Abstract
In this study, a maize F2:3 population and a population of RILs were constructed from a cross between Dong156 and Dong237, and SSR markers were then used to map QTL. A phenotypic index of the traits (N = 6) commonly related to seed storability, such as the germination energy, germination percentage, germination index, vigor index, seedling length, and simple vigor index generated under artificial aging treatment. Two consistent regions, cQTL-7 on chromosome 7 and cQTL-10 on chromosome 10, were identified by comparing QTL analysis results from these two populations. The four SSR markers (umc1671, phi328175, umc1648 and phi050) linked to cQTL-7 and cQTL-10 were selected using a Chi-squared test. Marker umc1648 exhibited the highest selection efficiency value of 88.89%. The four SSR markers were used to identify genomic regions related to storability in 141 maize inbred lines from NCRPIS. Finally, seven lines were identified that included the two regions consistently associated with superior seed storability. These results indicated that the four SSR markers screened in this study could be used for selection of maize germplasm with a high degree of seed storability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize is cultivated worldwide in a range of agroecological environments for food, livestock feed, and as an industrial feedstock (Conelly and Chaiken 2000). Maize seed must be stored from harvest until the next planting season, and sometimes much longer, because seed production usually considerably exceeds consumption each year (Rajjou and Debeaujon 2008). Seed storability or longevity is an important aspect of seed quality that is controlled by many genes and influenced by the environment (Bewley et al. 2013).
Over time, seeds lose viability due to seed aging or deterioration, even when stored at low temperatures, likely due to their physiological, cellular, biochemical, and metabolic activities. The reactive oxygen species (ROS) that accumulate in seeds during storage can oxidize lipids and proteins and consequently modify the structure and function of membranes (Bailly 2004). Lipid peroxidation results in high concentrations of H2O2 and malondialdehyde (MDA), which have been considered critical causes of seed deterioration (Zacheo et al. 2000; Debeaujon et al. 2000; He et al. 2015). Other factors have also been implicated in seed storability, including DNA and RNA degradation, mitochondrial activity, and redox homeostasis states (Parkhey et al. 2012; Xu et al. 2015; Yin et al. 2016).
To further understand the genetic mechanisms controlling seed storability, researchers have performed proteomic analyses after artificial seed aging treatments in Arabidopsis, maize, and rice (Rajjou et al. 2008; Wu et al. 2010; Xin et al. 2011; Yan et al. 2018), which have shown that changes in proteome of dry seeds of, for example, Arabidopsis, are associated with decreased seed viability (Rajjou et al. 2008). Compared to inviable seeds, the expression of small HSPs (heat shock proteins), late embryogenesis abundant (LEA) proteins, and antioxidant enzymes increases strongly in the high-viability seeds of maize hybrid Zhendan958 (Wu et al. 2010). Proteomic analyses revealed that proteins involved in metabolism and energy were the largest downregulated protein group, suggested the important roles of the mobilization of stored carbohydrates and energy supply in seed ageing and seed vigor (Xin et al. 2011).
Several redox regulation proteins (mainly glutathione-related proteins) and some disease/defense-related proteins (proteins related to repair or tolerance of DNA-damage, and a putative LEA protein) also might be correlated with seed storability (Yan et al. 2018).
QTL in several plant species have been linkage mapped after subjecting seeds to natural or artificial conditions that result in seed aging (Bettey et al. 2000; Rehman et al. 2011; Xie et al. 2014). In soybean, 34 QTL affecting seed storability were identified on 11 chromosomes in two RIL populations. Twenty-one of those QTL clustered into five regions on four chromosomes carrying many QTL and 13 other QTL for seed storability were detected in two other QTL-rich regions (Zhang et al. 2019). In rice treated with both natural and artificial aging conditions, 13 QTL affecting seed storability were identified on eight rice chromosomes. Two of these QTL were detected more than once under both natural and artificial aging treatments. Four other QTL were detected once under natural aging conditions and seven QTL were detected once under artificial aging conditions (Hang et al. 2015).
In maize, using an F2:3 population and a RIL population derived from the inbred lines X178 and I178, 13 QTL for six traits related to seed vigor above were mapped in five chromosome regions (Liu et al. 2019). Han et al. (2018) detected 74 QTL and 20 meta-QTL (mQTL) related to seed vigor in two related RIL populations derived from the crosses Yu82 × Shen137 and Yu537A × Shen137 subjected to three aging treatments. The chromosome regions containing four key mQTL (mQTL2-2, mQTL5-3, mQTL6, and mQTL8) located near other important QTL might represent regions containing many QTL for traits associated with seed storability. Earlier, Han et al. (2014) used single-nucleotide polymorphism (SNP) markers to detect 65 QTL for four seed vigor-related traits in two RIL populations derived from the same crosses under various artificial seed aging conditions. They also identified another 18 meta-QTL associated with at least two QTL across two populations.
