Abstract
The Mlo gene was discovered in barley because the mutant ‘mlo’ allele conferred broad-spectrum, non-race-specific resistance to powdery mildew caused by Blumeria graminis f. sp. hordei. The Mlo genes also play important roles in growth and development of plants, and in responses to biotic and abiotic stresses. The Mlo gene family has been characterized in several crop species, but only a single legume species, soybean (Glycine max L.), has been investigated so far. The present report describes in silico identification of 18 CcMlo and 20 PvMlo genes in the important legume crops Cajanus cajan (L.) Millsp. and Phaseolus vulgaris L., respectively. In silico analysis of gene organization, protein properties and conserved domains revealed that the C. cajan and P. vulgaris Mlo gene paralogs are more divergent from each other than from their orthologous pairs. The comparative phylogenetic analysis classified CcMlo and PvMlo genes into three major clades. A comparative analysis of CcMlo and PvMlo proteins with the G. max Mlo proteins indicated close association of one CcMlo, one PvMlo with two GmMlo genes, indicating that there was no further expansion of the Mlo gene family after the separation of these species. Thus, most of the diploid species of eudicots might be expected to contain 15–20 Mlo genes. The genes CcMlo12 and 14, and PvMlo11 and 12 are predicted to participate in powdery mildew resistance. If this prediction were verified, these genes could be targeted by TILLING or CRISPR to isolate powdery mildew resistant mutants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mlo (mildew resistance locus o) gene family is one of the largest seven transmembrane (TM) domain protein encoding gene families in plants. This gene family is reported in few mosses, but is ubiquitous in all higher plants (Devoto et al. 1999), indicating that the family evolved at the time when the first land plants appeared and, that it performs some essential functions in these plants. An extensive search for resistance to powdery mildew led to the discovery of mutant ‘mlo’ gene (Hvmlo1; Hordeum vulgare Mlo) in barley (Büschges et al. 1997). This recessive Hvmlo1 allele represents a loss of Mlo function, and is associated with broad-spectrum, non-race-specific resistance against powdery mildew disease in barley caused by Blumeria graminis f. sp. hordei (Jørgensen 1992; Piffanelli et al. 2002). The Hvmlo1-mediated resistance is durable under the field conditions, and has been used to develop powdery mildew resistant barley varieties that have been cultivated in the disease prone areas (Jørgensen 1992).
The recessive mlo allele leads to the formation of cell wall appositions (Jørgensen 1992) and spontaneous cell death, resulting in physical obstruction to the development of the fungal pathogen (Jørgensen 1992; Humphry et al. 2011). Thus, the expression of dominant Mlo allele is essential for the development of powdery mildew disease (Panstruga 2005a). Gene expression analysis in grapevine (Vitis vinifera) showed that four of the 17 VvMlo (V. vinifera Mlo) genes were over-expressed in response to infection by powdery mildew (Erysiphe necator), strengthening the conclusion that Mlo function is required for susceptibility to the pathogen (Feechan et al. 2009). These results are consistent with the observation that an over-expression of the Mlo gene leads to hyper-susceptibility to powdery mildew (Kim et al. 2002a, b). The Mlo gene product forms channels in the plasma membrane, through which the powdery mildew fungal hyphae penetrate into the host cells (O’Connell and Panstruga 2006) during a compatible interaction.
Mlo genes are thought to be reminiscent of G-protein coupled receptors (GPCRs) with respect to their TM topology and plasma membrane localization (Devoto et al. 1999), and they may be involved in G-protein activation (Kim et al. 2002a). In case of Arabidopsis thaliana, there was modulation of mutant AtMlo2-mediated powdery mildew resistance by the Gβ and Gγ subunits of the G-protein and that of callose deposition by the Gγ1 subunit. But the roles of Mlo proteins do not entirely match those of typical G-proteins (Lorek et al. 2013). Mlo genes are implicated in regulation of the Ca2+ ion-mediated channel for inter- and intra-cellular trafficking involved in diverse plant developmental and physiological processes, cellular signaling, and several other biotic and abiotic stress responses (Piffanelli et al. 2002; Zielinski 1998). The Mlo proteins associate with ‘calmodulin’ (CaM), the protein with calcium binding domain, and have a conserved CaM binding domain (CaMBD) at the proximal ends of their seventh TM domain (Kim et al. 2002a, b). The Mlo gene family has recently been shown to be involved in responses to hormone treatment and metal ion stress (Cheng et al. 2012; Lim and Lee 2014).
Büschges et al. (1997) identified a novel protein of 60.4 kDa encoded by a Mlo gene with ‘hairpin structure’ of seven TM domains that is regarded as the ‘topological signature’ (Devoto et al. 1999) of this family. Confocal microscopy studies using GFP-tagged Mlo proteins have shown that these proteins are localized in the plasma membrane with their seven helices spanning across the membrane (Devoto et al. 1999, 2003; Kim and Hwang 2012). Patterns of protease digestion revealed that the membrane spanning Mlo proteins are 60 % exposed into the cytoplasm and only 25 % of their amino acid residues are embedded in the plasma membrane (Devoto et al. 1999). The N-termini of Mlo proteins project out into the extracellular matrix, while their C-termini are located in the cell cytoplasm. Further, deletion of C-terminal regions of Mlo proteins reduces half of their ability to confer susceptibility to the powdery mildew pathogen in barley (Elliott et al. 2005).
