Abstract
The dispersal tendencies of individuals can provide an important adaptive basis to counter environmental and ecological variation to increase fitness and benefit populations. We evaluated whether indirect genetic-based measures of dispersal of a large monitor lizard (Varanus varius) were sex-biased and further covaried with putative demographic and ecological differences across a forest ecotone in Southern Australia. The mean corrected assignment index (i.e., mAIC) estimated that lace monitors had significant male-biased dispersal. There was strong evidence that common dispersal promoting influences, including lace monitor density and arboreal mammal prey availability, but not the overall male-biased sex ratio, differed with forest type across the ecotone. These different forest types influenced the extent of sex-biased dispersal, with lace monitors captured in banksia woodland having a significant male bias in dispersal, whilst lace monitors sampled in adjacent eucalypt forest showed no difference in assignment values indicating no sex-biased dispersal. However, individual-based spatial autocorrelation analyses and mixed effect models indicated no spatial mediated genetic structuring nor evidence of sex or habitat-related effects. Estimates of recent migration (i.e., < 3 generations) indicated strong and symmetrical migration between banksia woodland and eucalypt forest patches suggesting limited habitat resistance to gene flow across an ecotonal landscape. Not discounting method-specific evidence for male- and forest-type-biased dispersal, the absence of spatial genetic structuring suggests lace monitors retain a high dispersal capacity. An absence of landscape-scale genetic structure is consistent with this species’s high vagility. This landscape-scale result is further supported because only the most significant biogeographic barriers (e.g., mountain ranges) impede gene flow within the species’ extensive range distribution, allowing for genetic structuring among populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dispersal, the movement of an individual and its genes, can strongly influence ecological dynamics at many biological levels (Gadgil 1971; Garant et al. 2007; Bonte et al. 2012; Clobert 2012; Jessop et al. 2018). Individuals differ considerably in their dispersal capacities because of complex interactions among adaptive processes (i.e., evolution and phenotypic plasticity) and environmental (e.g., habitat quality), demographic (e.g., population density) or phenotypic influences (Greenwood 1980; McPeek and Holt 1992; Baguette 2003; Hendry et al. 2004; Massot et al. 2008; Clobert et al. 2009). For example, kin avoidance is a major cue that individuals can use to inform dispersal (Clobert et al. 2009; Stevens et al. 2014; Cayuela et al. 2018). Additionally, ecological factors such as low-quality habitats may induce animals to disperse into more favourable habitats, promoting source-sink dynamics in landscape-scale population dynamics (Hanski 1999). Such examples highlight the importance of informed, compared to random, animal dispersal, which potentially allows individuals to respond to environmental, demographic, life-history or phenotypic variation (Ronce 2007; Clobert et al. 2009).
Sex-biased dispersal can result from multiple selective pressures and is a typical example of non-random dispersal in animals (Greenwood 1980; Pusey 1987; Trochet et al. 2016). Sex-biased dispersal is favoured if males or females experience different fitness costs associated with site philopatry. For example, the sex of an individual can influence fitness costs from intraspecific resource competition (Greenwood 1980), local mate competition (Dobson 1982) or inbreeding (Pusey 1987). Parental care and sexual dimorphism are additional forces that govern the gender and extent of sex-biased dispersal (Trochet et al. 2016). The fitness cost of dispersal can be higher for the more dispersive sex than if it remained resident in its natal habitat (Hendry et al. 2004; Bonte et al. 2012; Jessop et al. 2018). Additionally, landscape features (topography, habitat resource differences) can promote or suppress resistance to dispersal-related gene flow (Slatkin 1985). These processes may also interact with sex-specific drivers of dispersal to allow for spatially mediated variation in the extent of sex-biased dispersal within or among populations (Lane and Shine 2011; Tucker et al. 2017).
