Abstract
This study was conducted to characterize the energy metabolism level and the features of muscle growth regulation during the development of Atlantic salmon (Salmo salar) inhabiting the Indera River (Kola Peninsula, Russia). The activities of aerobic and anaerobic enzymes (cytochrome c oxidase and lactate dehydrogenase) and carbohydrate metabolism enzymes (glucose-6-phosphate dehydrogenase, glycerol-3-phosphate dehydrogenase, and aldolase) were measured in muscle and liver tissue. Gene expression levels of myosin heavy chain (MyHC), myostatin (MSTN-1a), and myogenic regulatory factors (MRFs—MyoD1a, MyoD1b, MyoD1c, Myf5, myogenin) were measured in the white muscles of salmon parr of ages 0+, 1+, 2+, and 3+ and smolts of ages 2+ and 3+. Multidirectional changes in the activity of enzymes involved in aerobic and anaerobic energy metabolism with age were shown in the white muscles of the parr. The cytochrome c oxidase activity was higher in muscles of underyearlings (0+) and yearlings (1+) and decreased in 2+ and 3+ age groups. The activity of lactate dehydrogenase, in contrast, increased with age. The patterns of changes in expression levels of MyoD1a, MyoD1b, myogenin, MyHC, and MSTN-1a at different ages of the parr were similar. Particularly, the expression of these genes peaked in the yearling parr (1+) and then decreased in elder groups. The differences were revealed in parameters studied between the parr and smolts. The level of aerobic and anaerobic metabolism enzyme activities was higher in the white muscles of smolts than in parr. The activity of carbohydrate metabolism enzymes was decreased in the smolts’ livers. The expression levels of MyHC, MyoD1a, MyoD1b, and myogenin were lower in smolts at age 2+ compared to parr. These findings expand our knowledge of age-related and stage-related features of energy metabolism and muscle development regulation in young Atlantic salmon in their natural habitat. The results might be used for monitoring of the salmon population during restoration and rearing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Atlantic salmon (Salmo salar L.) is an important animal model to investigate the regulation of growth and development processes in fish during ontogenesis. The life cycle of salmon includes various developmental stages with a complex system of adaptations (Atlantic salmon ecology 2011). Individuals of the same generation can differ significantly based on their size and growth rates, which influences the onset of parr–smolt transformation (smoltification) at different ages (Pavlov et al. 2007; McCormick 2013). The process resulted in the formation of complex age structure of salmon population on duration of river life period. Each age group of young salmon is characterized by features in metabolism related to changes of stream location, feeding type and regime, activity, and beginning of smoltification.
The most important parameters which have age-related variations and determine the state of individuals and the population as a whole are energy metabolism and growth. A sufficient level of adenosine triphosphate (ATP) synthesis determines active growth and development of fish, especially during early ontogenesis and in the first years of life when high energy costs are needed for the synthesis of structural, functional, and storage compounds (Ozernyuk 2011). The level of energy metabolism can be assessed by measurement of the activities of key enzymes of respiratory chain and glycolysis. Cytochrome c oxidase (COX), the enzyme of the respiratory chain of mitochondria, is used as an indicator of the aerobic capacity (Gauthier et al. 2008). Lactate dehydrogenase (LDH) is the terminal enzyme of anaerobic glycolysis and is used as an indicator of anaerobic capacity (Somero and Childress 1980).
The enzymes involved in the pathways of carbohydrate oxidation could be applied to characterize the level of carbohydrate use in energy metabolism and biosynthesis. Aldolase, an enzyme of glycolysis, converts fructose 1,6-bisphosphate into dihydroxyacetone phosphate (DHAP) and glyceraldehydes 3-phosphate, which are precursors to glycolysis, gluconeogenesis, and lipid formation (Johansen and Overturf 2006). Glycerol-3-phosphate dehydrogenase (GPDH) catalyzes the process of formation of glycerophosphate from DHAP, a precursor of structural and storage lipids (Treberg et al. 2002). Glucose-6-phosphate dehydrogenase (G6PDH) is the key enzyme involved in the pentose phosphate pathway, which leads to the formation of pentoses and reduction of nicotine amide dinucleotide phosphates used in nucleic acids and fatty acid synthesis (Gauthier et al. 2008). Previous researches have shown that the activity of enzymes involved in energy and carbohydrate metabolism are associated with the processes of growth in fish, indicating age-related and seasonal metabolic changes, and correlate with size-weight characteristics of individuals, which have been demonstrated in various fish species (Imsland et al. 2006; Davies and Moyes 2007; Gauthier et al. 2008; Koedijk et al. 2010; Churova et al. 2010, 2015).
According to the study of COX, LDH, GPDH, and G6PDH activities in Atlantic salmon parr, it was found that energy and carbohydrate metabolism differed between groups of underyearling salmon living in the mainstream of the subarctic Varzuga River and its tributary. It was assumed that the noticeable differences in parameters of energy metabolism of juvenile salmon in the first year of life in the river and in its tributary make a basis for subsequent smoltification of parr at different ages 2+, 3+, or 4+ (Pavlov et al. 2007). Changes in respiratory and glycolytic enzyme activities in organs have been demonstrated during smoltification of several salmonid species (Leonard and McCormick 2001; Mizuno et al. 2012). These studies indicated the alteration in energy status during parr–smolt transformation in salmonids.
In most fish, skeletal muscles comprise the larger part of body (about 60% of weight) and therefore play a great role in metabolism of the entire organism and determine total growth rate (Houlihan et al. 1993). The rate and mechanisms of muscle growth in juvenile and adult fish depend on numerous factors, such as genotype, photoperiod, temperature, water flow characteristics, hypoxic, and food availability (Johnston 2006).