Maize seed storability is a seriously under-researched problem considering its importance in terms of crop yield, and food quality. Our previous study demonstrated that two maize inbred lines bred by our group, Dong156 and Dong237, showed significant differences in seed storability under natural storage conditions. In the present study, an F2:3 and RILs derived from these two lines were subjected to an artificial aging treatment to (1) map QTL for traits related to seed storability, (2) examine the consistency of QTL regions across two populations, and (3) develop and verify SSR markers located within consistent QTL.
Materials and methods
Plant materials
Dong156 and Dong237, the two inbred lines used in the present study, were developed at Northeast Agricultural University (NEAU) in Harbin, China, and have good and poor seed storability, respectively. A total of 300 F2 progeny were produced from the F1 progeny of Dong156 × Dong237. The F2:3 population was grown at the NEAU experimental farm in Harbin, Heilongjiang Province (N45°46′ and E126°54′ and situated at an elevation of 126 m above mean sea level) along with the parental lines in 2012. We harvested seeds from 267 F3 ears on F2 plants. F2 plants were genotyped with SSR markers, and F3 seeds from each F2 plant were phenotyped for seed storability. A RIL population consisting of 288 lines was developed from five successive generations of self-pollination of F2 individuals in 2016. The F7 RIL lines were then genotyped using SSR markers, and the harvested F8 seeds were used for assess phenotypes for the component traits of seed storability. All the harvested seeds were dried in the sun (water content was lower than 14%) and then stored at – 20 °C in a low temperature freezer (DW-25W518, Qingdao Haier Co. LTD) before testing seed storability under an artificial aging treatment.
We grew out a set of 141 inbred maize lines obtained from the North Central Regional Plant Introduction Station (NCRPIS) to evaluate the genotypes of SSR markers developed in this study (Supplementary table 1).
The parents Dong156 and Dong237, 267 F2 individuals, the 288 RIL lines, and the 141 inbred lines were planted in three-row plots during April to September in 2012 and 2016 respectively. Each plot was 3.0 m long and 0.65 m wide, with an interval of 0.20 m between plants. We followed field management practices typically used for producing maize commercially in the Harbin region. The effective accumulated temperature at above 10 °C was in the range of 2700–3000 °C, and water content of soil was 60–70% the content of field holding water.
SSR assays
Genomic DNA was extracted from the two fresh youngest leaves (about 5 g) of 20-day-old plants using the CTAB protocol (Murray and Thompson 1980). Two hundred and fifty-seven polymorphism SSR markers distributed on the 10 chromosomes of maize (MaizeGDB (http://www.maizegdb.org/) were used to genotype individuals in the F2 and RIL populations in different years (Supplementary table 2). SSR analyses including PCR, polyacrylamide gel electrophoresis, and silver staining were performed following the protocols described by Di et al. (2015a).
Artificial seed aging treatment
Seeds were sterilized by soaking for 20 min in 10% sodium hypochlorite/water v/v, and then washed three times in deionized water. We performed an artificial aging germination (AG) treatment as follows (Groot et al. 2008): 50 seeds were enclosed in a mesh bag, which was then placed in a thermostatic water bath at 58 °C and 100% relative humidity for 0 h or 1 h. The 0 h treatment was used as a control. After the artificial aging treatment, seeds were dried at room temperature (~ 25 °C) for 2–3 d. We then performed a germination test in a bed of sand (following the standard procedures of the International Seed Testing Association (ISTA 2015), with three independent replications per line. After high temperature sterilization, the sand less than 0.20 mm was added to the germination trays. The fifty seeds were placed on top of 3.0 cm of moist sand in a germination tray and then covered with 1.5 cm of sand. The germination trays were placed in a growth chamber at 25 °C, 65% RH, illumination of 4000 lx and a 14:8 h (day/night) photoperiod. Germination was recorded each day and used to calculate the germination index (GI). After holding seeds at 25 °C for 4 d, germination energy (GE) was calculated. At 7 d, germination percentage (GP) was calculated. Roots were then removed from the seedlings, and seedling length (SL) and seedling weight (SW) were measured. A vigor index (VI) and a simple vigor index (SVI) were then calculated based on the above values. The calculation method of each indices was shown in the Table 1. The relative aging indices were calculated as the ratio of the above values of GI, GE, GP, SL, SW, VI, and SVI for the artificially aged group to those of the control group, which were the relative germination energy (RGE), relative germination percentage (RGP), relative germination index (RGI), relative seedling length (RSL), relative seedling weight (RSW), relative vigor index (RVI), and relative simple vigor index (RSVI).
