Abstract
The subsequent human migrations that dispersed out of Africa, both prehistoric and historic and colonization of India by modern humans is unanimous, and phylogeny of major mitochondrial DNA haplogroups have played a key role in assessing the genetic origin of people of India. To address more such events, complete mitogenomes of 113 Melakudiya tribe of Southern India were sequenced and 46 individuals showed the presence of west Eurasian autochthonous haplogroups HV14 and U7. Phylogenetic analysis revealed two novel subclades HV14a1b and HV14a1b1 and sequences representing haplogroup U7 were included under previously described subclade U7a3a1a2* specific to India. Moreover, the present analysis on complete mtDNA reveals addition information of the spread and distribution of west Eurasian haplogroups in southern India, in tracing an unexplored genetic link between Melakudiya tribe with the people of Iranian Plateau, South Caucasus, and Central Asia. Coalescence ages of HV14 and U7a3a1a2* trees in the present study dates ~ 16.1 ± 4.3 and ~ 13.4 ± 5.6 kya respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anatomically modern humans (Homo sapiens) originated in East Africa during Middle Paleolithic ~ 200 thousand years ago (kya) and dispersed ‘Out-of-Africa’ to populate the world (Lewin 1987; Stringer et al. 1989; Cavalli-Sforza et al. 1994; Lahr and Foley 1998; Kivisild et al. 1999a, b). During the Neolithic period ~ 10 kya, a major agricultural event along with extensive immigration by demic diffusion played a major role in recolonizing human population in Europe, which expanded with genetic crossroads (Zvelebil 1980; Cavalli-Sforza 1996; Thorpe 1999; Quintana-Murci et al. 2004; Sahoo et al. 2006; Alizadeh et al. 2010). It served as a reservoir of genetic variation for particular lineages, which would have subsequently expanded to other regions (De Fanti et al. 2015).
The subcontinent of India serves as a major corridor for human dispersal with multiple waves of migration and admixture, representing a sizeable fraction of global genetic diversity (Cann 2001; Basu et al. 2003), and the present mitochondrial DNA (mtDNA) gene pool of India was shaped by initial settlers and was instigated by minor events of gene flow from the East and West (Chandrasekar et al. 2009). Autosomal genetic evidence indicates that most of the ethnolinguistic groups in India have descended from a mixture of two divergent ancestral populations: Ancestral North Indians (ANI) related to People of West Eurasia, the Caucasus, Central Asia and the Middle East, and Ancestral South Indians (ASI) distantly related to indigenous Andaman Islanders (Reich et al. 2009). It is presumed that proto-Dravidian language, most likely originated in Elam province of South Western Iran, and later spread eastwards with the movement of people to the Indus Valley and later the subcontinent India (McAlpin et al. 1975; Cavalli-Sforza et al. 1988; Renfrew 1996; Derenko et al. 2013). West Eurasian haplogroups are found across India and harbor many deep-branching lineages of Indian mtDNA pool, and most of the mtDNA lineages of Western Eurasian ancestry must have a recent entry date less than 10 Kya (Kivisild et al. 1999a). The frequency of these lineages is specifically found among the higher caste groups of India (Bamshad et al. 1998, 2001; Basu et al. 2003) and many caste groups are direct descendants of Indo-Aryan immigrants (Cordaux et al. 2004). These waves of various invasions and subsequent migrations resulted in major demographic expansions in the region, which added new languages and cultures to the already colonized populations of India. Although previous genetic studies of the maternal gene pools of Indians had revealed a genetic connection between Iranian populations and the Arabian Peninsula, likely the result of both ancient and recent gene flow (Metspalu et al. 2004; Terreros et al. 2011). Therefore, studies involving uncovered Indian tribal population are warranted to understand the genetic connection with the neighboring gene pool in the evolution of modern human beings.
To know the mtDNA lineages in Indian population many studies were carried out covering both tribe and caste communities for the last one and half decade or so (Bamshad et al. 1996, 1998, 2001; Kivisild et al. 1999a, 2003; Majumder 2001, 2010; Roychoudhury et al. 2000, 2001; Edwin et al. 2002; Basu et al. 2003; Cordaux et al. 2003; Palanichamy et al. 2004, 2015; Metspalu et al. 2004; Rajkumar et al. 2005; Sun et al. 2005; Thangaraj et al. 2005, 2006, 2008, 2009; Kumar et al. 2008; Chaubey et al. 2008; Chandrasekar et al. 2009). The present study on the complete mitochondrial genome of Melakudiya tribal population of southern India provides a comprehensive structure of mtDNA phylogenetic distribution and molecular classification of haplogroups of the Indian matrilineal gene pool. The data obtained from this study are compared with previously published data on the phylogenetic distribution of the West Eurasian lineages.