Postnatal muscle growth in fish occurs by both hyperplasia (generation of new myotubes) and hypertrophy (increase in myotube size) which are controlled by sequential expression of certain transcription factors. The main role in myogenesis regulation belongs to specific miogenic regulatory factors (MRFs), the transcription factors of bHLH family: MyoD, Myf5, myogenin, and MRF4 (Watabe 2001). These factors have highly conserved basic helix-loop-helix (bHLH) domain which is linked to the DNA sequence E-box that is found in the promoting region of many skeletal muscle-specific genes. Transcription factors MyoD and Myf5 play a key role in specification and proliferation of myoblasts and myogenin, and MRF4 mediate their differentiation (Watabe 2001). MRF gene expression could be used in description of juvenile and adult myogenesis of fish (Johansen and Overturf 2005). The quantitative expression of MRF in reference to hypretrophic and hyperplastic muscle growth mechanisms were studied in rainbow trout Oncorhyncus mykiss (Johansen and Overturf 2005) and pacu Piaractus mesopotamicus (Almeida et al. 2008, 2010) at different developmental stages. MRF expression depends on different factors. Studies on myogenesis during salmon embryogenesis have shown that the MRF expression level was affected by incubation temperature, which would define the postnatal muscle growth mechanisms (Johnston et al. 2000; Macqueen et al. 2007). The studies on farmed salmonidae revealed that MRF messenger RNA (mRNA) levels depend on changes in farmed conditions such as feeding regime and feed composition (Johansen and Overturf 2006; Bower et al. 2009; Overturf et al. 2010).
A key regulator of myogenesis is myostatin (MSTN), a member of transforming growth factor-β (TGF-β). MSTN is a negative regulator of muscle growth and inhibits proliferation and differentiation of muscle cells in mammals (Gabillard et al. 2013). Studies in salmonids have shown that MSTN regulation of muscle growth mechanisms is dependent on the muscle type, developmental stage, and nutritional conditions (Østbye et al. 2001; Johansen and Overturf 2005, 2006).
Myosin, the most abundant protein in muscle, amounts to 25% of total protein in the whole organism and 50% of the muscle proteins (Watabe and Ikeda 2006); thus, it is an advantageous protein for growth studies. It was revealed that myosin heavy chain (MyHC) mRNA level correlated with the growth rate of rainbow trout O. mykiss (Overturf and Hardy 2001), wolfish Anarhichas minor (Imsland et al. 2006), and walleye Sanders vitreus (Dhillon et al. 2009). The study in Atlantic salmon indicated that MyHC mRNA expression can be a useful marker for evaluating growth and protein accretion (Hevroy et al. 2006). A positive correlation between MyHC expression level and weight was revealed in Atlantic salmon parr within different age groups (Churova et al. 2015).
There are numerous studies devoted to metabolism and myogenesis of Atlantic salmon in ontogenesis. However, there is not enough information about age-related differences and alterations during smoltification in the energy status and muscle growth regulation of young salmon in wild. The complex study of energy metabolism and muscle growth processes in wild parr and smolts of different age groups allows for the determination of characteristics of Atlantic salmon development in the freshwater period. Based on this, the study was designed to evaluate the energy and carbohydrate metabolism enzyme activities and expression of genes that control muscle growth in young salmon. In particular, the activities of COX, LDH, G6PDH, GPDH, and aldolase in muscles and the liver and gene expression levels of MyHC, myostatin (MSTN-1a), and myogenic regulatory factors (MyoD1a, MyoD1b, MyoD1c, Myf5, myogenin) were studied in white muscles in salmon parr and smolts of different ages.
Materials and methods
Fish samples
Parr of Atlantic salmons (S. salar L.) at ages 0+, 1+, 2+, and 3+ and smolts at age 2+ and 3+ inhabited the Indera River (the Kola Peninsula, Russia) were studied. Samples were collected in June 2015 at a water temperature of 11.5 °C. The parr were caught by electric fishing gear (model Fa_2, Norway). To avoid the effect of electrofishing, fingerlings were held for 24 h in cages located in the mainstream portion of the river. Smolts were caught by smolt catches, located in the river 300 m from an estuary. Fish were individually measured and weighed. Tissue pieces were frozen in liquid nitrogen and kept at −80 °C prior to analysis. The mean body weight, total length, and number of fish are presented in Table 1.
This study was carried out at the Scientific Center of Collective Usage Platform of the Institute of Biology of the Karelian Research Centre of the Russian Academy of Sciences.
Enzyme analyses
The activities of enzymes were determined both in white muscles and the liver of parr at age 1+, 2+, and 3+ and smolts of 2+ and 3+ age groups. The enzyme activities in underyearlings (0+) were assayed only in muscles. The activity of liver enzymes in parr at age 0+ was not measured due to the low weight of organ, which was inadequate for sampling.
Tissue samples were weighed and homogenized in 0.05 M Tris-HCl buffer (pH 7.5) containing 40% sucrose and 0.1% Triton X-100. The enzyme activities were assayed spectrophotometrically. Specifically, the activity of cytochrome c oxidase (COX, E.C.1.9.3.1) was measured based on the oxidation of cytochrome c by COX, which was determined from the decrease in absorbance at 550 nm (Smith 1955). Activities of lactate dehydrogenase (LDH, E.C.1.1.1.27, Vassault 1983), glucose-6-phosphate dehydrogenase (G6PDH, E.C.1.1.1.49, Bergmeyer 1983), and glycerol-3-phosphate dehydrogenase (GPDH, E.C.1.1.1.8, Bergmeyer 1983) were assayed at 340 nm. One unit of enzyme activity was defined as the amount of enzyme necessary to elicit a change in absorbance of 1.0 U min−1 (1.0-cm light path). The extinction coefficient for NADH + H+ or NADPH was 6.22 cm−1 μmol−1. Aldolase (E.C.4.1.2.13, Bergmeyer 1965) activity was determined based on the colorimetric reaction of products of aldolase and 2,4-dinitrophenylhydrasin in alkaline medium and measured at 535 nm.
The activities of enzymes were expressed as international units per gram of tissue (IU g−1 tissue), where one activity unit equaled the formation of 1 μmol of product per minute.
Myf5, MyoD, myogenin, MSTN-1a, and MyHC mRNA gene expression
Total RNA was isolated from the dorsal white muscle samples using a kit “RNA-extran,” analog of TRIzol (Evrogen, Russia) according to the manufacturer’s protocol. Then, total RNA was treated with DNase (Sileks, Russia). RNA integrity and quality were assessed by 1% agarose gel electrophoresis and spectrophotometrically at 260/280 nm absorbance ratio (SmartSpec Plus, BioRad, USA). RNA was reverse transcribed using MMLV-reverse transcriptase and random hexamer primers (Evrogen, Russia).