Data were analyzed using SPSS 19.0 software (IBM Corp., Armonk, NY, USA) (https://www.ibm.com/analytics/spss-statistics-software) (Kishor et al. 2019), and Microsoft Excel 2010 (Zhang et al. 2010). Multiple comparisons with Student’s t-test were performed using GraphPad Prism Software (La Jolla, CA, USA), and the values for all measured parameters were expressed as mean ± SD. Differences were considered significant at P < 0.05.
Linkage mapping and QTL analysis
Comparing the phenotypic and genotypic data, we detected QTL by composite interval mapping (Zeng 1994) in the QTL analysis software IciMapping 4.1 (Lander et al. 1987). We calculated the LOD threshold with 1000 permutations P = 0.05 and a 5-cM window (Churchill and Doerge 1994). A t-test was used for multiple comparisons. Additional polymorphic SSR markers were added to the regions containing consistent QTL detected in the two populations, and QTL related to seed storability were reanalyzed.
SSR markers screening and identification
Thirty lines with very good or poor seed storability where chosen from the RIL population to create a pair of mixed pools, and the genotypes of the two parental lines and two pools were analyzed using the SSR markers linked to the consistent QTL for seed storability identified above. Chi-squared tests were conducted to analyze the genotyping results. SSR markers were thus identified that were polymorphic between the two parental lines and two pools.
Further, 141 maize inbred lines from the NCRPIS were used to evaluate the SSR markers screened in this study (Supplementary table 1). We used a relatively gentle artificial aging treatment, considering the complex pedigree of test materials. The seeds were sterilized and were incubated at 45 °C with 90% humidity for 0 h or 72 h (Liu et al. 2019). The methods then used to evaluate phenotypes and analyze SSR genotypes were as same as described above. Cluster analysis was also conducted to evaluate the seed storability of these 141 inbred lines using the system classification method (Cao et al. 2015) to validate these SSR markers by analyzing these phenotypic and genotypic results.
Results
Phenotypic performance for seed storability-related traits in parental lines, F2:3, and RILs
Dong156 and Dong237 exhibited significant differences (P < 0.01) in relative germination energy (RGE), relative germination percentage (RGP), relative germination index (RGI), relative vigor index (RVI), and relative simple vigor index (RSVI) after the artificial aging treatment, and significant differences (P < 0.05) in relative seedling length (RSL) (Table 2). The values of each of these compared traits in Dong156 exceeded those in Dong237. These results indicated highly dominant differences in seed storability between Dong156 and Dong237 and that these lines would be suitable for creating a segregating population for genetic analysis of seed storability.
The seed germination rate (GR) of the F2:3 population was above 99% before applying the artificial aging treatment. Significant differences were detected among the F2:3 lines for all traits tested. We found that the indices related to seed storability GI, GE, GP, SW, VI, and SVI are normally distributed and exhibit transgressive segregation, as expected for patterns of variability in quantitative traits (Fig. 1).
Distributions of and correlations between 6 relative phenotypic traits in an F2:3 population and their parental lines Dong156 and Dong237. The frequency distribution of each trait is shown on a central diagonal in the form of a histogram. Scatter plots of between every pair of traits are shown in the areas below the diagonal, and numerical correlation coefficients between every pair of traits are shown in the areas above in the diagonal. ***Indicate significance at P < 0.001. GE Germination energy, GP Germination percentage, GI Germination index, VI Vigor index, SW Seedling weight. The indices related to seed storability GE, GP, GI, VI, SW, and SVI are normally distributed and significantly correlated with each other
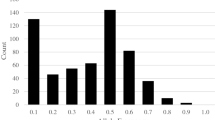
A great range of variability and significant differences became apparent among the traits related to seed storability RGE, RGP, RGI, RVI, RSL, and RSVI during the standard germination test in the RIL population of 288 lines before and after the artificial aging treatment. Variance coefficients for RGE, RGP, RGI, RVI, and RSVI exceeded 40% except for RSL, with the greatest value (57.07%) for RVI. Analysis of the frequency distributions revealed normal distributions for the relative values of RGE, RGP, RGI, RVI, RSVI, and RSL, as presented in Fig. 2 and Table 3. There are also significant differences in GR of these RIL lines at 7 d after artificial aging (Fig. 3).