Although legumes are crops of considerable economic importance, the lack of genome sequence data has limited the application of genomics-based approaches for their improvement. For example, Mlo genes have been studied in cereals like barley, wheat (Konishi et al. 2010; Wang et al. 2014), rice (Liu and Zhu 2008) and sorghum (Singh et al. 2012a), and in several eudicot species, viz., A. thaliana (Devoto et al. 1999), soybean (Shen et al. 2012), roses (Kaufmann et al. 2012), tomato, Vitis and Cucumis (Bai et al. 2008; Feechan et al. 2009; Zhou et al. 2013), apple, peach and strawberry (Pessina et al. 2014), tobacco (Appiano et al. 2015), etc. However, a comprehensive study of the Mlo gene family in legumes is limited to soybean. Therefore, this study was taken up to identify and characterize the putative members of Mlo gene family in the grain legumes like pigeonpea (Cajanus cajan L. Millsp.) and common bean (Phaseolus vulgaris L.), both members of the Milletoids clade (http://tolweb.org/Fabaceae/). Pigeonpea is the first grain legume whose genome sequence became available in the public domain (Singh et al. 2012b; Varshney et al. 2012; Kudapa et al. 2012) and the genome sequence of common bean was recently made available (Schmutz et al. 2014) for studies.
The present study identified 18 C. cajan Mlo (CcMlo) and 20 P. vulgaris Mlo (PvMlo) family members. Since these two species are closely related to G. max (Li et al. 2014), we carried out a comparative phylogenetic analysis of the CcMlo and PvMlo gene families with the previously identified 39 GmMlo (G. max Mlo; Deshmukh et al. 2014) members to investigate their evolutionary relationships. To the best of our knowledge this is the first report of genome-wide and comparative analysis of Mlo gene family in these two legume crop species. The results reported here may be helpful in designing studies on expression profiling of CcMlos and PvMlos, and the inferences drawn from this study may ultimately be useful in the development of Mlo gene mutations conferring resistance to the powdery mildew pathogens.
Materials and methods
Database search and comparative phylogeny
Protein sequences corresponding to the 15 AtMlo (A. thaliana Mlo) genes were downloaded from TAIR (The Arabidopsis Information Resource; http://www.arabidopsis.org/; Swarbreck et al. 2008). Each AtMlo sequence was used as query for tBLASTn similarity search (Altschul et al. 1990) in the genome sequence databases of C. cajan (assembly Cajanus_cajan_Asha_ver1.0) and P. vulgaris (assembly PhaVulg1_0) available at the NCBI (www.ncbi.nlm.nih.gov). Protein sequences with query coverage greater than 50 % and identity >40 % were considered to be putative Mlo members. The non-redundant hits were further analyzed through BLASTp across NR (non-redundant) protein database to detect truncated, partial or incorrect protein sequences. All the protein sequences thus obtained were confirmed for the presence of Mlo domain through Pfam (http://pfam.xfam.org/; Punta et al. 2012) and InterProScan (Quevillon et al. 2005).
A multiple sequence alignment of the full-length CcMlo and PvMlo members was done using ClustalW (Thompson et al. 1994) and Clustal Omega programs (Sievers et al. 2011) keeping the default parameters, and the alignment results were checked by the BioEdit (Hall 1999) tool. The partial protein sequences (CcMlo16, 17 and 18, and PvMlo18, 19 and 20) were not included in the alignment in order to obtain more precise results. The sequence alignment was visualized using CLC Sequence Viewer 6.8.1 program (www.clcbio.com). To get an insight into the evolutionary phylogeny, we constructed different phylogenetic trees for CcMlo and PvMlo families, and in combination with the previously reported GmMlo family members in view of the high genome synteny between G. max and C. cajan (Varshney et al. 2012) and the presence of ‘shared loci’ between P. vulgaris and G. max (McClean et al. 2010). The phylogenetic trees were constructed with MEGA6 suite (Tamura et al. 2011) using Maximum likelihood estimation (MLE; to infer ancestry) method with 1000 bootstrap replications. In addition, a comparative phylogenetic tree of the four eudicots (G. max, C. cajan, P. vulgaris and A. thaliana) with the Mlo homologues of several other monocot and eudicot species was also constructed to determine orthologous relationships among the members of leguminoseae and other eudicots and monocots.