Real-time PCR assay was conducted using the iQ5 real-time PCR detection systems (BioRad, USA). The primers for the MyHC, myogenin, MyoD1a, MyoD1b, MyoD1c, Myf5, MSTN-1a, and elongation factor-1 (Ef-1α) were selected using the Beacon Designer 5.0 software (Premier Biosoft, USA). The primer sequences are given in Table 2. Amplification of 2 μl complementary DNA (cDNA; 1:5 dilution of RT reaction) occurred using 5 μl qPCRmix-HS SYBR Green 5× (Evrogen, Russia) and 500 nM primers to yield a final volume of 25 μl. The real-time conditions were as follows: DNA denaturation for 5 min at 95 °C; repeat cycles (40): denaturation for 20 s at 95 °C, annealing for 30 s at 60 °C, and DNA elongation for 30 s at 72 °C. Melt curve analysis verified that the primer sets for each qPCR assay generated one single product and no primer–dimer artifacts.
The relative standard curve method was used for quantification of gene expression (Garikipati et al. 2006). A serial dilution of mixed cDNA from smolts and parr of different ages was used to construct a standard curve for each assay plate and each gene studied. This curve was used to calculate relative abundance of the transcript in each sample. Transcript abundance values were normalized to those of reference gene Ef-1α. Each sample was run in triplicate on a single plate. The data are presented as normalized gene expression.
Statistical analysis
Data were analyzed using the Kruskal–Wallis test. Differences between groups were evaluated by Mann–Whitney U test. Spearman’s correlation and multiple regression analyses were used to examine relationships between parameters studied and parr’s body mass. All results were considered significant at p < 0.05. All data are presented as the means ± SE.
Results
Enzyme activity in muscle and liver of parr and smolts of salmon
The results revealed significant age-related changes in enzyme activities in muscles of salmon parr. The COX activity was higher in muscles of underyearlings (0+) and yearlings (1+) and decreased in 2+ and 3+ age groups (p < 0.05) (Fig. 1a). The activity of LDH, in contrast, increased with age and was significantly higher in parr of ages 1+ (p < 0.05) and 2+ (p < 0.05) in comparison to the previous age group (Fig. 1b). The aldolase activity was over two times higher in yearlings than in underyearlings (p < 0.05) (Fig. 1c). This enzyme activity was higher at age 2+ compared to 1+ group (p < 0.05). The COX activity was negatively related to body mass in parr (p < 0.05) (Table 3). The activities of LDH and aldolase were positively correlated with parr’s body mass (p < 0.05) (Table 3).
Enzyme activity (IU g−1 tissue). COX (a), LDH (b), and aldolase (c) in muscles of parr (0+, 1+, 2+, 3+) and smolts (2+ smolt, 3+ smolt). Asterisks denote significant differences between parr in comparison with previous age group. Double asterisks denote significant differences between parr and smolts of the same age
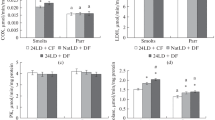
The data analysis on the liver enzyme activity of parr did not reveal any significant differences between the studied age groups (Fig. 2). There were no significant differences in enzyme activity in muscle and liver between age groups in smolts (Figs. 1 and 2).
Comparative analysis of results revealed the differences in enzyme activity between parr and smolts. In muscle, the activity of COX (2+, 3+, p < 0.05, Fig. 1a) and LDH (2+, 3+, p < 0.05, Fig. 1b) was higher in smolt than in parr of the same age. No differences in aldolase activities were observed between parr and smolts (Fig. 1c).
In the liver, there were no differences in COX activity (Fig. 2a). Low activity of LDH (Fig. 2b), G6PDH (Fig. 2c), and aldolase (Fig. 2d) was revealed in smolts compared to parr in both 2+ and 3+ age groups (p < 0.05). GPDH activity was lower in smolts than in parr at age 2+ (p < 0.05) (Fig. 2e).
Muscle-specific gene expression levels in parr and smolts
RT-qPCR results showed age-related and stage-related differences in gene expression levels. The highest MyHC (Fig. 3a), MyoD1a (Fig. 3b), and MyoD1b (Fig. 3c) mRNA levels were revealed for yearling salmon, p < 0.05. Their expression level decreased gradually in parr at age 2+ and then in 3+, p < 0.05. MyoD1c expression level was highest in 0+ parr then decreased at age 1+ and 3+ (Fig. 3d) (p < 0.05). The MyoD1c mRNA level (1.63 ± 0.21) was higher than MyoD1a (1.01 ± 0.04) and MyoD1b mRNA (1.11 ± 0.10) level in the 0+ group (p < 0.05).
Expression of growth-related genes. MyHC (a), MyoD1a (b), MyoD1b (c), MyoD1c (d), myogenin (e), Myf5 (f), and MSTN-1a (g) in parr (0+, 1+, 2+, 3+) and smolts (2+ smolt, 3+ smolt). Asterisks denote significant differences between parr in comparison with previous age group. Double asterisks denote significant differences between parr and smolts of the same age
The level of myogenin gene expression was higher in 1+ parr compared to the 0+ group, р < 0.05 (Fig. 3e). The expression level of myogenin was lower in 3+ parr compared to 2+, p < 0.05. No significant age-related changes were observed in Myf 5 expression (Fig. 3f).
The MSTN-1a mRNA level was eight times higher in yearlings than in underyearlings, p < 0.05 (Fig. 3g). Its expression was lower in groups 2+ and 3+ compared to 1+, p < 0.05.
According to comparative analysis, the expression level of MyHC, p < 0.01 (Fig. 3a), MyoD1a, p < 0.01 (Fig. 3b), MyoD1b, p < 0.01 (Fig. 3c), and myogenin, p < 0.05 (Fig. 3e), was lower in smolts at age 2+ compared to parr of the same age. There were no differences in Myf5 (Fig. 3f), MyoD1c (Fig. 3d), and MSTN-1a mRNA (Fig. 3g) levels between smolts and parr at age 2+. No differences were observed between parr and smolt in gene expression at age 3+ (Fig. 3).
Discussion
Metabolic enzyme activity in muscles of parr of different age groups
Multidirectional changes in activities of enzymes of aerobic and anaerobic energy metabolism in white muscles in parr with age were shown. Results revealed differences in COX activity in white muscle that are due to changes of intensity of aerobic processes in ontogenesis. The level of COX activity was the highest for yearlings and underyearlings and decreased in older groups. Thus, it can be concluded that aerobic capacity is high in parr at age 0+ and 1+. The high level of aerobic synthesis of ATP is of great importance at the early ontogenesis and at first years, when the growth rate is highest and intensive processes of biosynthesis take place (Ozernyuk 2011). It is known that with age and increasing weight, the intensity of oxygen consumption, tissue respiration, and standard metabolism in whole decreased (Davies and Moyes 2007; Ozernyuk 2011). The growth rate decreased with age, and therefore energy consumption causing a biosynthesis decline.
