Abstract
The Kho population speaking Khowar language reside since long ago in Chitral District of North-western Pakistan. So far, no report is available about their genetic structure and relationship with surrounding population groups. We partially sequenced the mitochondrial DNA control region from 16 unrelated Kho male and female individuals of different ages. The D-Loop region sequences of Kho were aligned and compared with the revised Cambridge Reference Sequence (rCRS). The genetic data of Kho was compared with surrounding north-western Pakistani population groups including Pathan, Kashmiri, and Hazara. Comparison with rCRS identified overall 49 different haplotypes for Kho samples. Among these 21 haplotypes were shared by more than one Kho individuals. The genetic diversity and power of discrimination observed for Kho group were 0.215 and 0.202 respectively indicating the Kho tribe as a least differentiated group among north-western Pakistani populations. The haplogroup mapping, phylogenetic and haplotype network analysis revealed the nearby maternal ancestral relationship between Kho and Kashmiri populations. The haplogroups analysis demonstrates the western Eurasian ancestral origin of Kho samples. However, the appearance of a few South Asian haplogroups with low frequency speculate the Kho tribe as an admixed population of western Eurasian and South Asian genetic components.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pakistan located in South Asia with two hundred million estimated human population. Pakistani population is comprised of diverse ethnicities. This region is characterized by the co-occurrence of several language families including, Indo-European, Sino-Tibetan as well as linguistic outliers (Qamar et al. 2002). The genetic diversity of these ethnolinguistic groups from Pakistan are represented in the worldwide DNA variation project of the Human Genome Diversity Cell Line Panel (Cann et al. 2002). The endogamy is widely practicing in this region which generated a high degree of genetic differentiation and structuring among different ethnic groups (Singh et al. 2016). A previous report about the mitochondrial DNA (mtDNA) variation in this region sketched sharp boundaries and revealed that population located at the west of the Indus basin of Pakistan mainly consist of western Eurasian mtDNA lineages with a limited South Asian contribution (Bhatti et al. 2017).
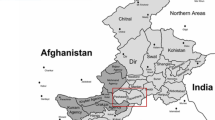
The Kho people are living in north-western Chitral district of Pakistan and speak a distinct Dardic sub-branch, Indo-Aryan language known as Khowar. This ethnic group is not presented in previous population genetic studies. The district Chitral of Pakistan is an area of 14,850 km2 populated by approximately 0.45 million Kho population inhabitants. The region is surrounded by Kunnar District of Afghanistan in the west, while in north and north-western sides the district Chitral meets Upper Dir, Swat, and Gilgit Baltistan regions of Pakistan (Fig. 1) (Nusser and Dickore 2002). The area of Chitral have come under the influences of several invaders in past include the Alexander of Macedonia, Tamerlane, Tartars Genghis Khan, and Arabs. In addition to strong endogamy been practiced since long, an extreme isolate location of Chitral may have generated a varying genetic composition of Kho group in the context of surrounding populations.
The mtDNA has been found suitable to articulate the population evolutionary genetics and molecular ecology (Grasgruber et al. 2017). Particularly, the mtDNA control sequences also known as D-loop region has been found most suitable to profile history and geographic information of a population (Quintana-Murci et al. 2004). The D-loop region is the large noncoding segment in mitochondrial genome involves in initiation and regulation of mtDNA transcription and replication. The polymorphic sites are present in D-loop and therefore this mtDNA region is an object of many studies to find the origin, history, and evolution of population (Beha et al. 2008). The haplogroup typing based on this region is remained as a hallmark to study human evolutionary history and ancestry determination (Hagelberg et al. 1999). In the present study, we sequenced the mtDNA D-loop region of Kho population samples from Chitral to elucidate its maternal lineage distribution. Besides, we performed comparative population genetic analyses of Kho samples with other north-western Pakistani groups to understand the Kho genetic structure in the context of surrounding populations.
Materials and methods
Ethical approval and samples collection
The blood samples were collected from 16 unrelated Kho male and female volunteers following the institutional ethical guidelines. Informed consent was obtained from all the study participants. The sample donors were living in three different regions of Chitral KP i.e. Mastuj (MT), Torkhow (TK) and Mulkhow (MK). The participants were identified by maternal language, ethnicity and parent’s birthplace (Table S1; Supplementary Material). The blood samples were preserved in EDTA tubes.
DNA extraction
The DNA was extracted from the blood samples via the TIANamp Blood DNA Kit (Cat.no.DP318) according to the manufacturer’s instruction.