Distributions of and correlations between 6 relative phenotypic traits in a RIL population derived from Dong156 × Dong237 and parental lines Dong156 and Dong237. The frequency distribution of each trait is shown on a central diagonal in the form of a histogram. Scatter plots of between every pair of traits are shown in the areas below the diagonal, and numerical correlation coefficients between every pair of traits are shown in the areas above in the diagonal. ***Indicate significance at P < 0.001. RGP Relative germination percentage, RGE Relative germination energy, RGI Relative germination index, RVI Relative vigor index, RSL Relative seedling length, RSVI Relative simple vigor index. The indices related to seed storability RGP, RGE, RGI, RVI, RSL, and RSVI are normally distributed and significantly correlated with each other
Germination of parental lines Dong156 and Dong237and a RIL line 201-7 derived from Dong156 × Dong237 at 7 d after treatment in control and artificial seed aging treatment groups. A: parental line Dong156 with good storability; B: RIL line 201-7 with medium storability; C: parental line Dong237 with poor storability
Linkage map construction and QTL analysis
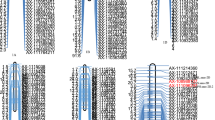
The order and positions of 192 polymorphic SSRs in our molecular linkage map for the F2:3 population agreed with those at the MaizeGDB (http://www.maizegdb.org/ssr.php). We detected a total of 11 QTL for seed storability on maize chromosomes 1, 2, 4, 5, 7, 8, and 10 (Fig. 4, Table 3) in the F2:3 population subjected to the artificial aging treatment. The alleles conferred by Dong156 had positive additive effects that increased seed storability for all QTL detected. qSVI-10, which is located between bnlg162 and umc2043 on chromosome 10, explains 24.35% of the phenotypic variation in SVI in this F2:3 population, with a LOD score of 5.34. Between markers umc2063 and umc2400 on chromosome 5, qSW-5 explains 22.68% of the phenotypic variation in SW in this F2:3 population, with a LOD score of 3.31. We also found that qSVI-7-2 between markers umc1545 and umc2333 on chromosome 7 explains 20.87% of the phenotypic variation in SVI in this F2:3 population, with a LOD score of 2.63.
The molecular linkage map for the RIL population includes 226 polymorphic SSRs with an average spacing of 15.34 cM. Analysis of the phenotypes and genotypes after artificial seed aging in the RIL population revealed 17 QTL affecting seed storability located on chromosomes 1, 2, 7, 9, and 10 (Fig. 5, Table 3) that explain between 2.33 and 20.11% of the phenotypic variation observed in RGE, RGP, RGI, RVI, RSVI, and RSL in the RIL population. Finally, qRGE-10, which is located between phi054 and umc2043 on chromosome 10, explains 20.11% of the phenotypic variation in RGE in the RIL population with a LOD score of 5.81. The alleles that increase seed storability for nine QTL detected in the RIL population were conferred by Dong156.
QTL for seed storability-related traits on the linkage map for RILs derived from Black filled star refers to locus controlling relative germination percentage RGP; black filled downward triangle refers to locus controlling relative germination energy RGE; black filled upward triangle refers to locus controlling relative vigor index RVI; black filled circle refers to locus controlling relative germination energy RGE; black filled square refers to locus controlling relative seedling length RSL; black filled diamond refers to locus controlling relative simple vigor index RSVI
Consistent QTL identified in the F2:3 and RIL populations
A comparison of the results of QTL analysis for the F2:3 and RIL populations revealed two consistent QTL region, cQTL-7 and cQTL-10 on maize chromosomes 7 and 10 containing the major QTLs with the phenotypic contribution rate above 10.0%, respectively, with positive additive effects on seed storability. cQTL-7, a 9.73-Mb region on chromosome 7 between markers umc1295 and umc2333, contains three QTL, qRGP-7, qRGE-7, and qRGI-7, which explain 10.52%, 12.23%, and 14.34% % of the phenotypic variation in RGP, RGE, and RGI, respectively. cQTL-10, a 93.13-Mb region located between the phi054 and umc2043 markers on chromosome 10, contains five QTL (qRGP-10, qRGE-10, qRGI-10, qRVI-10, and qRSVI-10), which explain 12.43%, 20.11%, 14.33%, 10.08%, and 10.85% of the phenotypic variation in RGP, RGE, RGI, RVI, and RSVI, respectively.
After adding 9 and 22 SSR markers near the boundaries and internal regions of cQTL-7 and cQTL-10, respectively, QTL analyses were repeated to refine the molecular linkage maps of chromosomes 7 and 10 (Fig. 6). cQTL-7 was remapped to a 7.97-Mb region between umc1671 and phi328175 on chromosome 7 and cQTL-10 was remapped to a 39.15-Mb region on chromosome 10 between umc1648 and phi050.