In contrast to the COX activity, the LDH activity in muscles increased with age and body mass. The LDH activity in the white muscles of fish is related mainly to its involvement in anaerobic glycolysis (Somero and Childress 1980). This process is the main energy supply of the white muscle during intensive contractions. Therefore, with age and body mass gain, the rate of parr anaerobic metabolism increases. It was shown that the level of LDH activity reflects the degree of physical activity in fish and their swimming characteristics. Many fish species were found to have an age-related and size-related increase in the intensity of anaerobic energy metabolism, as well as in the activity of LDH and other glycolytic enzymes (Somero and Childress 1980; Davies and Moyes 2007). Atlantic salmon parr at age 1+, 2+, and 3+ are characterized by high physical activity as they inhabit areas with high current velocity and have to actively resist the current. Therefore, an increase in the LDH activity of the muscles with age and body mass may be aimed to increase the energy supply for swimming activity and retention of parr in the current. The aldolase activity in the muscles increased in age groups along with LDH activity. It indicates that changes in the level of carbohydrate utilization correspond to the tendency of the level of anaerobic metabolism changes in the muscles in the age groups.
Similar studies on enzyme activity in parr were conducted in the summer period of the previous year. The pattern of COX, LDH, and aldolase activity changed in the muscle of parr at ages 1+ and 2+ corresponding to the present results (Churova et al. 2015). The present and previous results indicate the age-dependent variation of energy metabolism in muscles, which are characteristic for the parr inhabiting the Indera River. These results might be used in monitoring and health assessment of the salmon population.
Muscle-specific gene expression in parr of different age groups
MRF play the key role in postembryonic myogenesis as regulate specification, proliferation, and differentiation of myogenic precursor cells (MPCs). Transcription factors MyoD and Myf5 are required for myoblast determination, and myogenin is involved in the differentiation and fusion of myoblasts to form myofibers (Watabe 2001). According to studies on rainbow trout and pacu, they are differently expressed in postembryonic development (Johansen and Overturf 2005; Almeida et al. 2010). The results of the present study have shown specific patterns of age-dependent changes in muscle-specific gene expression involved in myogenesis regulation in salmon parr.
It was noticeable that paralogs of MyoD1 were differently expressed in salmon age groups. In the result of tetraploidization, salmonids expressed three MyoD1 paralogs—MyoD1a, MyoD1b, and MyoD1c—that have subfunctionalized and exhibited distinct expression patterns during development and in different fiber types (Macqueen and Johnston 2006; Macqueen and Johnston 2008). In salmon primary cell culture, MyoD1 paralogs are differentially expressed during myotube maturation, and that suggests that Myod1b and Myod1c are primarily expressed in proliferating cells and MyoD1a in differentiating cells. Based on this, it is possible that Myod1b and Myod1c regulate cell cycle and MyoD1a is involved in terminal differentiation (Bower and Johnston 2010). According to our results, the MyoD1a and MyoD1b gene expression peaked in 1+ parr, but the MyoD1c expression was highest in underyearlings and then decreased. Moreover, the MyoD1c mRNA level was higher than MyoD1a and MyoD1b mRNA level in the 0+ group. In our study on brown trout Salmo trutta L. (Churova et al. not published), the similar result on the highest MyoD1c expression level in underyearlings was revealed. It is possible that paralogs differently expressed in the ontogenesis and MyoD1c is upregulated at the earlier stages (in parr with mass 0.14 g). It was shown that MyoD paralogs differently responded to meal distribution after fasting; most especially, Myod1c expression level was elevated after refeeding in rainbow trout (Valente et al. 2012). The MyoD paralogs differently respond to amino acids after starvation that was shown on myogenic cell culture. In response to amino acids, MyoD1c and MyoD1b increased significantly and MyoD1a did not respond markedly (Bower and Johnston 2010). On juvenile and adult salmon (with average mean mass 304.8 and 4297 g), it was shown that MyoD1a was upregulated in fast muscles in comparison to MyoD1b/c, but MyoD1c was upregulated in slow muscles (Macqueen and Johnston 2006).
The pattern of myogenin expression levels in different age groups was similar to MyoD1a and MyoD1b expression levels and peaked in 1+ parr and then decreased with ages. As in our study, the concurrent increase in TMyoD2 (that is referred as Myod1b by Macqueen and Johnston 2008) and myogenin gene expression at the same development stage was shown in rainbow trout O. mykiss (Johansen and Overturf 2005) and MyoD and myogenin in pacu P. mesopotamicus (Almeida et al. 2010). Coupled changes of myogenin and MyoD expression levels are described by the mechanisms of regulation of muscle-specific genes. It is known that MyoD activation leads to robust expression of target genes, such as myogenin, M-cadherin, MyHC, and muscle creatin kinase. MyoD activates myogenin expression by directly binding to its regulatory elements (Berkes and Tapscott 2005). In a study on rainbow trout, it was shown that myogenin expression was restricted to small-diameter muscle fibers and was therefore indicative of muscle hyperplasia (Rescan et al. 1995; Rescan et al. 2015; Montfort et al. 2016). Gene expression profiling of trout regenerating muscle shows that MyoD1b, MyoD1c, and myogenin were upregulated during muscle regeneration and in hyperplastic growth zones (Montfort et al. 2016). That is why, it is possible to suggest that high expression of Myod1 and myogenin expression in yearlings indicated the hyperplastic muscle growth.
In contrast to MyoD1a and MyoD1b mRNA levels, the Myf5 gene expression did not differ between age groups. It is likely that the changes in expression level of MyoD1 are more significant for myogenesis regulation in parr at different ages. According to the study of Johansen and Overturf (2005), expression of MyoD and Myf5 peaked independently from each other at different stages of rainbow trout development. It was suggested by the authors that MyoD and Myf5 might complementarily control muscle growth processes.