Characterization of Mlo genes
The Fgenesh (http://linux1.softberry.com/) and GENSCAN (http://genes.mit.edu/; Burge and Karlin 1997) online servers were used to detect/confirm the Mlo gene structures and the positions of transcription start sites and polyA tails in the Mlo members from the two species. Each incomplete/partial protein sequence was extended on the genome till the complete protein coding sequence with a start and a stop codon was obtained. The protein sequences predicted by Fgenesh in C. cajan were submitted to Pfam and InterProScan to confirm the presence of the signature Mlo domain (Pfam- PF03094 and InterProScan- IPR004326). Since the Mlo protein prediction for P. vulgaris had already been submitted at the NCBI, we did not attempt to predict the PvMlo protein sequences using Fgenesh, and full length protein sequences from NCBI were used for further analyses. However, the partial PvMlo proteins were extended on the genome to fetch complete sequences through Fgenesh. Fgenesh was also used to identify the putative coding sequences (mRNAs) in the Mlo genes. The visualization of exon and intron boundaries was done using the Gene Structure Display Server (GSDS; http://gsds.cbi.pku.edu.cn/; Guo et al. 2007). The TM domains were identified using the HMMTOP 2.0 server (http://www.enzim.hu/hmmtop/; Tusnády and Simon 2001) and the sub-cellular localization of the Mlo proteins was predicted using CELLO v.2.5 (http://cello.life.nctu.edu.tw/; Yu et al. 2006).
The molecular weight, theoretical pI and aliphatic indices of the identified full-length CcMlo and PvMlo proteins were computed using Expasy based ProtParam server (http://web.expasy.org/protparam/). The cis-acting elements present in the 1,000 bp upstream regions, i.e., the putative promoter regions, of the CcMlo and PvMlo genes were predicted by the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/; Lescot et al. 2002) program. We also performed a STRING (Search Tool for the Retrieval of Interacting Genes/Proteins database version 9.1; Franceschini et al. 2013) analysis to study the protein–protein interaction of CcMlo and PvMlo proteins. The functionally conserved motifs present in the Mlo proteins were investigated using MEME (Multiple Expectation Maximization for motif Elicitation; http://meme.nbcr.net/; Bailey et al. 2006) tool at a motif width of 25–40 with maximum motif number of 40. A comparative combined motif analysis of GmMlo, PvMlo and CcMlo members at similar motif width was also performed to retrieve the conserved motifs universally present in the three species.
Results
Identification of CcMlo and PvMlo genes
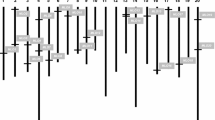
Genome-wide BLAST search using the AtMlo proteins as query against the C. cajan and P. vulgaris genome sequences resulted in 29 and 30 hits, respectively. After removal of redundant scaffolds and incomplete scaffold data, a total of 18 CcMlo and 20 PvMlo paralogs were obtained, but the sequences of three genes in each of the CcMlo and PvMlo families were incomplete. As the chromosomal locations of the CcMlo and PvMlo genes were not known, we used the following method to name these members. The protein sequences of the full-length CcMlo and PvMlo proteins were used for multiple sequence alignment and construction of phylogenetic trees by MLE. The genes were numbered from CcMlo1 to CcMlo15 and PvMlo1 to PvMlo17, beginning at the bottom of the tree (Fig. 1) on the basis of their corresponding orthologous pairing since the bulk of CcMlo and PvMlo members formed 1:1 terminal pairs (Table 1; Fig. 1). The designation of CcMlo15 to CcMlo17 and of PvMlo15 to PvMlo19 was essentially arbitrary, since CcMlo15, PvMlo15, and PvMlo16 were present as additional members with different CcMlo-PvMlo pairs. PvMlo17 formed a distinct terminal branch, and CcMlo16, CcMlo17, CcMlo18, PvMlo18, PvMlo19 and PvMlo20 were partial genes, and were excluded from the alignment.
Phylogenetic classification of CcMlo and PvMlo members along with the gene organization. The phylogenetic tree was constructed using Maximum likelihood estimation by MEGA 6.0 software with 1000 bootstrap replications, and the bootstrap support is indicated at each node. The members were grouped into three major clusters. The branch lengths indicate number of substitutions per site. The gene organization was visualized through GSDS server. The green blocks represent exons, while thin lines denote introns. The blue lines in the beginning and at the end of the gene indicate the 5′ and 3′ UTR regions, respectively. 0, 1 and 2 signify intron phase, and the intron and exon positions are depicted in kb scale. The gene scale is drawn immediately below the individual Mlo genes
Transmembrane domain conservation and orthologous relationships
A comparative multiple sequence alignment of the predicted CcMlo and PvMlo proteins visualized by CLC sequence viewer (Online resource Fig. 1) showed wide gaps in the inter-transmembrane (inter-TM) regions TM2–TM3, TM4–TM5, and TM5–TM6 in four CcMlo and one PvMlo members. For example, CcMlo4 had an insertion of about 88 amino acids between positions 533 and 621 (Online resource Fig. 1) and PvMlo1 had 35 amino acid residues inserted between positions 465 and 514 (Online resource Fig. 1). In addition, there were numerous relatively short deletions concentrated in the TM2-TM3 inter-TM and the CaMBD regions. But some large deletions were also observed, e.g., the deletion in CcMlo12 involved amino acid residues between positions 88 and 287. In contrast, there was overall sequence conservation in the TM regions. Most of the invariant residues detected by Elliott et al. (2005) were present in PvMlo and GmMlo members, but the CcMlos had few invariant residues (Table 2).