Materials and methods
Ethics statement, sample collection, and complete mtDNA sequencing
The Institutional Ethical Committee of the Anthropological Survey of India and the University of Mysore approved the protocol and ethical clearance of the study. Written informed consent was obtained from all the 113 healthy unrelated subjects belonging to Melakudiya tribal population, a Dravidian speaking tribe from the Kodagu district of Karnataka, Southern India. Genomic DNA from whole blood was extracted using a standard phenol–chloroform method (Sambrook et al. 1989). Complete mtDNA genome was amplified with 24 standard primers (Rieder et al. 1998), and were checked on 2% agarose gels and were directly sequenced using Big Dye Terminator v3.1 Cycle Sequencing Ready Reaction Kit and ABI PRISM 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA). The resulting sequences were analyzed with the Seqscape v2.5 software (Applied Biosystems, Foster City, CA, USA). Mutations were scored by comparing the sequences with the revised Cambridge Reference Sequence (rCRS) (Andrews et al. 1999) and aligned with MEGA v7 (Kumar et al. 2016) and BioEdit Sequence Alignment program (Hall 1999). The 46 complete mtDNA sequences reported in this study have been submitted to the GenBank database (http://www.ncbi.nlm.nih.gov/Genbank/, accession numbers: MG649324–MG649328; MH368695–MH368735) (Supplementary File Table S1).
Phylogenetic analysis
Putative haplogroup identification was done using Mitomaster (Brandon et al. 2009), based on PhyloTree Build 17 (Van Oven and Kayser 2009). Maximum parsimonious trees of the complete mtDNA sequences were reconstructed manually by computing the median-joining network algorithm using NETWORK5.1 (Bandelt et al. 1999). For the tree reconstruction, a total of 46 complete mitogenome sequences from the present study and previously published literature by Palanichamy et al. (2004, 2015), Derenko et al. (2013), Khan et al. (2013), Vyas et al. (2016), Margaryan et al. (2017), Sahakyan et al. (2017) and Peng et al. (2018) were utilized for analysis, excluding insertion sites (Supplementary File Table S2).
Molecular dating
The age estimates of the coding-region at positions 577–16,023 (Andrews et al. 1999) with 95% confidence intervals were estimated using the rho (ρ) statistic and standard errors (σ), and the variances of rho-based dating were calculated according to Saillard et al. (2000) using two previously described mutation rates. The first mutation rate based on substitution rate of the entire coding region 1.26 × 10−8 mutations per nucleotide per year, which yields 5.39 years for each mutation (Mishmar et al. 2003). The second mutation rate based on substitution rates for protein-coding synonymous changes of 3.5 × 10−8 mutations per nucleotide per year, which yields 7884 years per synonymous mutation (i.e., transition or transversion) (Soares et al. 2009).