The high MyoD1a, Myod1b, and myogenin mRNA levels in parr at age 1+ and decrease in the older age groups corresponded with the high expression levels of MyHC. It was shown that mRNA MyHC levels, which is the most abundant muscle protein, correlate with the growth rate of several fish species (Overturf and Hardy 2001; Imsland et al. 2006; Dhillon et al. 2009) as well as protein accretion (Hevroy et al. 2006). But there are controversial studies that did not show the link between MyHC expression level and rapid growth in atlantic cod (Koedijk et al. 2010) and rainbow trout following growth hormone treatment (Biga et al. 2004) and fed different diet (Alami-Durante et al. 2010). The correlation of MyHC mRNA level with growth rate would depend on different factors, such as size of fish, type of diet (feed content), protein synthesis, and degradation rate (Koedijk et al. 2010; Biga et al. 2004; Alami-Durante et al. 2010). The study on protein degradation system in salmon parr from River Indera (the same research as present) revealed the decrease in proteasome activity in parr 1+ and 2+ in comparison to the previous age group (Kantserova et al. 2017). In the study of Overturf and Gaylord (2009), proteasome enzymatic activity decreased significantly in the muscle of rainbow trout as fish size increased. It was suggested by authors that muscle proteasome activity decrease with trout size would benefit muscle accretion and weight gain and that the fish is becoming more efficient in building muscle as it grows. It allows us to conclude that high level of MyHC expression along with MRF high expression in yearling salmon reflects the intensive growth processes and muscle weight gain. In the previous study on Atlantic salmon from River Idera (Churova et al. 2015), the decrease of MyHC mRNA levels at age 2+ compared to 1+ was observed. Moreover, in that study, it was revealed that the RNA/DNA ratio and protein concentration were highest at age 1+ and decreased at age 2+. Both study results indicate that there is a high protein synthesis level and growth rate in salmon yearlings (1+). The low MyHC mRNA levels along with decreased COX activity at age 2+ and 3+ indicate the decline in muscle growth rate.
The high levels of MyHC, MyoD1a, MyoD1b, and myogenin expression in yearlings were accompanied by high MSTN-1a mRNA level, the negative regulator of muscle growth. Simultaneous increase of expression of MyoD and myogenin with MSTN on the same development stages was shown in rainbow trout (Johansen and Overturf 2005). One potential explanation is that MSTN is expressed in response to high MRF expression levels, which are the necessary regulation mechanism to attenuate hyperplasia and hypertrophy and control muscle growth (Johansen and Overturf 2005, 2006). In mammals, MSTN inhibits myoblast proliferation and differentiation through a tumor growth factor-B pathway involving the activin receptor, Act RIIB, and the phosphorylation of Smads 2 and 3. Myogenin is probably one from the major physiological targets of MSTN (Joulia et al. 2003; Johnston et al. 2008). The MyoD binding sites were found in MSTN promoter (Johnston et al. 2008). However, according to recent studies, the mechanism of MSTN action on growth in fish probably differs from mammals. MSTN is conserved as two genes in most teleost and four genes in salmonids and is found to be expressed in muscles and other tissues. Consequently, it may possess different functions (Østbye et al. 2001; Gabillard et al. 2013). Studies using rainbow trout myosatellite cells have shown that MSTN inhibits proliferation and has no effect on differentiation (Seiliez et al. 2012) or, on the contrary, consequently stimulates it (Garikipati and Rodgers 2012). The study on comparison of wild, domesticated, and growth hormone transgenic coho salmon showed high expression levels of both MSTN and MyoD with myogenin in fast-growing fish (Overturf et al. 2010). It was concluded by the authors that MSTN expression increases in faster growing fish when there is sufficient energy but is not negatively acting to regulate muscle growth. The feeding experiments in fish have shown that the MSTN gene is differently expressed in periods of fasting and refeeding. In rainbow trout, the high MSTN mRNA levels during the refeeding period was associated with high myogenin expression that possibly is a mechanism to attenuate the muscle growth (Johansen and Overturf 2006). In a study on pacu during a fasting period, high MSTN gene expression indicated lower satellite cell proliferation activity, which was confirmed by low MyoD mRNA levels (Nebo et al. 2013). Thus, the MSTN expression is affected by several factors, but precise mechanisms of action require further investigation.
Comparative analysis of enzyme activity and muscle-specific genes expression in parr and smolts
The parr–smolt transformation, or smoltification, is a developmental process in salmon to prepare for life in the sea. Significant biochemical, physiological, morphological, and behavioral changes occur in the fish throughout this period. The alterations affect lipid and carbohydrate metabolism, osmoregulation, oxygen transport, growth, and rheotaxis (Stefansson et al. 2008; McCormick 2013). In particular, the intensification of oxidative processes is revealed in smolts. In Atlantic salmon, standard and active metabolic rates are 50% higher in smolts than in parr (Maxime et al. 1989), suggesting that this elevation might be related with the increase in respiratory enzyme activity and mitochondrial proliferation caused by an increase in thyroid hormone levels (Leonard and McCormick 2001). Decrease in hepatic glycogen content was observed in masu salmon Oncorhyncus masou (Mizuno et al. 2012), in Atlantic salmon (Wendt and Saunders 1973), and coho salmon Oncorhynchus kisutch (Sheridan et al. 1985), that could be related with decreased glycogen synthesis and increased glycogenolysis in the liver (Sheridan et al. 1985). It is known that in salmon smolts, total body and muscle lipid content and fatty acid synthesis decreased and lipolysis increased (Sheridan et al. 1985; Stefansson et al. 2008; Pavlov et al. 2009). That is why smolts at the period of downstream migration and ocean entry are “energy deficient” (Sheridan et al. 1985; Stefansson et al. 2008).
According to data analysis from the present study, there were differences in aerobic and anaerobic metabolism in muscles between parr and smolts at age 2+. The higher activity of COX in smolts than in parr indicates that the increase in energy costs during smoltification is necessary for supporting adaptive reactions of metabolism (Stefansson et al. 2008).
The increase in LDH activity in the muscle of smolts in comparison to parr is possibly related to their migration activity. The LDH activity indicates the activation of glycolysis which is necessary for energy supply, allowing for physical activity during the migration downstream the river (Stefanson et al. 2008). Higher activity of the glycolysis enzyme, phosphofructokinase, in muscle was observed during smoltification in smolts than in parr (Leonard and McCormick 2001).