The phylogenetic trees constructed using the MLE showed strong bootstrap support. All the phylogenetic trees comprised three major clusters (Figs. 1, 2). The cluster I showed greater expansion than the other two clusters, suggesting that these paralogs might be involved in diverse essential cellular functions. Alternatively, the incidence of duplications in the genomic regions harboring the cluster I genes might have been much higher than those in the genomic regions having clusters II and III genes. In general, one CcMlo and one PvMlo proteins formed closely related pairs (Fig. 1). In the combined tree of CcMlo, PvMlo and AtMlo proteins (Online resource Fig. 2a), the CcMlo and PvMlo members generally showed one-to-one close association, while the AtMlo proteins showed much greater divergence from them; this is consistent with the known taxonomic relationships of the three species.
The comparative phylogenetic analysis of the Milletoids Mlo proteins, viz., CcMlo, PvMlo and GmMlo proteins, showed association of one CcMlo, one PvMlo and two GmMlo members in nearly all the terminal groupings (Fig. 2). G. max is known to have undergone two rounds of whole genome duplication (WGD). The first WGD occurred in the papilionoids clade around 54 Myr ago in all the legumes. But G. max underwent a second WGD around 13 Myr ago, which led to the doubling of chromosomes followed by diploidization and stabilization at the present 2n = 40. Hence, the terminal grouping pattern of CcMlo, PvMlo and GmMlo proteins is consistent with the occurrence of the second WGD event in G. max after its divergence from C. cajan and P. vulgaris (Schmutz et al. 2010). In a majority (six) of the groups, the CcMlo proteins were more divergent than the PvMlo proteins from the GmMlo members. But in three terminal groups, the PvMlo members were more divergent than the CcMlo members from the GmMlo proteins, and in two groups both CcMlo and PvMlo members were equally divergent from the GmMlos. In one of the clusters, one CcMlo, two PvMlo and three GmMlo members were remarkably close to each other and showed very little divergence. These observations are not entirely consistent with the reported evolutionary history of these three species (Li et al. 2014), and would suggest differential rates of divergence of the Mlo orthologs in the different species. The CcMlo, PvMlo and GmMlo members showing homology with the corresponding AtMlos have been presented in Table 1, based on the clade classification of the four eudicots (Online resource Fig. 2b). The comparative analysis of CcMlo and PvMlo with several other eudicots and monocots also resulted in similar clade classification (Online resource Fig. 2b).
Relative resemblance of gene structure and protein topology
The gene structure studied through Fgenesh and GENSCAN showed that Mlo genes have mostly 14 exons (range 12–16; Tables 3, 4). The total gene length in C. cajan varied from 3813 bp (CcMlo9) to 8432 bp (CcMlo11; Table 3). Similarly, the total gene length for PvMlo varied from 3803 bp (PvMlo2) to 6533 bp (PvMlo3; Table 4). On an average, it appeared that PvMlo genes were shorter than the orthologous CcMlo genes. The gene structure visualized through GSDS showed variable arrangement of exons and introns in both CcMlo and PvMlo genes (Fig. 1), but the number of exons did not vary widely. This conservation was also observed among the Mlo genes of G. max, A. thaliana, rice, wheat and sorghum (Liu and Zhu 2008; Konishi et al. 2010; Singh et al. 2012a). The P. vulgaris chromosome 9 carried 4 PvMlo genes; chromosomes 5 and 11 carried three genes each, while chromosome 8 did not have any PvMlo gene. The chromosomes 1, 3, 7 and 10 carried only one PvMlo member each, and the remaining three chromosomes had two PvMlo genes each. The PvMlo genes could not be mapped due to the non-availability of their relative positions in the concerned chromosomes. The chromosome organization data for C. cajan is not available (as on 02/09/2015), due to which the chromosomal locations of the CcMlo genes could not be determined.
The protein topology studied through HMMTOP 2.0 predicted 6–10 TM domains (Tables 3, 4), but only 7 TM domains are marked in the multiple sequence alignment. The subcellular localization of the CcMlo and PvMlo proteins predicted through CELLO indicated them to be localized in the plasma membrane as integral membrane proteins. The analysis of amino acid composition revealed that most of the CcMlo (13 out of 15 full length proteins) and PvMlo (16 out of 17 full length proteins) members were rich in leucine (8.9–12.2 %; Online resource Table 1a–b). The GRAVY (grand hydropathicity index) values indicated that the Mlo protein family consists of both hydrophobic and hydrophilic members, which can be expected due to their TM localization. The molecular weights of CcMlo members ranged from 56132.0 to 67643.3 Da, while those for the PvMlos varied from 55061.0 to 68770.8 Da. The CcMlo proteins formed two molecular weight groups; five proteins were of 56–59 kDa, while another six were of 61–63 kDa. But the PvMlos had one major cluster of 10 proteins of 61–64 kDa.