Result and discussion
The spread of mtDNA haplogroup HV14 in South India
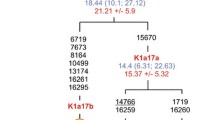
The mtDNA haplogroups detected, together with their frequencies are illustrated in (Table 1), haplogroup assignment for each individual according to the nomenclature of PhyloTree Build17 (Van Oven and Kayser 2009). In the present study we have reconstructed the phylogeny of haplogroup HV14 based on 49 compete mitogenomes which include 11 previously published sequences (Palanichamy et al. 2004, 2015; Khan et al. 2013; Derenko et al. 2013; Vyas et al. 2016; Margaryan et al. 2017 and; Peng et al. 2018) and 38 newly generated sequences from Melakudiya samples. With the addition of a substantial set of Melakudiya sequences to the tree, gives a branching point at HV14a1 node, now defined by a coding and control region transitions at 146 and 3834, which was defined as a novel subclade HV14a1b (Fig. 1). Further, two Melakudiya sequences shared a transition at 16,274, which allowed us to reveal another novel subclade HV14a1b, where additional coding region insertion at 5642.1 T and transition at 1842 was detected in the two individual sequences (Fig. 1). mtDNA haplogroup HV14 has prominence in North/Western Europe, West Eurasia, Iran, and South Caucasus to Central Asia (Malyarchuk et al. 2008; Schonberg et al. 2011; Derenko et al. 2013; De Fanti et al. 2015). Although Palanichamy identified haplogroup HV14a1 in three Indian samples (Palanichamy et al. 2015), it is restricted to limited unknown distribution. In the present study, by the addition of considerable sequences from the Melakudiya population, a unique novel subclade designated as HV14a1b was found with a high frequency (43%) allowed us to reveal the earliest diverging sequences in the HV14 tree prior to the emergence of HV14a1b in Melakudiya. Furthermore, four sequences from Pamiris of Tajikistan and one sequence from Artsakh of South Caucasus did not resolve within the HV14a tree and was branched as HV14* (Fig. 1). The coalescence age for haplogroup HV14 in this study is dated ~ 16.1 ± 4.2 kya and the founder age of haplogroup HV14 in Melakudiya tribe, which is represented by a novel clade HV14a1b is ~ 8.5 ± 5.6 kya (Table 2).
Maximum Parsimonious tree of complete mitogenomes constructed using 38 sequences from Melakudiya tribe and 11 previously published sequences belonging to haplogroup HV14 [Supplementary file Table S2] Suffixes @ indicate back mutation, a plus sign (+) an insertion. Control region mutations are underlined, and synonymous transitions are shown in normal font and non-synonymous mutations are shown in bold font. Coalescence ages (Kya) for complete coding region are shown in normal font and synonymous transitions are shown in Italics
A likely in-situ origin of subhaplogroup U7a3a1a2 in India
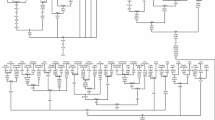
Unlike, haplogroup HV14, haplogroup U7, has a geographic distribution ranging from Europe to India and is prominent in the Near East, Central Asia to South Asia (Quintana-Murci et al. 2004; Metspalu et al. 2004; Kim et al. 2010; Li et al. 2010; Sahakyan et al. 2017). Focusing on the Melakudiyas, the reconstructed phylogeny of haplogroup U7 based on 26 complete mitogenomes which includes 18 sequences from previous study (Palanichamy et al. 2015 and; Sahakyan et al. 2017) and eight newly generated sequenced samples from Melakudiya revealed the complete mtDNA sequences form a distinct clade U7a3a1a2, of which a consortium of 11 sequences belonging to the people of Tamil Nadu, Kerala, Karnataka (both tribe and castes) of Dravidian linguistic family, five sequences of the Indo-European speaking people from Gujarat, Maharashtra and Uttar Pradesh, one Sri Lankan Tamil, and one Kuwaiti sequence represented the reconstructed U7 tree (Fig. 2). The coalescence age of haplogroup U7a3a1a2 dates to ~ 13.3 ± 4.0 kya. The eight Melakudiya sequences were defined by transition at 10,238, which was classified within the U7a3a1a2* node, additional three coding region mutations (5147–10,245–13,745) and one control region mutations (16,189) was detected in four sequences (Fig. 2) and the founder age of haplogroup U7a3a1a2* in Melakudiyas tribe is dated ~ 12.8 ± 9.6 kya. Studies on mtDNA control region sequences have also detected the presence of U7 lineages with haplotypes (viz. 151–16,069–16,260–16,274–16,318 T) in Afghanistan, Tajikistan (Irwin et al. 2010); Sri Lankan (Ranaweera et al. 2014); in Dravidian (Andhra Brahmans) and Indo-European (Gujarat) speakers of India (Metspalu et al. 2004) (Supplementary File Table S3). Coalescence ages estimated for the complete coding region for the main and sub-branches of HV14 and U7a3a1a2 trees and coalescence times based on synonymous mutations are inferred in Table 1.