Estimating sex-biased dispersal in animals can be achieved by both direct (e.g., mark-recapture or biotelemetry) or indirect methods (e.g., molecular approaches) (Slatkin 1985). Analyses of multilocus genotypes may offer a practical methodology for identifying sex-biased dispersal in animal populations (Banks and Peakall 2012). Multiple statistical approaches can be used to evaluate sex-biased dispersal between or within populations using molecular markers (Mossman and Waser 1999; Prugnolle and De Meeˆus 2002; Peakall and Smouse 2012). In particular, individual-based statistical analyses such as assignment indices and spatial autocorrelation analyses are often used to evaluate processes affecting the contemporary dispersal behaviours of organisms (Smouse and Peakall 1999; Goudet et al. 2002). Frequentist and Bayesian “assignment tests” can be used to estimate the probability that an individual’s genotype is drawn from either immigrants or residents within the population in which it was captured (Paetkau et al. 1995; Favre et al. 1997; Pritchard et al. 2000). Here it is expected that individuals of the most dispersive sex will exhibit frequency differences in their genotypes compared to more philopatric individuals because they will have a higher frequency of rare genotypes that belong to recent immigrants (Mossman and Waser 1999; Goudet et al. 2002; Banks and Peakall 2012). Another approach is to use tests that evaluate sex-related differences between pairwise individual genetic relatedness and geographic distance (Smouse and Peakall 1999). If one sex exhibits sufficiently restricted gene flow (i.e., an absence of migrants) it can produce positive spatial autocorrelation (i.e. genetic structuring) because relatedness among genotypes decrease with geographic distance (Banks and Peakall 2012). There is good evidence from studies that concurrently used direct and indirect methods to indicate that genetic-based methods can accurately infer sex-biased dispersal (Goudet et al. 2002).
We used biparentally inherited markers (eight microsatellite loci), analyzed with molecular tests of sex-biased dispersal to explore differences in the gene flow (i.e., indirect dispersal) of the lace monitor (Varanus varius) in a temperate coastal forest ecosystem of Southern Australia. Lace monitors are a sizeable predatory lizard (< 10 kg) distributed across Eastern and Southern Australia (Jessop et al. 2012, 2015; Smissen et al. 2013). Previous research shows that this species has limited genetic (i.e., mitochondrial and nuclear markers) structure over its vast distribution (Smissen et al. 2013). Indeed lace monitors appear able to maintain gene flow (i.e., an absence of population genetic structure) across vast geographic scales (100 s-1000 s of km) (Smissen et al. 2013). Nevertheless, little is known about the drivers of lace monitor dispersal related gene flow at the landscape scale (i.e., 10 s of km). Aside from sex, lace monitors, at this spatial scale, are likely exposed to multiple cues or selective pressures that might affect their dispersal (Anson et al. 2014). Such processes could include spatial variation in habitat types, disturbance regimes and associated ecological resources that influence population densities that may select for dispersal differences among individuals. Thus evaluating how sex and or environmental factors might influence dispersal is essential to better understand what key processes affect lace monitor spatial population dynamics at this landscape scale.
For our first aim, we examined whether there is sex dispersal bias in lace monitors by estimating genetic differentiation and spatial genetic structure at a landscape scale. Most reptiles exhibit male-biased dispersal (Tucker et al. 1998; Keogh et al. 2006; Dubey et al. 2008; Ujvari et al. 2008). However, some reptiles can have female-biased dispersal (Olsson and Shine 2003; Ryberg et al. 2004) or lack sex-biased dispersal altogether (Lukoschek et al. 2008). The much larger body size of an adult male compared to a female lace monitor suggests the presence of intrasexual competition (e.g. “local mate competition hypothesis”), which could promote male-biased dispersal in these lizards (Dobson 1982; Trochet et al. 2016).
We measured if the two adjacent dominant forest types, banksia woodland and eucalypt forest, differed in lace monitor population density, sex ratio, and prey availability for our second aim. We measured these three factors because they are widely recognised as critical drivers of animal dispersal-related responses (Bohonak 1999; Clobert et al. 2009; Bonte et al. 2012). Suppose these processes indeed differ among forest types. In that case, it could provide a basis to test if key habitat associated differences also influenced mean or sex-related gene flow tendencies within this lace monitor population. For example, our study area occurred in an ecotone where the dominant vegetation communities transitioned from banksia woodland to mixed eucalypt forest. Such substantial habitat differences can also provide a basis for producing landscape-scale ecological resource gradients. Elsewhere, ecotones have significant ecological consequences for individuals, populations and communities (Kark 2013), but their influence on landscape-scale animal dispersal is less obvious.
