th Int Grass Land Congr 1:277–283
Burton CW, De Vane EH (1953) Estimating heritability in tall Fescue (Festuca arundinacea) from replicated clonal material. Agron J 45:1476–1481
Chander S, Bhat KV, Kumari R et al (2017) Analysis of spatial distribution of genetic diversity and validation of Indian foxtail millet core collection. Physiol Mol Biol Plants 23(3):663–673
Chaudhary VS, Singh BB (1982) Heterosis and genetic variability in relation to genetic diversity in soybean. Indian J Genet 42:324–328
Cho YG, Ishii T, Temnykh S et al (2000) Diversity of microsatellites derived from genomic libraries and Genbank sequences in rice (Oryza sativa L.). Theor App Gene 100:713–722
Cho YI, Chung JW, Lee GA et al (2010) Development and characterization of twenty-five new polymorphic microsatellite markers in proso millet (Panicum miliaceum L.). Genes Genom 32:267–273
Darmency H, Zangre GR, Pernes J (1987) The wild-weed-crop complex in Setaria: a hybridization study. Genetica 75(2):03–107
De Wet JMJ, OestryStidd L, Cubero JI (1979) Origins and evolution of foxtail millet. J Agric Trop Bot Appl 26:54–64
Dekker J (2003) Evolutionary biology of the foxtail (Setaria) species-group. In: Inderjit K (ed) Weed Biology and Management. Kluwer Academic Publishers, The Netherlands, pp 65–114
Gupta S, Kumari K, Sahu PP et al (2012) Sequence-based novel genomic microsatellite markers for robust genotyping purposes in foxtail millet [Setaria italica (L.) P. Beauv.]. Plant Cell Rep 31(2):323–337
Gupta S, Kumari K, Muthamilarasan M et al (2013) Development and utilization of novel SSR s in foxtail millet [Setaria italica (L.) P. Beauv.]. Plant Breed 132(4):367–374
Gupta S, Kumari K, Muthamilarasan M et al (2014) Population structure and association mapping of yield contributing agronomic traits in foxtail millet. Plant Cell Rep 33(6):881–893
Hanson PM, Sitathani K, Sadashiva AT et al (2007) Performance of Solanum habrochaites LA1777 introgression line hybrids for marketable tomato fruit yield in Asia. Euphytica 158:67–178
Heerwaarden JV, Doebley J, Briggs WH et al (2011) Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc Natl Acad Sci USA 108:1088–1092
Huang X, Kurata N, Wei X et al (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490:497–501
Hubbard FT (1915) A taxonomic study of Setaria and its immediate allies. Am J Bot 2:169–198
Jia G, Shi S, Wang C et al (2013) Molecular diversity and population structure of Chinese green foxtail [Setaria viridis (L.) Beauv.] revealed by microsatellite analysis. J Exp Bot 64:3645–3655
Johnson HW, Robinson HF, Comstock RE (1955) Estimate of genetic and environmental variability in Soybeans. J Agron 47:314–318
Kim E, Sa K, Park KC et al (2012) Study of genetic diversity and relationships among accessions of foxtail millet [Setaria italica (L.) P. Beauv.] in Korea, China, and Pakistan using SSR markers. Genes Genom 34:529–538
Kimura M, Crow JF (1964) The number of alleles that can be maintained in a finite population. Genetics 49(4):725
Kölliker R, Herrmann D, Boller B, Widmer F (2003) Landraces and distinct and diverse genetic resource of red clover (Trifolium pratense L.). Theor Appl Genet 107:306–315
Lata C, Gupta S, Prasad M (2013) Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses. Crit Rev Biotechnol 33(3):328–343
Layton DJ, Kellogg EA (2014) Morphological, phylogenetic, and ecological diversity of the new model species Setaria viridis (Poaceae: Paniceae) and its close relatives. Am J Bot 101(3):539–557
Li HW, Li CH, Pao WK (1945) Cytological and genetic studies of interspecific cross of the cultivated foxtail millet, Setaria italica P. Beauv. and the green foxtail millet, S. viridis L. J Amer Soc Agron 37:32–54
Li Y, Jia J, Wang Y, Wu S (1998) Intraspecific and interspecific variation in Setaria revealed by RAPD analysis. Genet Resour Crop Evol 45:279–285
Linnaeus C (1753) Species plantarum. Stockholm
Mondal S, Sutar SR, Badigannavar AM (2009) Assessment of genetic diversity in cultivated groundnut (Arachis hypogaea L.) with differential responses to rust and late leaf spot using ISSR markers. Indian J Genet Plant Breed 69(3):219–224
Morrone O, Aliscioni SS, Veldkamp JF et al (2014) Revision of the old world species of Setaria (Poaceae: Panicoideae: Paniceae). System Botany Monogr:1–161