PCR amplification and mtDNA sequencing
Amplification of the target mtDNA region i.e., 1122 bp was performed by PCR primers set reported by Hayat et al. (2015). The 1–2 ng of genomic DNA was used during PCR amplification. The 0.05 µM primers concentration was kept during each PCR reaction. The PCR reaction was performed in 30 µL total volume using Ampi Taq Gold Master Mix. The PCR amplification condition was set according to the previously published report (Hayat et al. 2015). The Sanger sequencing of the PCR amplified products was carried using ABI 3130 genetic analyzer. The primers set used during PCR amplification were reused in DNA sequencing chain termination cycle sequencing reaction. All the samples were sequenced in triplicates (3x) to confirm the sequence variants.
Data analyses
DNA sequence reads quality check
The DNA sequence reads trace quality check was performed using the Staden Package (Staden et al. 2000). The high-quality sequence reads assembling was performed using Lasergene v. 7. 1 package (DNASTAR Inc. USA).
Nucleotide sequence polymorphism and comparative population genetic analysis
The good quality filtered sequence data was aligned and compared with the same region of the Revised Cambridge Reference Sequence rCRS. A software package DnaSP 5.10 was used to estimate parsimony informative sites and mutations (Rozas et al. 2017). The mtDNA sequences of the other north-western Pakistani ethnic groups (i.e. Pathan, Kashmiri, and Hazara) were obtained from the Genbank, National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) (Table S2; Supplementary Material). The sequences acquired from Genbank was trimmed according to the sequence data of Kho samples and downstream comparative population genetic analyses were performed.
Haplogroup and haplotype analysis
A bioinformatics tool MITOMASTER was used to allocate haplogroups and haplotypes to the population’s mtDNA sequences data. The MITOMASTER resource makes efficient and accurate analysis of human mtDNA sequences to determine the haplogroups and haplotypes (Brandon et al. 2009).
Phylogenetic analysis
The neighbor-joining (NJ) method based phylogenetic tree analysis of Kho mtDNA sequences in comparison with surrounding populations (i.e. Pathan, Kashmiri, and Hazara) was performed via MEGA7.0.9 resource with 1000 bootstrap support replicates (Kumar et al. 2016).
Haplotypes network analysis
The haplotype network paradigm for Kho mtDNA sequences in comparison with surrounding population groups was generated via NETWORK 5.0 (Gehring et al. 2016). The median-joining method implemented in NETWORK 5.0 was adopted which primarily processed by star contraction (SC) and post-processed by maximum parsimony (MP) calculations. The star radius in SC was kept to be 5. The data generated from the MP calculations was then subjected to networking. The network was then refined and each population was specified by a separate color.
Samples sequences submission to GenBank
The high-quality mtDNA draft sequences of Kho samples were submitted to GenBank, NCBI under the accession IDs; MK124589-MK124604.
Results and discussion
The nucleotide sequence date of 706 bp was generated for 16 Kho individuals. The sequence data spanning the nucleotide position 16110-247 of rCRS mtDNA D- loop region. The polymorphic sites observed during the analysis of these mtDNA sequences is represented as haplotypes and haplogroups (Table 1). Total 49 haplotypes and 13 haplogroups of mtDNA were observed for Kho samples sequences. The 21 of these haplotypes were found only once, while 28 haplotypes were found shared by more than one Kho individuals. The haplotypes C16221d (30.61%), T16519C (26%), A73G (22.44%), and T16311C (10.20%) were found frequently. The remaining all haplotypes were present with a frequency of less than 2%. The observed mutations in these sequences compared to rCRS were; transition (69.4%), transversion (5.34%), deletion (24.4%), and Insertion (0.7%) (Table S3; Supplementary Material).
The western Eurasian haplogroups were observed predominantly and mostly shared in Kho samples with overall frequency of 50%. These include HV8, H19, H57, H24, C and, C4a haplogroups. The South Asian haplogroups and its relevant subgroups including U4, U4c, U6, U5a, and W were also found in Kho samples with overall 37.5% frequency (Table 2). The haplogroups and haplotypes specify the origin and linkages of an individual and population. The haplogroup analysis eventually demonstrates the western Eurasian ancestral origin of Kho samples. The previous study also inferred western ancestral origin for other Pakistani populations include Pathan and Hazara residing in the west of Indus basin of Pakistan (Ilyas et al. 2015). The haplogroup W found with 12.5% frequency in Kho samples has been reported in northern India with a frequency of 6%. Its coalescence age is younger, i.e. about 17,000 years ago. This haplogroup is though not predominant but still observed at low frequency in European populations (Derenko et al. 2007). In addition, another South Asian haplogroup, i.e. M30 was also identified for Kho samples with frequency of 6.2%. This indicates the genetic affiliation of Kho with the South Asian populations.