Our third aim evaluated if differences in lace monitor dispersal were evident between eucalypt forest and banksia woodland. Habitat mediated differences in gene flow could arise if this ecotone produces a spatial resource gradient or differs spatially in other dispersal selective processes. In that case, we may see increased gene flow in the more dispersive lace monitor sex occurring within low resource habitats compared to high resource habitats. We may also see asymmetrical rates of gene flow between habitat types consistent with underlying habitat mediated source-sink dynamics (Cure et al. 2017; Edelaar et al. 2017; Lowe and Addis 2019). Here we would predict that if there are significant resource differences in prey between forest types, lace monitor gene flow might increase from low into high-prey density habitat types. However, such asymmetrical gene flow could be opposed if high-prey density habitats also support greater densities of lace monitors. In this latter scenario opposing density-dependent could counter asymmetrical gene flow to promote otherwise similar rates of gene flow across the study area.
Materials and methods
Study area
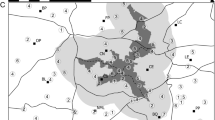
This study was conducted in Cape Conran Coastal Park (37° 49’S, 148° 44’E) and the adjacent Murrungowar State Forest (37° 37’S, 148° 44’E) in East Gippsland, Victoria, Australia (Fig. 1). Annual rainfall for the region averaged approximately 846 mm, and mean maximum and minimum temperatures ranged from 27.0 °C (January) to 4.7 °C (July) from 2007 to 2011 (Bureau of Meteorology 2017). Climatic conditions were generally uniform across the study area, and elevation ranged from sea level to 350 m. The study area comprised 42,000 hectares of coastal lowland vegetation communities consisting mainly of two widespread vegetation types: coastal woodland dominated by Banksia serrata and Banksia integrifolia, with emergent eucalypts and a shrub-rich understorey; and lowland forest dominated by Eucalyptus sieberi and Eucalyptus globoidea, with a diverse understorey of shrubs, grasses and herbs (see Anson et al. 2013).
Map of study area comprising coastal forest in East Gippsland, Victoria. This study area is overlaid onto eucalypt forest-banksia woodland ecotone. The yellow polygon represents individuals captured within coastal eucalypt forest, and the green polygon represents individuals captured in banksia woodland. Points represent the individual capture locations of lace monitors with males represented by black points and females by pink points
Study species
Lace monitors are common semi-arboreal predators in the study area and reach considerable length (2.1 m) (Jessop et al. 2012; Smissen et al. 2013). This species has a generalist diet, where typical prey items include insects in juveniles and arboreal mammals, birds and eggs in adults (Jessop et al. 2010; McCurry et al. 2015). This species is very active during the summer months, and adult males use large home range areas (i.e. an average of 65 ± 34 ha) (Weavers 1993; Jessop et al. 2013).
Lace monitor sampling design, capture and genetic sampling
Three annual summer (December to January) surveys were conducted to capture lace monitors at or between 76 fixed trapping sites across the study area. Each site (a 1 ha forest area) was spaced at a 2 km interval. Lace monitors were captured using traps (methods detailed below) or a lasso pole in banksia woodland (N = 63) and eucalypt forest (N = 48) distributed across the study site (Fig. 1). Captures in each summer (commencing in November 2007 and ending in January 2010) were centred around 76 fixed trapping locations. Across the three summers, we obtained 111 genetic samples from adult lace monitors of mixed-sex (N = 35 F: 76 M; sex determination explained below). There was no evidence of spatial sampling differences between male and female lace monitors across the sampling area (Fig. 1).
We maintained a consistent survey effort (6 days) and sampling design over the study duration, resulting in 35, 36 and 40 lace monitor genetic samples obtained in each annual sampling period.