Murray MG, Thompson TF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Pandey G, Misra G, Kumari K et al (2013) Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]. DNA Res 20(2):197–207
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Prasada Rao KE, De Wet JJ, Brink DK et al (1986) Intraspecific variation and systematics of cultivated Setaria italica, foxtail millet (Poaceae). Econ Bot 41:108–116
Reddy MP, Sarla N, Siddiq EA (2002) Inter-simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 128:9–17
Sakamoto S (1987) Origin and dispersal of common millet and foxtail millet. Japan Agr Res Quart 21:84–89
SAS (2012) SAS, JMP Statistical discovery 10.0. SAS Institute Inc Cary
Semagn K, Bjørnstad Å, Ndjiondjop MN (2006) An overview of molecular marker methods for plants. Afr J Biotechnol 5(25):2540–2568
Simioniuc D, Uptmoor R, Friedt W (2002) Genetic diversity and relationships among pea cultivars revealed by RAPDs and AFLPs. Plant Breed 121:429–435
Smýkal P, Hýbl M, Corander J et al (2008) Genetic diversity and population structure of pea (Pisum sativum L.) varieties derived from combined retrotransposon, microsatellite and morphological marker analysis. Theor Appl Genet 117:413–424
Song ZP, Xu X, Wang B et al (2003) Genetic diversity in the northernmost Oryza rufipogon populations estimated by SSR markers. Theor Appl Genet 107:1492–1499
Spooner DM, Mclean K, Ramsay G et al (2005) A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proc Natl Acad Sci USA 102:14694–14699
Virk PS, Newbury HJ, Jackson MT et al (2000) Are mapped markers more useful for assessing genetic diversity? Theor Appl Genet 100:607–613
Wang R-L, Wendel JF, Dekker JH (1995) Weedy adaptation in Setaria ssp. I. Isozyme analysis of genetic diversity and population genetic structure in Setaria viridis. Am J Bot 82:308–317
Wang C, Chen J, Zhi H et al (2010) Population genetics of foxtail millet and its wild ancestor. BMC Genet 11:90
Wang YW, Samuels TD, Wu YQ (2011) Development of 1030 genomic SSR markers in switchgrass. Theor Appl Genet 122:677–686
WangYQ Zhi H, Li W et al (2007) Chromosome number identification of some wild Setaria markers more useful for assessing genetic diversity? Theor Appl Genet 100:607–613
Willweber-Kishimoto E (1962) Interspecific relationships in the genus Setaria. Control Biol Kyoto Univ 14:1–41
Yeh FC, Yang RC, Boyle T (1999) POPGENE Version 1.32: microsoft window-based freeware for population genetics analysis. University of alberta Edmonton
Yu F, Wang BH, Feng SP et al (2011) Development, characterization, and cross-species/genera transferability of SSR markers for rubber tree (Hevea brasiliensis). Plant Cell Rep 30:335–344
Zhao W, Lee GA, Kwon SW et al (2012) Development and use of novel SSR markers for molecular genetic diversity in Italian millet (Setaria italica L.). Genes Genom 34:51–57
Acknowledgements
First author greatly acknowledge CSIR UGC of India, for providing the financial support to this study. ACS and PCOR in YVU, acknowledge the partial utilization of the financial support for consumables (No. CRG./2018/003280 dated 30th May, 2019) from Department of Science and Technology, Science and Engineering Research Board (DST-SERB), New Delhi, India. We thank to Dr Manoj Prasad, NIPGR, NewDelhi for providing one Setaria species and few SSR markers.
Author information
Authors and Affiliations
Contributions
ACS conceived the idea; ACS, RPCO and RP collected samples; BSK and AMR validated the samples taxonomically and maintained all vocher specimens; RP conducted the experiments; RP, YP, JNV and RCVCM performed statistical and marker analysis; RP wrote the original draft. ACS, RPCO, RP, YP and JNV edited manuscript, and finalized the manuscript; ACS and RPCO contributed consumables. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Palakurthi, R., Poli, Y., Juturu, V.N. et al. Molecular genetic and taxonomical relationship among selected Setaria species using inter simple sequence repeat (ISSR’s) and microsatellite (SSRs) markers. Genet Resour Crop Evol 70, 903–917 (2023). https://doi.org/10.1007/s10722-022-01474-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-022-01474-8