Remapping QTL for seed storability-related traits on chromosomes 7 and 10. Black filled star refers to locus controlling relative germination percentage (RGP0; black filled downward triangle refers to locus controlling relative germination energy (RGE); black filled upward triangle refers to locus controlling relative vigor index (RVI); black filled circle refers to locus controlling relative germination energy (RGE); black filled square refers to locus controlling relative seedling length (RSL); black filled diamond refers to locus controlling relative simple vigor index (RSVI)
SSR markers linked to the detection of major QTL
We tested nine SSR markers at the boundaries and interior regions of cQTL-7 and cQTL-10, including umc1671, phi328175, phi050, umc1648, umc1295, phi082, umc1367, phi054, and umc2043, for selection efficiency (Supplementary figure 1). The markers umc1671, phi328175, phi050, and umc1648 showed polymorphisms between both Dong156 and Dong237 and the two resistant or susceptible pools. Individual lines from the two pools were then immediately genotyped using the above nine SSR markers. Chi-squared tests revealed a correlation between major QTL linked to seed storability. The markers umc1671, phi328175, phi050, and umc1648, with χ2 values all less than 3.84 (P = 0.05, n = 1), could thus be used for molecular marker assisted selection (MAS) for seed storability (Table 4). Selection efficiency when using these four markers was then tested in 85 RIL lines with good seed storability. On average, the correlation between genotypes and phenotypes was 83.94%, with the highest correlation between genotype and seed storability of 88.89% for the marker umc1648 (Table 5).
Application of SSR markers in 141 maize inbred lines
The phenotypic data for 141 American maize inbred lines indicated substantial variation and significant differences in RGE, RGP, RGI, RVI, RSVI, and RSL and significant differences among the lines we analyzed (P < 0.001).
Variance coefficients for RGE, RGP, RGI, RVI, and RSVI exceeded 35%, except for RSL, with the greatest value (68.03%) for RGE. Arranged according to seed storability from high to low, the 141 inbred lines were divided to five groups consisting of 21, 35, 32, 33, and 20 lines during cluster analysis (Table 5, and Supplementary figure 2).
Genotypic results for the SSR markers umc1671, phi328175, phi050, and umc1648 showed that seven lines including ND248, ND252, ND246, SD65, N532, N209, and Tx110 contained the two consistent QTLs cQTL-7 and cQTL-10. The lines ND248, ND252, ND246, and SD65 clustered into the first group of 21 lines with the best seed storability, and the lines N532, N209 and Tx110 clustered into the second group of 33 lines with the second-best seed storability in our study (Supplementary figure 3). These results indicated that the four SSR markers developed in this study could be used to select for and develop maize germplasm with superior seed storability (Table 6).
Discussion
Comparison of artificial seed aging methods
Sometimes during extensive natural aging of seeds, seeds can become prone to mildew and deterioration that can affect germination tests. In order to study the storability of seeds, scientists have artificially accelerated aging of plant seeds, but so far, ISTA seed evaluation procedures have only included a method for artificial aging of soybeans (ISTA, 2015). For other crops such as maize, there had previously been no standardized methods for artificially accelerating seed aging. Treatments used to accelerate aging in crop seeds include exposure to high temperature or high humidity (Liu et al. 2019), saturated salt (Bennett et al. 2004), hot water (Groot et al. 2008), or methanol (Addai and Safo-Kantanka 2006).
In our previous study (Di et al. 2015b), the two maize inbreds Dong156 (good seed storability) and Dong237 (poor seed storability) were used to compare the four artificial seed aging treatments mentioned above. The 58 °C hot water aging treatment resulted in the greatest number of significant differences in seed storability between Dong156 and Dong237 for GP, GR, GI, VI, RGR, RGR, RGI, and RVI. Therefore, we chose a 58 °C hot water treatment as the most suitable artificial aging method for maize seeds in the present study (Di et al. 2015b).
Considering the different pedigree of test materials, sample seeds from the 141 American maize inbred lines were treated by a relatively gentle artificial aging treatment, 45 °C and 90% relative humidity for 3 days. The phenotypic data related to seed storability for these inbred lines showed substantial variation and significant (P < 0.001), which indicated validity of the aging method. Using phenotypic selection combined with molecular marker selection, the seven lines including ND248, ND252, ND246, SD65, N532, N209, and Tx110 were picked out. They should be used in maize breeding and basic genetic research related seed storability in the future.
Screening of SSR markers for mining consistent QTL related to seed storability
Different QTL can be identified for a particular trait by using different genetic materials, genotypes, or phenotype detection methods. Integration and optimization of related QTL to obtain consistent QTL is very important for analyzing the genetic basis of complex traits. The physical locations of molecular markers in the genome can be determined and then used to explore other genes located within consistent QTL regions.