We performed additional analysis to compare the mtDNA partial control region sequences of Kho samples in the context of nearby north-western Pakistani groups (i.e. Pathan, Kashmiri, and Hazara) mtDNA sequences. Among surrounding populations, the Kho samples were found to share few haplotypes and haplogroups with Kashmiri population and do not share significant number of haplogroups and haplotypes with Pathan and Hazara populations. This somehow assumed that Kho population have preserved distinct western antecedent ancestry (Table 3). The comparative analysis revealed the Kho as a least diverse group with less genetic diversity compare to rest of north-western Pakistani populations (Table 4). This represents a high rate of endogamy in Kho tribe.
The Neighbor-joining (NJ) method based phylogenetic tree analysis was carried out for Kho mtDNA sequences along with the surrounding population groups to know their phylogenetic relationship. The geographically distinct population groups generated separate tree clades. The Kho samples were found to develop three different clusters in the phylogenetic tree. In addition, the Kho samples shared the phylogenetic tree clades with Kashmiri mtDNA sequences with significant bootstrap values, i.e. 93%, 80% and 46% (Fig. 2). The analysis of the present study demonstrates that three individuals of Kho population showed common maternal lineages with Kashmiri population, though significant regional differences are present between these two populations in Pakistan. The presence of shared mtDNA haplogroups (i.e., C4a and W) between Kho and Kashmiri reinforcing their close phylogenetic relationship and eventually speculate that Kho holds close maternal lineages relationship with Kashmiri population (Table 3; Fig. 2).
We performed haplotype networking analysis to further elucidate the genetic relationship of Kho with surrounding north-western Pakistani populations. The Kho haplotypes were found in three well supported, share nodes with Kashmiri mtDNA sequences haplotypes. A Kashmiri sample based haplotype was found in mid as an ancestral haplotype for few Kho nodes. Hazara population`s haplotypes were found in a small distance of relatedness to Kho, while the Pathan population haplotypes were found scattered in the periphery of network plot and showed no close relationship with Kho samples (Fig. 3).
The phylogenetic network paradigm of Kho and nearby population groups. The haplotypes of each population in the plot are presented with different colors; Red: Kho Green: Kashmiri; Black: Pathan and Blue: Hazara. Each node represents one mtDNA sequence, the size of nodes depended on haplotypes frequency. The length of the lines connecting the haplotypes refer to the distance of relatedness
The predominant prevalence of western mtDNA haplogroups in Kho samples indicate the western Eurasian origin of Kho group. However, the presence of few South Asian haplogroups with a minor proportion revealed that Kho might be an admixed population of south and western Asian genetic components. This is congruent to the geographical location of Kho people currently residing at the crossroad of the south and central Asian corridors. The shared haplogroups, haplotype network nodes and close phylogenetic tree branches of Kho mtDNA with Kashmiri population represent a common maternal lineage between Kho and Kashmiri.
The analyses conducted led to useful information about the genetic structure of Kho ethnic group. The data generated and analyses performed will provide a framework for future forensics investigation. The genome-wide population genetic analyses of Kho individuals in the context of worldwide major ethnic groups will infer useful information about demography, origin, and natural selection events of this population tribe.