Probable functions of CcMlo and PvMlo members
The analysis of 1000 bp upstream regions of the CcMlo and PvMlo genes detected the following response elements: circadian, GCN4-motif, RY-element, skn-1 motif, etc. (involved in development), AT-rich and Box-W1 (for fungal elicitor response), ABRE, ERE, GARE-motif, TATC-box, etc. (concerned with hormone response), and HSE, LTR, MBS, etc. (for abiotic stress response; Online resource Tables 2a–c and 3a–c). The most common regulatory elements detected were related to hormone and stress (biotic/abiotic) responses. Surprisingly, only two CcMlo and two PvMlo genes had pathogen and fungal elicitor response elements. The protein interaction study of individual CcMlo and PvMlo proteins with respect to their homology with the AtMlo proteins was studied using STRING v 9.1. The AtMlo homolog with the highest score was used to study the interaction. Most of the CcMlo and PvMlo members showed interaction similarity with the AtMlo members 1, 4, 9, 6, 8, 11, 12 and 13 (Online resource Table 3).
The identification of conserved motifs through MEME at a minimum and maximum motif widths of 25 and 40, respectively, for CcMlo, PvMlo and GmMlo members (Fig. 3a) revealed the highly conserved motif 3 and a less conserved motif 7 in all the Mlos (Fig. 3b). The motif 7 had a highly conserved LEETPTW sequence within the TM domain 1 (Fig. 3c). This sequence was also conserved in the Mlo proteins of soybean, Arabidopsis, wheat, maize and rice (data not shown; Elliott et al. 2005; Shen et al. 2012). The L, T, P, T and W residues were either invariant or highly conserved, and they were substituted in only 4, 3, 4, 1 and 12 Mlo proteins, respectively. Further, only one of these substitutions (L replaced by H in GmMlo22) was such that would affect protein structure. In contrast, the EE residues were much less conserved and few of their replacements would affect protein structure (Online resource Table 4). This LEETPTW sequence was searched in the CDD (conserved domain database), Pfam, Prosite and InterProScan, and was found to be present exclusively in Mlo proteins.
Determination of conserved motifs. a A combined block diagram of conserved motifs, obtained through MEME suite, of the 15 full-length CcMlos and the 17 full-length PvMlos compared with the 39 GmMlo proteins at a width of 25–40 amino acids. b The highly and evenly conserved motif 3 of Mlo proteins. c The motif block 7 of the CcMlo and PvMlo members showing the LEETPTW consensus sequence
Discussion
The Mlo gene family lineage
The Mlo gene family has recently gained importance due to its ubiquitous presence in higher plants and their classification into the plant defense-related gene family. The Mlo genes have provided durable resistance in barley without being counteracted by the powdery mildew pathogen (Atzema 1998). BLAST search identified 18 Mlo genes in C. cajan and 20 genes in P. vulgaris, which are higher than those in A. thaliana and approximately half of the number reported in G. max (Deshmukh et al. 2014). This is as expected, since C. cajan and P. vulgaris are diploid species, while G. max is diploidized tetraploid (Schmutz et al. 2010). Thus, it seems that there was no further expansion of the Mlo gene family after G. max, C. cajan and P. vulgaris diverged from their common ancestor. Further, the model plant A. thaliana has 15 Mlo genes (Devoto et al. 2003), and 17 and 14 Mlo genes have been reported in the diploid Vitis vinifera and Cucumis sativus, respectively (Feechan et al. 2009; Zhou et al. 2013). Similarly, peach and strawberry are reported to have 19 and 18 Mlo genes, respectively. Apple genome is reported to harbor 21 Mlo genes, of which MdMlo11 and 19 are known to have been generated due to a recent duplication event (Pessina et al. 2014). The tobacco and potato genomes are reported to have 15 and 13 Mlo genes, respectively (Appiano et al. 2015). Hence, most of the diploid eudicot species might be expected to contain between 15 and 20 Mlo genes.
Sequence analysis and phylogenetics of CcMlo and PvMlo proteins
The amino acid sequences of the 7 TM domains in the Mlo proteins from monocots like rice (OsMlo), wheat (TaMlo) and sorghum (SbMlo) (data not shown), and from the eudicots A. thaliana, G. max, C. cajan and P. vulgaris showed much more conservation than the inter-TM regions, suggesting that the TM sequences are necessary for Mlo protein function. The TM regions 1, 3, 4 and 7 showed greater conservation than the other three TM domains, and there were more insertions/deletions in monocots than in legumes. Further, we identified the ‘LEETPTW’ motif present in TM1 of Mlo proteins; the L, T, P, T, and W residues of this motif were highly conserved in all the species. Similarly, Panstruga (2005b) reported a four amino acids motif D/E–F-S/T-F to be present in the C-terminal region of the Mlo proteins involved in powdery mildew resistance. We analyzed the Mlo proteins from several species and noted that the Mlo proteins from the different species showed the predominance of different amino acid combinations of this motif. For example, G. max, C. cajan, P. vulgaris and Lotus japonicus Mlos had predominance of E–F-S-F sequence. The motif D/E–F-S/T-F was present in CcMlo8, 11, 12 and 13, and in PvMlo8, 11, 12, 13, 14 and 15, suggesting that they might be involved in powdery mildew resistance (Online resource Table 5). Mutants of these genes may be produced by suitable techniques like TILLING (Targeting Induced Local Lesion in Genome) or CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) and the disease reaction of the mutants may be assessed to verify the above predictions. Alternatively, these genes may be cloned and used for genetic transformation to determine their roles in the powdery mildew disease (Acevedo-Garcia et al. 2014).