Studies attempting to identify consistent QTL in maize have so far focused on yield, plant type, and disease resistance, among other important traits (Wang et al. 2016b; Wang et al. 2019). For example, Semagn et al. (2013) identified 68 mQTL that could affect anthesis-silking interval (ASI) and grain yield (GY) in 18 populations of maize derived from crosses between two parental lines. Xu et al. (2012) surveyed the literature and collected 202 reported QTL related to flowering time and photoperiod in maize and identified 25 consensus QTL and four genomic regions containing many QTL by synthesizing the results of those reports. Ali et al. (2013) combined information about 389 disease-resistance QTL from 36 studies and identified a total of 10 meta-QTL for three major foliar diseases on five maize chromosomes. However, few reports so far have focused on mapping of QTL and development of molecular markers related to maize seed storability (Han et al. 2014, 2018; Liu et al. 2019).
In the present study, we compared the detection of QTL for seed storability using F2:3 and RIL populations and observed two consistent QTL that were located separately on chromosomes 7 and 10. When we remapped cQTL-7, which includes qRGP-7, qRGE-7, and qRGI-7 between umc1671 and phi328175 (7.97 Mb), we noted four previously detected QTL nearby that are related to the seed vigor traits germination energy (GE), germination percentage (GP), germination index (GI), and vigor index (VI), and that could explain more than 10% of the phenotypic variances in these traits (Liu et al. 2011). The QTL qGI7a, which influences the seed germination index (Wang et al. 2016a), is close to the SSR marker phi328175 near cQTL-7. Further, Liu et al. (2019) had identified two QTL controlling GR in a 155-Mb region on chromosome 7. Han et al. (2014) detected another six QTL in the same region for seed vigor-related traits in two RIL populations. Thus, this region of maize chromosome 7 appears to contain genetic variants related to several seed storability-related traits.
Another consistent QTL region related to seed storability, cQTL-10, which includes qRVI-10 and qRSVI-10, was remapped to a 39.15-Mb region between umc1648 to phi050 in which other QTL had previously been identified (Liu et al. 2011; Han et al. 2018). According the results of the present study and several previous reports, clusters of seed vigor-related QTL might reside on both chromosomes 7 and 10. Compared with the results of previous reports, we refined the location of cQTL-7 cQTL-10 to a smaller region and wanted to take the further fine mapping and gene cloning.
The four SSR markers at the boundaries of the two regions were screened based on the reliable QTL analysis results and identified in the RIL population and 141 American maize inbred lines, which should be used in the selection of maize germplam related seed storability.
Conclusions
The four SSR markers located on the boundaries of qRGE-7, qRVI-10, umc1671, phi328175, phi050, and umc1648 were screened and applied in the selection of maize germplsm with superior seed storability. Seven maize inbred lines from the present study have now been designated as germplasm that will be useful for improving maize seed storability by MAS and should be further verified for use in applied maize breeding and theoretical research.
References
Addai IK, Safo-Kantanka O (2006) Evaluation of screening methods for improved storability of soybean seed. Intl J Bot 2:152–155. https://doi.org/10.3923/ijb.2006.152.155
Ali F, Pan Q, Chen G, Zahid KR, Yan J (2013) Evidence of multiple disease resistance (MDR) and implication of meta-analysis in marker assisted selection. PLoS ONE 8:e68150. https://doi.org/10.1371/journal.pone.0068150
Bailly C (2004) Active oxygen species and antioxidants in seed biology. Seed Sci Res 14:93–107. https://doi.org/10.1079/ssr2004159
Bennett MA, Grassbaugh EM, Evans AF, Kleinhenz MD (2004) Saturated salt accelerated aging (SSAA) and other vigor tests for vegetable seeds. Seed Technol 26:67–74. https://doi.org/10.2307/23433494
Bettey M, Finch-Savage WE, King GJ, Lynn JR (2000) Quantitative genetic analysis of seed vigour and pre-emergence seedling growth traits in Brassica oleracea. New Phytol 148:277–286. https://doi.org/10.1046/j.1469-8137.2000.00760.x