In the liver, LDH activities were decreased in smolts compared to parr at the same ages. This may indicate a decline in intensification of lactate–pyruvate conversion and consequently reduction in lactate-dependent gluconeogenesis (Leonard and McCormick 2001). Such a decrease in liver LDH was revealed in Atlantic salmon (Leonard and McCormick 2001) and masu salmon Onchorhinkus masou (Mizuno et al. 2012). The activity of G6PDH, GPDH, and aldolase in the liver decreased, that indicated the decrease of carbohydrate use in biosynthesis (pentose phosphate pathway and synthesis of lipids from glycerophosphate). These changes can be related with a decrease in glycogen synthesis and intensification of glycogenolysis (Sheridan et al. 1985).
As for muscle growth regulation, smolts at age 2+ differ from parr by low MyHC, MyoD1a, MyoD1b, and myogenin expression levels. It is an evidence of decrease in muscle growth. The MRF expression levels depend on a variety of factors, such as food quantity and quality and characteristics of total metabolism (Johansen and Overturf 2006; Bower et al. 2009; Alami-Durante et al. 2010; Nebo et al. 2013). In the present research, the smolts in the period of downstream migration were studied, and differences were observed which were caused by changes in metabolism during migration. Firstly, differences in MRF expression could be related to changes in food composition and quantity. This is supported by a study of Atlantic salmon from the Varzuga River (Kola Peninsula, Russia); this study revealed that during downstream migration, smolts do feed not intensively compared to parr (Shustov and Belyakova 2012). Taking in consideration the low level of energy reserves in smolts during migration (Stefansson et al. 2008), it is obvious that the delivered nutrients are used first of all for energy requirements then on growth processes. Moreover, the changes in protein turnover take place in smolts. It was revealed that in contrast to lipids and carbohydrates, there were no changes in protein content in salmon smolts and postsmolts during migration (Stefansson et al. 2008). Our colleagues observed the decrease in protein degradation rate in migrating smolts compared to parr of salmon in the Indera River (Kantserova et al. 2017). These changes could also affect on the muscle-specific gene expression.
Conclusion
This study demonstrated age-related and stage-related features of aerobic and anaerobic capacity and muscle growth regulation in parr and smolt of Atlantic salmon in the wild. Each age group differs from one another by energy enzyme activities and MRFs, MSTN and MyHC expression levels. Alteration in energy metabolism in smolts during downstream migration was associated with elevation of aerobic and anaerobic capacities in the muscles and the decline of carbohydrate oxidation in the liver. Smolts differed from parr by having a decline in myogenesis regulation and muscle growth processes in relation to metabolic alterations during smoltification and migration. These findings expand our knowledge on the mechanisms of growth and development in wild salmon. The results might be used for monitoring the salmon population during restoration.
References
Alami-Durante H, Wrutniak-Cabello C, Kaushik SJ, Médale F (2010) Skeletal muscle cellularity and expression of myogenic regulatory factors and myosin heavy chains in rainbow trout (Oncorhynchus mykiss): effects of changes in dietary plant protein sources and amino acid profiles. Comp Biochem Physiol A Mol Integr Physiol 156(4):561–568. doi:10.1016/j.aquaculture.2010.03.012
Almeida FLA, Carvalho RF, Pinhal D, Padovani CR, Martins C, Dal Pai-Silva M (2008) Differential expression of myogenic regulatory factor MyoD in pacu skeletal muscle (Piaractus mesopotamicus Holmberg 1887: Serrasalminae, Characidae, Teleostei) during juvenile and adult growth phases. Micron 39:1306–1311. doi:10.1016/j.micron.2008.02.011
Almeida FLA, Pessoti NS, Pinhal D, Padovani CR, Leitão NJ, Carvalho RF, Martins C, Portella MC, Dal Pai-Silva M (2010) Quantitative expression of myogenic regulatory factors myoD and myogenin in pacu (Piaractus mesopotamicus) skeletal muscle during growth. Micron 41:997–1004. doi:10.1016/j.micron.2010.06.012
Atlantic Salmon Ecology (2011) Aas Ø, Klemetsen A, Einum S, Skurdal J (Eds). Oxford: Blackwell Publishing Ltd.
Bergmeyer HU (1965) Methods of enzymatic analysis. Academic Press, New York
Bergmeyer HU (1983) Methods of enzymatic analysis, 3rd edn, vol. 2, Verlag Chemie, Weinheim.
Berkes CA, Tapscott SJ (2005) MyoD and the transcriptional control of myogenesis. Seminars in cell & developmental biology Academic Press 16(4):585–595. doi:10.1016/j.semcdb.2005.07.006
Biga PR, Cain KD, Hardy RW, Schelling GT, Overturf K, Roberts SB, Ott TL (2004) Growth hormone differentially regulates muscle myostatin1 and-2 and increases circulating cortisol in rainbow trout (Oncorhynchus mykiss). General and comparative endocrinology 138(1): 32-41. doi:10.1016/j.ygcen.2004.05.001
Bower NI, Johnston IA (2010) Paralogs of Atlantic salmon myoblast determination factor genes are distinctly regulated in proliferating and differentiating myogenic cells. Am J Phys Regul Integr Comp Phys 298(6):R1615–R1626. doi:10.1152/ajpregu.00114.2010
Bower NI, Taylor RG, Johnston IA (2009) Phasing of muscle gene expression with fasting-induced recovery growth in Atlantic salmon. Front Zool 6:1. doi:10.1186/1742-9994-6-18
Churova MV, Meshcheryakova OV, Nemova NN, Shatunovskii MI (2010) The correlation between fish growth and several biochemical characteristics with reference to the steelhead Parasalmo mykiss Walb. Biol Bull 37(3):236–245. doi:10.1134/S1062359010030040