Following lace monitor capture, we restrained each lizard to collect 0.5 ml of blood from the ventral coccygeal vein using a 22G needle attached to a 3 ml syringe (Scheelings and Jessop 2011). Blood was transferred into tubes and stored in blood lysis buffer to preserve DNA until subsequent genetic analyses. All lizards were marked with a passive integrated transponder to prevent duplication in blood samples among years and released at their capture point.
Microsatellite screening and genetic sex determination
Genomic DNA was extracted from blood (~ 20 µl) in lysis buffer using DNeasy® Blood and Tissue Kit (250), following the manufacturer’s protocol (Qiagen Inc., Valencia). Eight polymorphic microsatellite loci were used to genotype individuals sampled across the study site. The eight loci were successfully cross amplified from five microsatellite loci developed for V. komodoensis (K24, K23, K22, K20, K9; (Ciofi et al. 2011)), and three loci developed for V. acanthurus (Va34, Va25, Va7; (Fitch et al. 2005)).
DNA amplifications were done in a 20µL total volume, containing ~ 10 ng DNA, 1 unit GoTaq (Promega), 0.25 µM of each fluorescently labelled primer and mqH2O to make up a total volume of 20µL. A touchdown PCR protocol was used as follows: initial denaturation of 94 °C for 5 min, followed by two cycles of 94 °C for 30 s, an annealing step of 65 °C for 30 s and 72 °C for 90 s; this cycle was repeated with annealing temperatures of 60 °C, 55 °C and 50 °C (two cycles each) and a final annealing temperature of 48 °C for 30 cycles. Then a final extension step at 72 °C for ten minutes. Negative controls were included in each reaction, and amplifications were checked on a 1.2% agarose gel or on the QIAxcel System (Qiagen). Fragment analysis of PCR products was undertaken at the Australian Genome Research Facility on an ABI3730 DNA Analyser and analysed using ABI GeneMapper software.
Null alleles were tested using MICRO-CHECKER v. 2.2.3 (Van Oosterhout et al. 2004). GENEPOP v. 4.1.0 (Rousset 2008) was used to test for linkage disequilibrium between pairs of loci. Heterozygote deficit and excess were tested using exact tests in GENEPOP v. 4.1.0, and a posthoc Bonferroni test (Rice 1989) was applied. Bias-corrected FIS were calculated, with the Robertson & Hill’s value preferential for populations with low genetic diversity (Raufaste and Bonhomme 2000). GENALEX v. 6.5 (Peakall and Smouse 2006) was used to calculate the effective number of alleles (AE), observed (HO) and expected (HE) heterozygosity, and departures from Hardy–Weinberg equilibrium (HWE). We present these results in the supplementary material (Table S1). The absence of sex-linked markers was confirmed by the presence of heterozygous males at each loci (Table S1) in a species with a ZW female/ZZ males sex determination system (Iannucci et al. 2019). There was also no statistical evidence supporting that male and female lace monitors significantly differed in their allelic distributions at any of the eight loci (Tables S2-S9; Figures S1-S8).
The sex of lace monitors is challenging to infer from external morphology, and consequently, we used genetic sexing techniques to assign sex to each individual captured (Jessop et al. 2012). Lizards were genetically sexed using 2 sets of PCR primers that amplified sex-specific alleles (Halverson and Spelman 2002). Amplifications were performed in a 20µL total volume, containing 2µL of DNA (diluted 1:10 in TE buffer), 10µL Gotaq (Promega), 0.5µL of each primer (10 µM) and 7µL of H2O. PCR amplifications were performed on a Corbett Palm-Cycler using a touchdown thermal cycle program with the following parameters: initial denaturation @ 94 °C for 5 min, followed by two cycles of 94 °C for 30 s, an annealing step @ 65 °C for 30 s and 72 °C for 90 s; 2 cycles each with annealing temperatures of 60 °C, 55 °C and 50 °C; 30 cycles with an annealing temperature of 48 °C; then a final extension step of 10 min @ 72 °C. Amplified PCR products were run on a 1.2% agarose gel, and amplification patterns were compared to those of a male and female whose sex had been verified anatomically. One bright band indicated female, and multiple fainter bands indicated male for both primer combinations.