Maximum Parsimonious tree of complete mitogenomes constructed using 08 sequences from Melakudiya tribe and 19 previously published sequences belonging to haplogroup U7a3a1a2 (Supplementary file Table S2) Suffixes (FS) indicate Frame shift mutation, (T) in 16,318 T is a transversion, Control region mutations are underlined, and synonymous transitions are shown in normal font and non-synonymous mutations are shown in bold font. Coalescence ages (Kya) for complete coding region are shown in normal font and synonymous transitions are shown in italics
The complete mitogenome sequence analysis revealed the genetic diversity of the Melakudiya tribe, has an origin from a common maternal ancestral gene pools with the people of Iran, South Caucasus, and Central Asia, with the influence of west Eurasia component haplogroups HV14 and U7, suggest the migration might have occurred by a major agricultural event in Europe to the Near East during the Neolithic, which was accompanied by extensive immigration by vast demic population diffusions resulting in branching out of founder lineages into many daughter clades which explored the variation of West Eurasian haplogroup HV14 in the Melakudiya tribal population, which also supports the Elamo-Dravidian linguistic connections, with uniquely shared ancestry with populations of Iran (Derenko et al. 2013), also clustering with the Central Asia populations (Margaryan et al. 2017; Peng et al. 2018). Although, haplogroup U7 has its origin from the Near East and is widespread from Europe to India, the phylogeny of Melakudiya tribe with subclade U7a3a1a2 clusters with populations of India (caste and tribe) and neighboring populations (Irwin et al. 2010; Ranaweera et al. 2014; Sahakyan et al. 2017), hint about the in-situ origin of the subclade in India from Indo-Aryan immigrants. Furthermore, the newly branched recent subclades within haplogroup HV14 and U7 with most recent common ancestor split after the Last Glacial Maximum (LGM) ~ 20 kya, apparently resulting from glacial bottleneck from which the largest fraction of surviving lineages are originated. With the subclades of HV14 and U7 lineages detected in the Melakudiya tribal population, put forth an idea about the earliest settlement of the tribal population before the origin of caste system in India, suggesting either dispersal of an out-group population from Iranian Plateau migrated to India as a demic migrant and later settled in Southern India realm, with change in time the Melakudiya tribe acquired new haplotypes which deep branch into novel subclades. Nevertheless, they still practice their traditional agriculture and genetically with West Eurasian lineages traced back to Iranian Plateau indicating Neolithic genetic continuity. This study provides a comprehensive history for reconstructing ancient migration events.
Change history
10 September 2018
Unfortunately, the original version of this article was published with an error in the second sentence of the ‘Molecular dating’ section.
References
Alizadeh A, Alden JR, Danti MD, Garthwaite G, Alizadeh A (2010) The rise of the Highland Elamite State in Southwestern Iran: “enclosed” or enclosing nomadism? Curr Anthropol 51(3):353–383
Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23(2):147
Bamshad M, Fraley AE, Crawford MH, Cann RL, Busi BR, Naidu JM, Jorde LB (1996) MtDNA variation in caste populations of Andhra Pradesh, India. Hum Biol 68(1):1–28
Bamshad MJ, Watkins WS, Dixon ME, Jorde LB, Rao BB, Naidu JM, Prasad BR, Rasanayagam A, Hammer MF (1998) Female gene flow stratifies Hindu castes. Nature 395:651–652
Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, Naidu JM, Prasad BR, Reddy PG, Rasanayagam A, Papiha SS (2001) Genetic evidence on the origins of Indian caste populations. Genome Res 11(6):994–1004