Churova MV, Meshcheryakova OV, Veselov AE, Nemova NN (2015) Activity of enzymes involved in the energy and carbohydrate metabolism and the level of some molecular-genetic characteristics in young salmons (Salmo salar L.) with different age and weight. Russ J Dev Biol 5:254–262. doi:10.1134/S1062360415050021
Davies R, Moyes CD (2007) Allometric scaling in centrarchid fish: origins of intra-and inter-specific variation in oxidative and glycolytic enzyme levels in muscle. J Exp Biol 210(21):3798–3804. doi:10.1242/jeb.003897
Dhillon RS, Esbaugh AJ, Wang YS, Tufts BL (2009) Characterization and expression of a myosin heavy–chain isoform in juvenile walleye Sander vitreus. J Fish Biol 75(5):1048–1062. doi:10.1111/j.1095-8649.2009.02376.x
Gabillard JC, Biga PR, Rescan PY, Seiliez I (2013) Revisiting the paradigm of myostatin in vertebrates: insights from fishes. Gen Comp Endocrinol 194:45–54 http://dx.doi.org/10.1016/j.ygcen.2013.08.012
Garikipati DK, Rodgers BD (2012) Myostatin inhibits myosatellite cell proliferation and consequently activates differentiation: evidence for endocrine-regulated transcript processing. J Endocrinol 215(1):177–187. doi:10.1530/JOE-12-0260
Garikipati DK, Gahr SA, Rodgers BD (2006) Identification, characterization, and quantitativeexpression analysis of rainbow trout myostatin-1a and myostatin-1b genes. J Endocrinol 190:879–888. doi:10.1677/joe.1.06866
Gauthier C, Campbell P, Couture P (2008) Physiological correlates of growth and condition in the yellow perch (Perca flavescens). Comparative Biochemistry and Physiology: Part A 151:526–532. doi:10.1016/j.cbpa.2008.07.010
Hevroy EM, Jordal A-EO, Hordvik I, Espe M, Hemre G-I, Olsvik PA (2006) Myosin heavy chain mRNA expression correlates higher with muscle protein accretion than growth in Atlantic salmon, Salmo salar. Aquaculture 252:453–461. doi:10.1016/j.aquaculture.2005.07.003
Houlihan DF, Mathers EM, Foster A (1993) Biochemical correlates of growth rate in fish. In Rankin JC, Jensen FB (eds). Fish Ecophysiology. London UK, pp 45–71. doi:10.1007/978-94-011-2304-4_2
Imsland AK, Le Francois NR, Lammare SG, Ditlecadet D, Sigurosson S, Foss A (2006) Myosin expression levels and enzyme activity in juvenile spotted wolfish (Anarhichas minor) muscle: a method for monitoring growth rates. Can J Fish Aquat Sci 63:1959–1967. doi:10.1139/F06-091
Johansen KA, Overturf K (2005) Quantitative expression analysis of genes affecting muscle growth during development of rainbow trout (Oncorhynchus mykiss). Mar Biotechnol 7(6):576–587
Johansen KA, Overturf K (2006) Alterations in expression of genes associated with muscle metabolism and growth during nutritional restriction and refeeding in rainbow trout. Comp Biochem Physiol B: Biochem Mol Biol 144(1):119–127. doi:10.1016/j.cbpb.2006.02.001
Johnston IA (2006) Environment and plasticity of myogenesis in teleost fish. J Exp Biol 209:2249–2264. doi:10.1242/jeb.02153
Johnston IA, McLay HA, Abercromby M, Robins D (2000) Early thermal experience has different effects on growth and muscle fibre recruitment in spring-and autumn-running Atlantic salmon populations. J Exp. Biol 203(17): 2553-2564
Johnston IA, Macqueen DJ, Watabe S (2008) Molecular biotechnology of development and growth in fish muscle. In: Tsukamoto K, Kawamura T, Takeuchi T, Beard Jr TD, Kaiser MJ (eds) Fisheries for Global Welfare and Environment: Memorial book of the 5th World Fisheries Congress. Terrapub, Tokyo, pp 241–262
Joulia D, Bernardi H, Garandel V, Rabenoelina F, Vernus B, Cabello G (2003) Mechanisms involved in the inhibition of myoblast proliferation and differentiation by myostatin. Exp Cell Res 286:263–275. doi:10.1016/S0014-4827(03)00074-0
Kantserova NP, Lysenko LA, Nemova NN (2017) Protein degradation in the skeletal muscles of parr and smolt Atlantic salmon Salmo salar L. Biology bulletin Biology 44(1):55–60
Koedijk RM, Le François NR, Blier PU, Foss A, Folkvord A, Ditlecadet D, Lamarre SG, Stefansson SO, Imsland AK (2010) Ontogenetic effects of diet during early development on growth performance, myosin mRNA expression and metabolic enzyme activity in Atlantic cod juveniles reared at different salinities. Comp Biochem Physiol A Mol Integr Physiol 156(1):102–109. doi:10.1016/j.cbpa.2010.01.007
Leonard JB, McCormick SD (2001) Metabolic enzyme activity during smolting in stream-and hatchery-reared Atlantic salmon (Salmo salar). Can J Fish Aquat Sci 58(8):1585–1593. doi:10.1139/f01-105
Macqueen DJ, Johnston IA (2006) A novel salmonid myoD gene is distinctly regulated during development and probably arose by duplication after the genome tetraploidization. FEBS Lett 580:4996–5002. doi:10.1016/j.febslet.2006.08.016
Macqueen DJ, Johnston IA (2008) An update on MyoD evolution in teleosts and a proposed consensus nomenclature to accommodate the tetraploidization of different vertebrate genomes. PLoS One 3(2):e1567. doi:10.1371/journal.pone.0001567
Macqueen DJ, Robb D, Johnston IA (2007) Temperature influences the coordinated expression of myogenic regulatory factors during embryonic myogenesis in Atlantic salmon (Salmo salar L.) J Exp Biol 210:2781–2794. doi:10.1242/jeb.006981
Maxime V, Boeuf G, Pennec JP, Peyraud C (1989) Comparative study of the energetic metabolism of Atlantic salmon (Salmo salar) parr and smolts. Aquaculture 82(1):163–171. doi:10.1016/0044-8486(89)90405-5
McCormick SD (2013) Smolt physiology and endocrinology. Fish physiology 32:199–251. doi:10.1016/B978-0-12-396951-4.00005-0
Mizuno S, Urabe H, Aoyama T, Omori H, Iijima A, Kasugai K, Torao M, Misaka N, Koide N, Ueda H (2012) Changes in activity and transcript level of liver and gill metabolic enzymes during smoltification in wild and hatchery-reared masu salmon (Oncorhynchus masou). Aquaculture 362:109–120. doi:10.1016/j.aquaculture.2010.10.034