References
Behar DM, Metspalu E, Kivisild T, Rosset S, Tzur S, Hadid Y, Yudkovsky G, Rosengarten D, Pereira L, Amorim A, Kutuev I (2008) Counting the founders: the matrilineal genetic ancestry of the Jewish Diaspora. Plos One 3(4):e2062. https://doi.org/10.1371/journal.pone.0002062
Bhatti S, Aslamkhan M, Abbas S, Attimonelli M, Aydin HH, de Souza EMS (2017) Genetic analysis of mitochondrial DNA control region variations in four tribes of Khyber Pakhtunkhwa, Pakistan. Mitochondrial DNA Part A 28(5):687–697. https://doi.org/10.3109/24701394.2016.1174222
Brandon MC, Ruiz-Pesini E, Mishmar D, Procaccio V, Lott MT, Nguyen KC, Spolim S, Patil U, Baldi P, Wallace DC (2009) MITOMASTER: a bioinformatics tool for the analysis of mitochondrial DNA sequences. Hum Mut 30(1):1–6. https://doi.org/10.1002/humu.20801
Cann HM, Toma C, Cazes L, Legrand MF, Morel V, Piouffre L et al (2002) A human genome diversity cell line panel. Science 296:261–262. https://doi.org/10.1016/s0764-4469(98)80774-9
Derenko M, Malyarchuk B, Grzybowski T, Denisova G, Dambueva I, Perkova M, Dorzhu C, Luzina F, Lee HK, Vanecek T, Villems R (2007) Phylogeographic analysis of mitochondrial DNA in northern Asian populations. Am J Hum Genet 81(5):1025–1041. https://doi.org/10.1086/522933
Gehring PS, Pabijan M, Randrianirina JE, Glaw F, Vences M (2012) The influence of riverine barriers on phylogeographic patterns of Malagasy reed frogs (Heterixalus). Mol Phylogenet Evol 64(3):618–632. https://doi.org/10.1016/j.ympev.2012.05.018
Grasgruber P, Popović S, Bokuvka D, Davidović I, Hřebíčková S, Ingrová P, Potpara P, Prce S, Stračárová N (2017) The mountains of giants: an anthropometric survey of male youths in Bosnia and Herzegovina. R Soc Open Sci 4(4):161054. https://doi.org/10.1098/rsos.170445
Hagelberg E, Goldman N, Lio P, Whelan S, Schiefenhöel W, Clegg JB, Bowden DK (1999) Evidence for mitochondrial DNA recombination in a human population of island Melanesia. Proc R Soc Lond B Biol Sci 266(1418):485–492. https://doi.org/10.1098/rspb.1999.0663
Hayat S, Akhtar T, Siddiqi MH, Rakha A, Haider N, Tayyab M, Abbas G, Ali A, Bokhari SYA, Tariq MA, Khan FM (2015) Mitochondrial DNA control region sequences study in Saraiki population from Pakistan. Leg Med 17(2):140–144. https://doi.org/10.1016/j.legalmed.2014.10.010
Ilyas M, Kim JS, Cooper J, Shin YA, Kim HM, Cho YS, Hwang S, Kim H, Moon J, Chung O, Jun J (2015) Whole genome sequencing of an ethnic Pathan (Pakhtun) from the north-west of Pakistan. BMC Genom 16(1):172. https://doi.org/10.1186/s12864-015-1290-1
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452. https://doi.org/10.1093/bioinformatics/btp187
Nusser M, And Dickore WB (2002) A tangle in the triangle: vegetation map of the eastern Hindukush (Chitral, northern Pakistan). Erdkunde 56(1):37–59. https://doi.org/10.3112/erdkunde.2002.01.03
Qamar R, Ayub Q, Mohyuddin A, Helgason A, Mazhar K, Mansoor A, Zerjal T, Tyler-Smith C, Mehdi SQ (2002) Y-chromosomal DNA variation in Pakistan. Am J Hum Genet 70(5):1107–1124. https://doi.org/10.1086/339929
Quintana-Murci L, Chaix R. Wells RS, Behar DM, Sayar H, Scozzari R, Rengo C, Al-Zahery N, Semino O, Santachiara-Benerecetti AS, Coppa A (2004) Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am J Hum Genet 74(5):827–845. https://doi.org/10.1086/383236
Rozas J, Ferrer-Mata A, Sánchez-Delbarrio JC, Guiro-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34(12):3299–3302. https://doi.org/10.1093/molbev/msx248
Singh G, Talwar I, Sharma R, Matharoo K, Bhanwer AJS (2016) Genetic differentiation and population structure of five ethnic groups of Punjab (North-West India). Mol Genet Genomics 291(6):2055–2063. https://doi.org/10.1007/s00438-016-1239-3
Staden R, Beal KF, Bonfield JK (2000) The staden package, 1998. In: Bioinformatics methods and protocols. Humana Press, Totowa, pp 115–130. https://doi.org/10.1385/1-59259-192-2:115
Acknowledgements
We would like to thanks all volunteers for their contribution to this study and provided blood samples. This research did not receive any specific grant from funding agencies in the public, commercial or nonprofit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Aziz, S., Nawaz, M., Afridi, S.G. et al. Genetic structure of Kho population from north-western Pakistan based on mtDNA control region sequences. Genetica 147, 177–183 (2019). https://doi.org/10.1007/s10709-019-00060-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-019-00060-8