Elliott et al. (2005) identified 30 invariant amino acid residues from analysis of 38 Mlo proteins belonging to A. thaliana, wheat, maize and rice. A comparison of the invariant residues in the Mlo proteins of the three legume species with those reported by Elliott et al. (2005) revealed that residues Q, F and F in the inter-TM3-TM4 region were the only invariant amino acids in the 99 Mlo proteins (71 legume, 15 Arabidopsis and 13 cereal Mlo members) from different plant species. Further, CcMlo proteins had considerably fewer invariant residues than the Mlo proteins of other legume species. The insertions observed in CcMlo members were not detected in the orthologous PvMlo proteins, indicating that these events had occurred after the separation of the two species. All the CcMlo and PvMlo proteins possessed the CaM binding domain, which is known to be a conserved domain among plant proteins involved in defense response and calcium signaling (Zielinski 1998).
A comparative phylogenetic analysis of the 15 full-length CcMlo members with the 17 full-length PvMlo members using the MLE method showed that each CcMlo member was much closer to one PvMlo member rather than to another CcMlo protein (Fig. 1). This would indicate that the PvMlo and CcMlo orthologs have evolved from common ancestral sequences, and that C. cajan and P. vulgaris diverged sometime after the last expansion event of the Mlo gene family in the ancestral species. The CcMlo14 had two PvMlo orthologs, the PvMlo14 and 15; these PvMlo paralogs seem to be the result of a recent duplication event involving the PvMlo ortholog of CcMlo14. The exon–intron organization of the PvMlo14 and 15 genes were similar. The divergence in Mlo gene family paralogs/orthologs have occurred mainly due to substitution mutations, but insertions and deletions also appear to be involved; amino acid insertions/deletions would confer structural and functional variation to the proteins (Vetter et al. 1996). Thus, the different CcMlo and PvMlo genes are likely to be involved in non-overlapping and important functions affecting fitness of the concerned species. The simultaneous conservation of paralogous members is sought to be explained by the protein–protein interactions of their gene products (Pereira-Leal et al. 2007).
Structural conservation among orthologs
The Mlo family members in the two legume species shared similar gene organization and had comparable number of exons as in Arabidopsis, rice and barley with a ‘conserved intron–exon junction’ (Devoto et al. 1999). The exon–intron organization seemed to be more similar among the orthologs than among the paralogs (Table 3; Fig. 1). The average size of PvMlo genes was smaller (4726 bp) than that of CcMlo genes (5738 bp) most likely due to differences in their intron sizes. However, the number of amino acids in the concerned proteins also showed some variation (range 503 to 562 in 12 of the CcMlos and from 556 to 585 in 14 of the PvMlos). The paralogous CcMlo and PvMlo members seemed to have greater diversity in terms of their molecular weights, amino acid lengths and physico-chemical properties than their orthologous mates. The paralogous members are proposed to survive through either sub-functionalization or neo-functionalization while retaining the basic function of the family (Sanchez-Perez et al. 2008). Being ancestrally related to GPCRs, Mlo proteins have been shown to be associated with some functions of GPCRs like regulation of hormonal signaling (Chen et al. 2009), participation in developmental pathways (seed-, shoot- and root-specific expression; Konishi et al. 2010), stress related mechanisms (Shen et al. 2012), and defense responses (Büschges et al. 1997; Bai et al. 2008). However, whether Mlo proteins function independent of GPCRs or participate in the same defense signaling cascade as GPCRs is not clear (Lorek et al. 2013).
The cis-acting regulatory element analysis of the putative promoter regions of the CcMlo and PvMlo members suggested a variety of functions of these proteins. These predictions need to be verified by gene expression analyses even though they tend to agree with the reported expression patterns for Mlo genes of G. max (Shen et al. 2012). The Mlo proteins are known to be involved in response to fungal pathogen attack (Kim and Hwang 2012; Feechan et al. 2009). They are also suspected to be among the developmentally and physiologically expressed gene families (Chen et al. 2009). For example, Mlo gene expression was found to increase on ABA (abscisic acid) treatment, and in response to several biotic and abiotic stresses (Kumar et al. 2001; Chen et al. 2006; Lim and Lee 2014). All the AtMlo genes are reported to be involved in cellular defense mechanisms, possibly through cell-death pathways in response to the entry of fungal pathogen in the host (www.arabidopsis.org), indicating the evolution of Mlo gene family in response to pathogen invasions.