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Basu A, Mukherjee N, Roy S, Sengupta S, Banerjee S, Chakraborty M, Dey B, Roy M, Roy B, Bhattacharyya NP, Roychoudhury S (2003) Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res 13(10):2277–2290
Brandon MC, Ruiz-Pesini E, Mishmar D, Procaccio V, Lott MT, Nguyen KC, Spolim S, Patil U, Baldi P, Wallace DC (2009) MITOMASTER: a bioinformatics tool for the analysis of mitochondrial DNA sequences. Hum Mutat 1:1–6
Cann RL (2001) Genetic clues to dispersal in human populations: retracing the past from the present. Science 291(5509):1742–1748
Cavalli-Sforza LL (1996) The spread of agriculture and nomadic pastoralism: insights from genetics, linguistics, and archaeology. In: The origins and spread of agriculture and pastoralism in Eurasia. University College London Press, London, pp 51–59
Cavalli-Sforza LL, Piazza A, Menozzi P, Mountain J (1988) Reconstruction of human evolution: bringing together genetic, archaeological, and linguistic data. Proc Natl Acad Sci 85(16):6002–6006
Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton
Chandrasekar A, Kumar S, Sreenath J, Sarkar BN, Urade BP, Mallick S, Bandopadhyay SS, Barua P, Barik SS, Basu D, Kiran U (2009) Updating Phylogeny of Mitochondrial DNA Macrohaplogroup M in India: dispersal of modern human in South Asian Corridor. PloS One 4(10):e7447
Chaubey G, Karmin M, Metspalu E, Metspalu M, Selvi-Rani D, Singh VK, Parik J, Solnik A, Naidu BP, Kumar A, Adarsh N (2008) Phylogeography of mtDNA haplogroup R7 in the Indian peninsula. BMC Evol Biol 8(1):227
Cordaux R, Saha N, Bentley GR, Aunger R, Sirajuddin SM, Stoneking M (2003) Mitochondrial DNA analysis reveals diverse histories of tribal populations from India. Eur J Hum Genet 11(3):253–264
Cordaux R, Aunger R, Bentley G, Nasidze I, Sirajuddin SM, Stoneking M (2004) Independent origins of Indian caste and tribal paternal lineages. Curr Biol 14(3):231–235
De Fanti S, Barbieri C, Sarno S, Sevini F, Vianello D, Tamm E, Metspalu E, Van Oven M, Hübner A, Sazzini M, Franceschi C (2015) Fine dissection of human mitochondrial DNA haplogroup HV lineages reveals Paleolithic signatures from European glacial refugia. PloS One 10(12):e0144391
Derenko M, Malyarchuk B, Bahmanimehr A, Denisova G, Perkova M, Farjadian S, Yepiskoposyan L (2013) Complete mitochondrial DNA diversity in Iranians. PloS One 8(11):e80673
Edwin D, Vishwanathan H, Roy S, Usha Rani MV, Majumder PP (2002) Mitochondrial DNA diversity among five tribal populations of southern India. Curr Sci 83(2):25
Hall TA (1999) Bio Edit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–8
Irwin JA, Ikramov A, Saunier J, Bodner M, Amory S, Rock A, O’Callaghan J, Nuritdinov A, Atakhodjaev S, Mukhamedov R, Parson W (2010) The mtDNA composition of Uzbekistan: a microcosm of Central Asian patterns. Int J Legal Med 124(3):195–204
Khan NA, Govindaraj P, Jyothi V, Meena AK, Thangaraj K (2013) Co-occurrence of m. 1555A>G and m. 11778G>A mitochondrial DNA mutations in two Indian families with strikingly different clinical penetrance of Leber hereditary optic neuropathy. Mol Vis 19:1282
Kim K, Brenner CH, Mair VH, Lee KH, Kim JH, Gelegdorj E, Batbold N, Song YC, Yun HW, Chang EJ, Lkhagvasuren G (2010) A western Eurasian male is found in 2000-year-old elite Xiongnu cemetery in Northeast Mongolia. Am J Phys Anthropol 142(3):429–440
Kivisild T, Bamshad MJ, Kaldma K, Metspalu M, Metspalu E, Reidla M, Laos S, Parik J, Watkins WS, Dixon ME, Papiha SS (1999a) Deep common ancestry of Indian and Western-Eurasian mitochondrial DNA lineages. Curr Biol 9(22):1331–1334
Kivisild T, Kaldma K, Metspalu M, Parik J, Papiha S, Villems R (1999b) The place of the Indian mitochondrial DNA variants in the global network of maternal lineages and the peopling of the Old World. In: Genomic diversity. Springer, Boston, pp 135–152
Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, Metspalu E, Adojaan M, Tolk HV, Stepanov V, Golge M (2003) The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet 72(2):313–332