Montfort J, Le Cam A, Gabillard JC, Rescan PY (2016) Gene expression profiling of trout regenerating muscle reveals common transcriptional signatures with hyperplastic growth zones of the post-embryonic myotome. BMC Genomics 17(1):810. doi:10.1186/s12864-016-3160-x
Nebo C, Portella MC, Carani FR, de Almeida FLA, Padovani CR, Carvalho RF, Dal-Pai-Silva M (2013) Short periods of fasting followed by refeeding change the expression of muscle growth-related genes in juvenile Nile tilapia (Oreochromis niloticus). Comp Biochem Physiol B: Biochem Mol Biol 164(4):268–274. doi:10.1016/j.cbpb.2013.02.003
Østbye T, Galloway T, Nielsen C, Gabestad I, Bardal T, Andersen Ø (2001) The two myostatin genes of Atlantic salmon (Salmo salar) are expressed in a variety of tissues. Eur J Biochem 268(20):5249–5257. doi:10.1046/j.0014-2956.2001.02456.x
Overturf K, Gaylord TG (2009) Determination of relative protein degradation activity at different life stages in rainbow trout (Oncorhynchus mykiss).Comp Biochem Physiol Part B: Biochem Mol Biol, 152(2): 150-160. doi:10.1016/j.cbpb.2008.10.01210.1016/j.cbpb.2008.10.012
Overturf K, Hardy R (2001) Myosin expression levels in trout muscle: a new method of monitoring specific growth rates for rainbow trout Oncorhynchus mykiss (Walbaum) on varied planes of nutrition. Aquat Res 32:315–322. doi:10.1046/j.1365-2109.2001.00582.x
Overturf K, Sakhrani D, Devlin RH (2010) Expression profile for metabolic and growth-related genes in domesticated and transgenic coho salmon (Oncorhynchus kisutch) modified for increased growth hormone production. Aquaculture 307(1):111–122. doi:10.1016/j.aquaculture.2010.06.010
Ozernyuk ND (2011) Adaptive specific features of energy metabolism in fish ontogenesis. Russ J Dev Biol 42(3):201–205. doi:10.1134/S1062360411030131
Pavlov DS, Meshcheryakova OV, Veselov AE, Nemova NN, Lupandin AI (2007) Parameters of energy metabolism in juveniles of Atlantic salmon Salmo salar living in the mainstream and in the tributary of the Varzuga River (the Kola Peninsula). J Ichthyol 47(9):774–781. doi:10.1134/S003294520709010X
Pavlov DS, Nefedova ZA, Veselov AE, Nemova NN, Ruokolainen TR, Vasil’eva OB, Ripatti PO (2009) Age dynamics of lipid status of juveniles of Atlantic salmon (Salmo salar L.) from the Varzuga River. J Ichtiology 49(11):1073–1080. doi:10.1134/S003294520911006X
Rescan PY, Gauvry L, Paboeuf G (1995) A gene with homology to myogenin is expressed in developing myotomal musculature of the rainbow trout and in vitro during the conversion of myosatellite cells to myotubes. FEBS Lett 362(1):89–92. doi:10.1016/0014-5793(95)00215-U
Rescan PY, Rallière C, Lebret V, Fretaud M (2015) Analysis of muscle fibre input dynamics using a myog: GFP transgenic trout model. J Exp Biol 218(8):1137–1142. doi:10.1242/jeb.113704
Seiliez I, Sabin N, Gabillard JC (2012) Myostatin inhibits proliferation but not differentiation of trout myoblasts. Mol Cell Endocrinol 351(2):220–226. doi:10.1016/j.mce.2011.12.011
Sheridan MA, Woo N, Bern HA (1985) Changes in the rates of glycogenesis, glycogenolysis, lipogenesis, and lipolysis in selected tissues of the coho salmon (Oncorhynchus kisutch) associated with parr-smolt transformation. J Exp Zool 236(1):35–44. doi:10.1002/jez.1402360106
Shustov YA, Belyakova EA (2012) Comparative study on the feeding of parrs and smolts of the Atlantic salmon (Salmo salar L.) in the subarctic Varzuga River basin. Russ J Ecol 43(6):462–465. doi:10.1134/S106741361205013X
Smith L (1955) Spectrophotometric assay of cytochrome c oxidase. Methods in Biochem Analysis 2:427–434. doi:10.1002/9780470110188.ch13
Somero GN, Childress JJ (1980) A violation of the metabolism-size scaling paradigm: activities of glycolytic enzymes in muscle increase in larger size fish. Physiol Zool 53:322–337. doi:10.1086/physzool.53.3.30155794
Stefansson SO, Björnsson BT, Ebbesson LOE, McCormick SD (2008) Smoltification. In: Finn RN, Kapoon BG (eds) Fish Larval Physiology. Science Publishers, Enfield, pp 639–681
Treberg JR, Lewis JM, Driedzic WR (2002) Comparison of liver enzymes in osmerid fishes: key differences between a glycerol accumulating species, rainbow smelt (Osmerus mordax), and a species that does not accumulate glycerol, capelin (Mallotus villosus). Comp Biochem Physiol A Mol Integr Physiol 132:433–438. doi:10.1016/S1095-6433(02)00083-1
Valente LM, Bower NI, Johnston IA (2012) Postprandial expression of growth-related genes in Atlantic salmon (Salmo salar L.) juveniles fasted for 1 week and fed a single meal to satiation. Br J Nutr 108(12):2148–2157. doi:10.1017/S0007114512000396
Vassault A (1983) Lactate dehydrogenase. In: Bergmeyer HU (ed) Methods of Enzymatic Analysis. Vol. 3. Weinheim: Verlag Chemie, pp 118–126
Watabe S (2001) Myogenic regulatory factors. In: Johnston IA (ed) Muscle Development and Growth. Acad. Press, London, pp 19–41. doi:10.1016/S1546-5098(01)18003-9
Watabe S, Ikeda D (2006) Diversity of the pufferfish (Takifugu rubripes) fast skeletal myosin heavy chain genes. Comp Biochem Physiol 1D:28–34. doi:10.1016/j.cbd.2005.12.001
Wendt CAG, Saunders RL (1973) Changes in carbohydrate metabolism in young Atlantic salmon in response to various forms of stress. Int At. Salmon Found Spec Publ Ser 4: 55-82
Acknowledgements
The study was supported by the grant of Russian Science Foundation (project no. 14-24-00102).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Churova, M.V., Meshcheryakova, O.V., Veselov, A.E. et al. Activity of metabolic enzymes and muscle-specific gene expression in parr and smolts Atlantic salmon Salmo salar L. of different age groups. Fish Physiol Biochem 43, 1117–1130 (2017). https://doi.org/10.1007/s10695-017-0357-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-017-0357-0