Bewley JD, Bradford K, Hilhorst H, Nonogaki H (eds) (2013) Longevity, storage, and deterioration. In: Seeds: physiology of development, germination and dormancy, 3rd ed. Springer, New York, pp. 341–376. https://doi.org/10.1007/978-1-4614-4693-4_8
Cao X, Jiang FL, Wang X, Zang YW, Wu Z (2015) Comprehensive evaluation and screening for chilling-tolerance in tomato lines at the seedling stage. Euphytica 205:569–584. https://doi.org/10.1007/s10681-015-1433-0
Chapter 5: the germination test (2015) In: International rules for seed testing. International Seed Testing Association (ISTA), Bassersdorf, pp. i-5–56. https://doi.org/10.15258/istarules.2019.05
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971. https://doi.org/10.1101/gad.8.21.2653
Conelly WT, Chaiken MS (2000) Intensive farming, agro-diversity, and food security under conditions of extreme population pressure in western Kenya. Hum Ecol 28:19–51. https://doi.org/10.1023/a:1007075621007
Debeaujon I, Leon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122(2):403–414. https://doi.org/10.1104/pp.122.2.403
Di H, Liu XJ, Wang QK, Weng JF, Zhang L, Li XH, Wang ZH (2015a) Development of SNP-based dCAPS markers linked to major head smut resistance quantitative trait locus qHS2.09 in maize. Euphytica 202:69–79. https://doi.org/10.1007/s10681-014-1219-9
Di H, Lv TT, Liu LL, Xia HM, Wang ZH (2015b) Comparative analysis on phenotypic measurement of maize seeds storability by artificial aging methods. J Maize Sci 23:71–75. https://doi.org/10.13597/j.cnki.maize.science.20150312. (in Chinese with English abstract)
Groot SPC, Birnbaum Y, Kromphardt C, Forsberg G, Rop N, Werner S (2008) Effect of the activation of germination processes on the sensitivity of seeds towards physical sanitation treatments. Seed Science & Technology 36:609–620. https://doi.org/10.15258/sst.2008.36.3.11
Han Z, Ku L, Zhang Z, Zhang J, Guo S, Liu H, Zhao R, Ren Z, Zhang L, Su H, Dong L, Chen Y (2014) QTLs for seed vigor related traits identified in maize seeds germinated under artificial aging conditions. PLoS One 9:e92535. https://doi.org/10.1371/journal.pone.0092535
Han Z, Bin W, Zhang J, Guo S, Zhang H, Xu L, Chen Y (2018) Mapping of QTLs associated with seed vigor to artificial aging using two RIL populations in maize (Zea mays L.). Agric Sci 9:397–415. https://doi.org/10.4236/as.2018.94028
Hang NT, Lin QY, Liu LL, Liu X, Liu SJ, Wang WY, Li LF, He NQ, Liu Z, Jiang L, Wan JM (2015) Mapping QTLs related to rice seed storability under natural and artificial aging storage conditions. Euphytica 203:673–681. https://doi.org/10.1007/s10681-014-1304-0
He Y, Cheng J, Li X (2015) Acquisition of desiccation tolerance during seed development is associated with oxidative processes in rice. Botanique 94:91–101. https://doi.org/10.1139/cjb-2015-0154
Kishor DS, Lee C, Lee D, Venkatesh J, Seo J, Chin JH, Jin Z, Hong SK, Ham JK, Koh HJ (2019) Novel allelic variant of Lpa1 gene associated with a significant reduction in seed phytic acid content in rice (Oryza sativa L.). PLoS ONE 14(3):e0209636. https://doi.org/10.1371/journal.pone.0209636
Lander ES, Green P, Abrahamson J, Barlow A (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181. https://doi.org/10.1016/0888-7543(87)90010-3
Liu JB, Fu ZY, Xie HL, Hu YM, Liu ZH, Duan LJ, Xu SZ, Tang JH (2011) Identification of QTLs for maize seed vigor at three stages of seed maturity using a RIL population. Euphytica 178:127–135. https://doi.org/10.1007/s10681-010-0282-0
Liu YN, Zhang HW, Li XH, Wang F, Lyle D, Sun LJ, Wang GY, Wang JH, Li L, Gu RL (2019) Quantitative trait locus mapping for seed artificial aging traits using an F2:3 population and a recombinant inbred line population crossed from two highly related maize inbreds. Plant Breed 138:29–37. https://doi.org/10.1111/pbr.12663
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4326. https://doi.org/10.1093/nar/8.19.4321
Parkhey S, Naithani SC, Keshavkant S (2012) ROS production and lipid catabolism in desiccating Shorea robusta seeds during aging. Plant Physiol Biochem 57:261–267. https://doi.org/10.1016/j.plaphy.2012.06.008
Rajjou L, Debeaujon I (2008) Seed longevity: survival and maintenance of high germination ability of dry seeds. CR Biol 331:796–805. https://doi.org/10.1016/j.crvi.2008.07.021
Rajjou L, Lovigny Y, Groot SP, Belghazi M, Job C, Job D (2008) Proteome-wide characterization of seed aging in Arabidopsis: a comparison between artificial and natural aging protocols. Plant Physiol 148:620–641. https://doi.org/10.1104/pp.108.123141