Currently, the STRING database maintains the functions and interactions of only AtMlo genes with other genes. STRING analysis of CcMlo and PvMlo members through AtMlo homology predicted their interaction with several other proteins involved in the defense pathway. Further, the various CcMlo and PvMlo members were grouped with individual AtMlo members suggesting possible similarities in the roles played by them. However, expression analyses and other molecular biology investigations are needed to assess the validity of the leads provided by the STRING analysis. As more and more plant species are analyzed for Mlo genes and the functions of their gene products are studied, a greater insight into the functions of these genes might be achieved. The findings from this study provide some additional information on Mlo gene family, which may be useful for studies on expression analyses of the Mlo genes of C. cajan and P. vulgaris, and selection of suitable candidate CcMlo and PvMlo members for detailed molecular investigations, including their cloning and functional characterization.
References
Acevedo-Garcia J, Kusch S, Panstruga R (2014) Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. doi:10.1111/nph.12889.2014
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Appiano M, Pavan S, Catalano D et al (2015) Identification of candidate MLO powdery mildew susceptibility genes in cultivated Solanaceae and functional characterization of tobacco NtMLO1. Transgenic Response. doi:10.1007/s11248-015-9878-4
Atzema JL (1998) Durability of mlo resistance in barley against powdery mildew caused by Erysiphe graminis f. sp. hordei. Dissertation, Swiss Federal Institute of Technology, Zurich
Bai Y, Pavan S, Zheng Z, Zappel NF, Reinstädler A, Lotti C, De Giovanni C, Ricciardi L, Lindhout P, Visser R, Theres K, Panstruga R (2008) Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of mlo function. Mol Plant Microbe Interact 21:30–39
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373
Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268:78–94
Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A et al (1997) The barley MLO gene: a novel control element of plant pathogen resistance. Cell 88:695–705
Chen Z, Hartmann HA, Wu MJ, Friedman EJ, Chen JG, Pulley M, Schulze-Lefert P, Panstruga R, Jones AM (2006) Expression analysis of the AtMLO gene family encoding plant-specific seven-transmembrane domain proteins. Plant Mol Biol 60:583–597
Chen Z, Noir S, Kwaaitaal M, Hartmann HA, Wu MJ, Mudgil Y, Sukumar P, Muday G, Panstruga R, Jones AM (2009) Two seven-transmembrane domain MILDEW RESISTANCE LOCUS O proteins cofunction in Arabidopsis root thigmomorphogenesis. Plant Cell 21:1972–1991
Cheng H, Kun W, Liu D, Su Y, He Q (2012) Molecular cloning and expression analysis of CmMlo1 in melon. Mol Biol Rep 39:1903–1907
Deshmukh R, Singh VK, Singh BD (2014) Comparative phylogenetic analysis of genome-wide Mlo gene family members from Glycine max and Arabidopsis thaliana. Mol Genet Genomics 289:345–359
Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, von Heijne G, Schulze-Lefert P (1999) Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J Biol Chem 274:34993–35004
Devoto A, Hartmann HA, Piffanelli P, Elliott C, Simmons C, Taramino G, Goh CS, Cohen FE, Emerson BC, Schulze-Lefert P, Panstruga R (2003) Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J Mol Evol 56:77–88
Elliott C, Müller J, Miklis M, Bhat RA, Schulze-Lefert P, Panstruga R (2005) Conserved extracellular cysteine residues and cytoplasmic loop–loop interplay are required for functionality of the heptahelical MLO protein. Biochem J 385:243–254
Feechan A, Jermakow AM, Dry IB (2009) Grapevine MLO candidates required for powdery mildew pathogenicity? Plant Signal Behav 4:522–523
Franceschini A, Szklarczyk D, Frankild S et al (2013) STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41:808–815
Guo AY, Zhu QH, Chen X, Luo JC (2007) GSDS: a gene structure display server. Yi Chuan 29:1023–1026
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Humphry M, Reinstädler A, Ivanov S, Bisseling T, Panstruga R (2011) Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol Plant Pathol 12:866–878
Jørgensen IH (1992) Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63:141–152
Kaufmann H, Qiu X, Wehmeyer J, Debener T (2012) Isolation, molecular characterization, and mapping of four rose MLO orthologs. Front Plant Sci. doi:10.3389/fpls.2012.00244
Kim DS, Hwang BK (2012) The pepper MLO gene, CaMLO2, is involved in the susceptibility cell-death response and bacterial and oomycete proliferation. Plant J 72:843–855
Kim MC, Lee SH, Kim JK, Chun HJ, Choi MS et al (2002a) MLO, a modulator of plant defense and cell death, is a novel calmodulin-binding protein. J Biol Chem 277:19304–19314
Kim MC, Panstruga R, Elliott C, Müller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P (2002b) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416:447–450