Kumar S, Padmanabham PB, Ravuri RR, Uttaravalli K, Koneru P, Mukherjee PA, Das B, Kotal M, Xaviour D, Saheb SY, Rao VR (2008) The earliest settlers’ antiquity and evolutionary history of Indian populations: evidence from M2 mtDNA lineage. BMC EvolBiol 8(1):230
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Lahr MM, Foley RA (1998) Towards a theory of modern human origins: geography, demography, and diversity in recent human evolution. Am J Phys Anthropol 107(S27):137–176
Lewin R (1987) Africa: cradle of modern humans. Science 237:1292–1296
Li C, Li H, Cui Y, Xie C, Cai D, Li W, Mair VH, Xu Z, Zhang Q, Abuduresule I, Jin L (2010) Evidence that a West-East admixed population lived in the Tarim Basin as early as the early Bronze Age. BMC Biol 8(1):15
Majumder PP (2001) Ethnic populations of India as seen from an evolutionary perspective. J Biosci 26(4):533–545
Majumder PP (2010) The human genetic history of South Asia. Curr Biol 20(4):R184–R187
Malyarchuk B, Grzybowski T, Derenko M, Perkova M, Vanecek T, Lazur J, Gomolcak P, Tsybovsky I (2008) Mitochondrial DNA phylogeny in Eastern and Western Slavs. Mol Biol Evol 25(8):1651–1658
Margaryan A, Derenko M, Hovhannisyan H, Malyarchuk B, Heller R, Khachatryan Z, Avetisyan P, Badalyan R, Bobokhyan A, Melikyan V, Sargsyan G (2017) Eight millennia of matrilineal genetic continuity in the south Caucasus. Curr Biol 27(13):2023–2028
McAlpin D, Emeneau MB, Jacobsen WH Jr, Kuiper FB, Paper HH, Reiner E, Stopa R, Vallat F, Wescott RW (1975) Elamite and dravidian: further evidence of relationship. Curr Anthropol 16(1):105–115
Metspalu M, Kivisild T, Metspalu E, Parik J, Hudjashov G, Kaldma K, Serk P, Karmin M, Behar DM, Gilbert MT, Endicott P (2004) Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet 5(1):26
Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI (2003) Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci 100(1):171–176
Palanichamy MG, Sun C, Agrawal S, Bandelt HJ, Kong QP, Khan F, Wang CY, Chaudhuri TK, Palla V, Zhang YP (2004) Phylogeny of mitochondrial DNA macrohaplogroup N in India based on complete sequencing: implications for the peopling of South Asia. Am J Hum Genet 75(6):966–978
Palanichamy MG, Mitra B, Zhang CL, Debnath M, Li GM, Wang HW, Agrawal S, Chaudhuri TK, Zhang YP (2015) West Eurasian mtDNA lineages in India: an insight into the spread of the Dravidian language and the origins of the caste system. Hum Genet 134(6):637–647
Peng MS, Xu W, Song JJ, Chen X. Sulaiman X, Cai L, Liu HQ, Wu SF, Gao Y, Abdulloevich NT, Afanasevna ME (2018) Mitochondrial genomes uncover the maternal history of the Pamir populations. Eur J Hum Genet 26(1):124
Quintana-Murci L, Chaix R, Wells RS, Behar DM, Sayar H, Scozzari R, Rengo C, Al-Zahery N, Semino O, Santachiara-Benerecetti AS, Coppa A (2004) Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am J Hum Genet 74(5):827–845
Rajkumar R, Banerjee J, Gunturi HB, Trivedi R, Kashyap VK (2005) Phylogeny and antiquity of M macrohaplogroup inferred from complete mtDNA sequence of Indian specific lineages. BMC Evol Biol 5:26
Ranaweera L, Kaewsutthi S, Win Tun A, Boonyarit H, Poolsuwan S, Lertrit P (2014) Mitochondrial DNA history of Sri Lankan ethnic people: their relations within the island and with the Indian sub-continental populations. J Hum Genet 59:28–36
Reich D, Thangaraj K, Patterson N, Price AL, Singh L (2009) Reconstructing Indian population history. Nature 461(7263):489–494
Renfrew C (1996) Languages families and the spread of farming. In: Harris DR (ed) The origins and spread of agriculture and pastoralism in Eurasia. Smithsonian Institution Press, Washington, DC, pp 70–92
Rieder MJ, Taylor SL, Tobe VO, Nickerson DA (1998) Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res 26:967–973
Roychoudhury S, Roy S, Dey B, Chakraborty M, Roy M, Prabhakaran A, Ramesh B, Vishwanathan H (2000) Fundamental genomic unity of ethnic India is revealed by analysis of mitochondrial DNA. Curr Sci 79(9):1182–1192