Rehman Arif MA, Nagel M, Neumann K, Kobiljski B, Lohwasser U, Börner A (2011) Genetic studies of seed longevity in hexaploid wheat using segregation and association mapping approaches. Euphytica 186:1–13. https://doi.org/10.1007/s10681-011-0471-5
Semagn K, Beyene Y, Warburton ML, Tarekegne A, Mugo S, Meisel B, Sehabiague P, Prasanna BM (2013) Meta-analyses of QTL for grain yield and anthesis silking interval in 18 maize populations evaluated under water-stressed and well-watered environments. BMC Genom 14:313. https://doi.org/10.1186/1471-2164-14-313
Wang B, Zhang ZH, Fu ZY, Liu ZH, Hu YM, Tang JH (2016) Comparative QTL analysis of maize seed artificial aging between an immortalized F2 population and its corresponding RILs. Crop J 4:30–39. https://doi.org/10.1016/j.cj.2015.07.004
Wang YJ, Xu J, Deng DX, Ding HD, Bian YL, Yin ZT, Wu YR, Zhou B, Zhao Y (2016) A comprehensive meta-analysis of plant morphology, yield, stay-green, and virus disease resistance QTL in maize (Zea mays L.). Planta 243:459–471. https://doi.org/10.1007/s00425-015-2419-9
Wang YJ, Wang YL, Wang X, Deng DX (2019) Integrated meta-QTL and genome-wide association study analyses reveal candidate genes for maize yield. J Plant Growth Regul. https://doi.org/10.1007/s00344-019-09977-y
Wu X, Liu H, Wang W, Chen S, Hu X, Li C (2010) Proteomic analysis of seed viability in maize. Acta Physiol Plant 33:181–191. https://doi.org/10.1007/s11738-010-0536-4
Xie L, Tan Z, Zhou Y, Xu R, Feng L, Xing Y, Qi X (2014) Identification and fine mapping of quantitative trait loci for seed vigor in germination and seedling establishment in rice. J Integr Plant Biol 56:749–759. https://doi.org/10.1111/jipb.12190
Xin X, Lin XH, Zhou YC, Chen XL, Liu X, Lu XX (2011) Proteome analysis of maize seeds: the effect of artificial ageing. Physiol Plant 143:126138. https://doi.org/10.1111/j.1399-3054.2011.01497
Xu J, Liu YX, Liu J, Cao MJ, Wang J, Lan H, Xu YB, Lu YL, Pan GT, Rong TZ (2012) The genetic architecture of flowering time and photoperiod sensitivity in maize as revealed by QTL review and meta analysis. J Integr Plant Biol 54:358–373. https://doi.org/10.1111/j.1744-7909.2012.01128.x
Xu H, Wei Y, Zhu Y, Lian L, Xie H, Cai Q, Chen Q, Lin Z, Wang Z, Xie H, Zhang J (2015) Antisense suppression of LOX3 gene expression in rice endosperm enhances seed longevity. Plant Biotechnol J 13:526–539. https://doi.org/10.1111/pbi.12277
Yan SJ, Huang WJ, Gao JD, Fu H, Liu J (2018) Comparative metabolomic analysis of seed metabolites associated with seed storability in rice (Oryza sativa L.) during natural aging. Plant Physiol Biochem 127:590–598. https://doi.org/10.1016/j.plaphy.2018.04.020
Yin G, Whelan J, Wu S, Zhou J, Chen B, Chen X, Zhang J, He J, Xin X, Lu X (2016) Comprehensive mitochondrial metabolic shift during the critical node of seed ageing in rice. PLoS ONE 11:e0148013. https://doi.org/10.1371/journal.pone.0148013
Zacheo G, Cappello MS, Gallo A, Santino A, Cappello AR (2000) Changes associated with post-harvest aging in almond seeds. Lebensm-Wiss U-Technol 33:415–423. https://doi.org/10.1006/fstl.2000.0679
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468. https://doi.org/10.1007/s00122-012-2032-2
Zhang Y, Huo M, Zhou J, Xie S (2010) PKSolver: an add-in program for pharmacokinetic and pharmacodynamic data analysis in Microsoft Excel. Comput Methods Prog Biomed 99:306–314. https://doi.org/10.1016/j.cmpb.2010.01.00
Zhang X, Hina A, Song SY, Kong JJ, Bhat JA, Zhao TJ (2019) Whole-genome mapping identified novel “QTL hotspots regions” for seed storability in soybean. (Glycine max L.). BMC Genom 20:499. https://doi.org/10.1186/s12864-019-5897-5
Acknowledgements
This research was supported by the National Natural Science Foundation of China (32072128), the National Key Research and Development Program China (2018YFD0100901), and Provincial Funding for Major National Science and Technology Projects and Key Research and Development Projects (GX18B001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest directly or indirectly and informed consent to publish this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Guo, X., Sun, X., Liu, S. et al. Screening and application of SSR markers related to seed storability traits in maize (Zea mays L.). Genet Resour Crop Evol 68, 2521–2535 (2021). https://doi.org/10.1007/s10722-021-01146-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01146-z