Konishi S, Sasakuma T, Sasanuma T (2010) Identification of novel Mlo family members in wheat and their genetic characterization. Genes Genet Syst 85:167–175
Kudapa H, Bharti AK, Cannon SB et al (2012) A comprehensive transcriptome assembly of pigeonpea (Cajanus cajan L.) using Sanger and second-generation sequencing platforms. Mol Plant 5:1020–1028
Kumar J, Hückelhoven R, Beckhove U, Nagarajan S, Kogel KH (2001) A compromised Mlo pathway affects the response of barley to the necrotrophic fungus bipolaris sorokiniana (Teleomorph: Cochliobolus sativus) and its toxins. Phytopathology 91:127–133
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acid Res 30:325–327
Li YH, Zhou G, Ma J et al (2014) De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat Biotechnol 32:1045–1052
Lim CW, Lee SC (2014) Functional roles of the pepper MLO protein gene, CaMLO2, in abscisic acid signaling and drought sensitivity. Plant Mol Biol 85:1–10
Liu Q, Zhu H (2008) Molecular evolution of the Mlo gene family in Oryza sativa and their functional divergence. Gene 409:1–10
Lorek J, Griebel T, Jones A, Kuhn H, Panstruga R (2013) The role of Arabidopsis heterotrimeric G-protein subunits in MLO2 function and MAMP-triggered immunity. Mol Plant Microbe Interact. doi: 10.1094/MPMI-03-13-0077-R
McClean PE, Mamidi S, McConnell M, Chikara S, Lee R (2010) Synteny mapping between common bean and soybean reveals extensive blocks of shared loci. BMC Genomics 11:184–193
O’Connell RJ, Panstruga R (2006) Tête à tête inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytol 171:699–718
Panstruga R (2005a) Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem Soc Trans 33:389–392
Panstruga R (2005b) Discovery of novel conserved peptide domains by ortholog comparison within plant multi-protein families. Plant Mol Biol 59:485–500
Pereira-Leal JB, Levy ED, Kamp C, Teichmann SA (2007) Evolution of protein complexes by duplication of homomeric interactions. Genome Biol 8:R51.1–R51.12
Pessina S, Pavan S, Catalano D, Gallotta A, Visser RG, Bai Y, Malnoy M, Schouten HJ (2014) Characterization of the MLO gene family in Rosaceae and gene expression analysis in Malus domestica. BMC Genomics 15:618–629
Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, Collins NC, Panstruga R, Schulze-Lefert P (2002) The barley MLO modulator of defence and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol 129:1076–1085
Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J (2012) The Pfam protein families database. Nucleic Acid Res. doi:10.1093/nar/gkr1065
Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acid Res 33:116–120
Sanchez-Perez G, Mira A, Nyiro G, Pasić L, Rodriguez-Valera F (2008) Adapting to environmental changes using specialized paralogs. Trends Genet 24:154–158
Schmutz J, Cannon SB, Schlueter J et al (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46:707–713
Shen Q, Zhao J, Du C, Xiang Y, Cao J, Qin X (2012) Genome-scale identification of MLO domain-containing genes in soybean (Glycine max L. Merr.). Genes Genet Syst 87:89–98
Sievers F, Wilm A, Dineen D et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539–544
Singh VK, Singh AK, Chand R, Singh BD (2012a) Genome wide analysis of disease resistance MLO gene family in sorghum (Sorghum bicolor L. Moench). J Plant Genomics 2:18–27
Singh NK, Gupta DK, Jayaswal PK, Mahato AK et al (2012b) The first draft of the pigeonpea genome sequence. J Plant Biochem Biotechnol 21:98–112
Swarbreck D, Wilks C, Lamesch P, Berardini TZ et al (2008) The Arabidopsis information resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36:D1009–D1014
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tusnády GE, Simon I (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850
Varshney RK, Chen W, Li Y, Bharti AK et al (2012) Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol 30:83–89
Vetter IR, Baase WA, Heinz DW, Xiong JP, Snow S, Matthews BW (1996) Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme. Protein Sci 5:2399–2415
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32:947–951
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64:643–651
Zhou SJ, Jing Z, Shi JL (2013) Genome-wide identification, characterization, and expression analysis of the MLO gene family in Cucumis sativus. Genet Mol Res 12:6565–6578
Zielinski RE (1998) Calmodulin and calmodulin-binding proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 49:697–725
Acknowledgments
This research was financially supported by the University Grant Commission, New Delhi (Junior Research Fellowship Grant to RD). The SUB-DIC Centre at School of Biotechnology, Banaras Hindu University, funded by the Department of Biotechnology, New Delhi provided the authors access to the different software used in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Deshmukh, R., Singh, V.K. & Singh, B.D. Comparative analysis of genome-wide Mlo gene family in Cajanus cajan and Phaseolus vulgaris . Genetica 144, 229–241 (2016). https://doi.org/10.1007/s10709-016-9893-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-016-9893-2