Roychoudhury S, Roy S, Basu A, Banerjee R, Vishwanathan H, Rani MU, Sil SK, Mitra M, Majumder PP (2001) Genomic structures and population histories of linguistically distinct tribal groups of India. Hum Genet 109(3):339–350
Sahakyan H, Kashani BH, Tamang R, Kushniarevich A, Francis A, Costa MD, Pathak AK, Khachatryan Z, Sharma I, Van Oven M, Parik J (2017) Origin and spread of human mitochondrial DNA haplogroup U7. Sci Rep 7:46044
Sahoo S, Singh A, Himabindu G, Banerjee J, Sitalaximi T, Gaikwad S, Trivedi R, Endicott P, Kivisild T, Metspalu M (2006) A prehistory of Indian Y chromosomes: evaluating demic diffusion scenarios. Proc Natl Acad Sci USA 103:843–848
Saillard J, Forster P, Lynnerup N, Bandelt HJ, Norby S (2000) MtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 67(3):718–726
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schonberg A, Theunert C, Li M, Stoneking M, Nasidze I (2011) High-throughput sequencing of complete human mtDNA genomes from the Caucasus and West Asia: high diversity and demographic inferences. Eur J Hum Genet 19(9):988
Soares P, Ermini L, Thomson N, Mormina M, Rito T, Röhl A, Salas A, Oppenheimer S, Macaulay V, Richards MB (2009) Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet 84(6):740–759
Stringer CB, Grun R, Schwarcz HP, Goldberg P (1989) ESR dates for the hominid burial site of Es Skhul in Israel. Nature 338(6218):756–758
Sun C, Kong QP, Palanichamy MG, Agrawal S, Bandelt HJ, Yao YG, Khan F, Zhu CL, Chaudhuri TK, Zhang YP (2005) The dazzling array of basal branches in the mtDNA macrohaplogroup M from India as inferred from complete genomes. Mol Biol Evol 23(3):683–690
Terreros MC, Rowold DJ, Mirabal S, Herrera RJ (2011) Mitochondrial DNA and Y-chromosomal stratification in Iran: relationship between Iran and the Arabian Peninsula. J Hum Genet 56(3):235–246
Thangaraj K, Chaubey G, Kivisild T, Reddy AG, Singh VK, Rasalkar AA, Singh L (2005) Reconstructing the origin of Andaman Islanders. Science 308(5724):996
Thangaraj K, Chaubey G, Singh VK, Vanniarajan A, Thanseem I, Reddy AG, Singh L (2006) In situ origin of deep rooting lineages of mitochondrial macrohaplogroup M in India. BMC Genom 7(1):151
Thangaraj K, Chaubey G, Kivisild T, Rani DS, Singh VK, Ismail T, Carvalho-Silva D, Metspalu M, Bhaskar LV, Reddy AG, Chandra S (2008) Maternal footprints of Southeast Asians in North India. Hum Hered 66(1):1–9
Thangaraj K, Nandan A, Sharma V, Sharma VK, Eaaswarkhanth M, Patra PK, Singh S, Rekha S, Dua M, Verma N, Reddy AG (2009) Deep rooting in-situ expansion of mtDNA Haplogroup R8 in South Asia. PloS One 4(8):e6545
Thorpe IJ (1999) The origins of agriculture in Europe. Psychology, Routledge
Van Oven M, Kayser M (2009) Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 30(2):E386–E394
Vyas DN, Kitchen A, Miro-Herrans AT, Pearson LN, Al-Meeri A, Mulligan CJ (2016) Bayesian analyses of Yemeni mitochondrial genomes suggest multiple migration events with Africa and Western Eurasia. Am J Phys Anthropol 159(3):382–393
Zvelebil M (1980) The rise of the nomads in Central Asia. In: Sherratt A (ed) The Cambridge encyclopedia of archaeology. Crown, New York, pp 252–256
Acknowledgements
We acknowledge the Director, Anthropological Survey of India, and also express our gratitude to Dr. C. R. Satyanarayanan, Deputy Director and Head of Office, Anthropological Survey of India, SRC, Mysore. The authors also thank the Chairperson, Department of studies in Zoology, University of Mysore for their assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sylvester, C., Krishna, M.S., Rao, J.S. et al. Neolithic phylogenetic continuity inferred from complete mitochondrial DNA sequences in a tribal population of Southern India. Genetica 146, 383–389 (2018). https://doi.org/10.1007/s10709-018-0030-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-018-0030-2