Analysis of sex effects on lace monitor dispersal
We used three complementary genetic statistical based tests to analyze sex-specific differences in lace monitor dispersal patterns. The first approach was to use an assignment index test implemented in FSTAT version 2.9.3 (Paetkau et al. 1995; Favre et al. 1997; Goudet 2001; Goudet et al. 2002). This software calculated the mean corrected assignment index (mAIC) and the mean–variance of the corrected assignment index (vAIC) of AIC and gene diversity (HS) for each sex. The assignment index in (Goudet et al. 2002) was calculated for each lace monitor genotype. This equation estimates allele frequencies to calculates the expected frequency of each individual’s genotype at each locus. Then by multiplying the expected frequency of each genotype across all loci it produces a probability value for each individual. This value is log transformed to give a log likelihood of occurrence and the AIc value is the calculated as the individual’s log likelihood minus the mean log likelihood from all individuals (i.e., the mean value) sampled within the study area (Favre et al. 1997). Statistical significance for sex related differences in these indices was determined using a Mann Whitney U-test with 10,000 randomizations.
A bias in dispersal between the sexes should be reflected in a statistically significant difference between males and females in the estimated parameters. Lace monitors with positive AIC values indicate individuals that were residents in the study area (i.e., highly philopatric). Individuals with negative AIC values indicate those genotypes that are less likely to occur randomly in the population (i.e., as expected from immigrants). Thus, on average, the dispersive sex should possess a negative mAIC compared to the more philopatric sex’s mAIC. This assignment test was selected because both empirical studies and simulated data have shown that this test is robust at estimating sex or other phenotypic biases in animal dispersal (Banks and Peakall 2012). For the same reason, if the dispersive sex possesses a higher proportion of immigrants and thus rarer genotypes (i.e., increased allelic composition and diversity), it would be expected that the vAIC and Hs is higher than that of the more philopatric sex (i.e. common genotypes) (Goudet et al. 2002; Wang et al. 2020).
Second, we used spatial autocorrelation analyses to compare the pairwise relationship between the genetic and spatial distance of male and female lace monitors using GENALEX v.6.5 (Peakall and Smouse 2006). We used 0.25, 0.5, 1, 2, 5 and 10 km distance classes for both sexes to capture the relevant ecological scale at which lace monitor dispersal processes might occur while maintaining an adequate sample size within each distance class. This method estimates the autocorrelation coefficient (r) at each predefined distance interval. Whereby r (range: -1 to 1) measures the genetic similarity between all pairs of individuals whose geographic separation falls within each distance class. Each analysis was conducted using a permutation procedure with 999 simulations to test for deviations from zero and 1000 bootstraps to estimate the confidence intervals around r. A heterogeneity test based on ω (Omega) was used to test for significant departures (α = 0.01) from the null hypotheses of no geographic pattern in the genetic structure of each sex (Peakall and Smouse 2006; Banks and Peakall 2012). To infer sex-biased dispersal, we assessed for a significantly positive relatedness coefficient (i.e. where the mean r and associated 95% credible intervals do not overlap zero) within the first distance interval of each sex to provide evidence of increased relatedness due to increased philopatry (i.e. restricted dispersal) among individuals (Banks and Peakall 2012).
Third, we tested associations between geographic distance and pairwise genetic distances for each sex using a linear mixed effect model (LMM) incorporating a maximum likelihood population effect (MLPE). These models were run using lme4 (Bates et al. 2015) in R. These models incorporate a random effect structure to control nonindependence among pairwise genetic and geographic data (Clarke et al. 2002; Peterman et al. 2014). Individual based pairwise genetic distance (i.e., codominant genotypic distance estimated in GENALEX v.6.5) was used as the dependent variable, and log-transformed Euclidean geographic distance was included as the dependent factor in models. For geographic distance to have a significant effect, we considered that the beta coefficient for genetic distance must produce 95% confidence intervals that did not overlap zero. We also calculated the Marginal R2 for further inference to assess the strength of the association between geographic and genetic distance for each lace monitor sex using the MuMIn package (Barton and Barton 2019).
Effects of forest type on lace monitor abundance, sex ratio, and preferred prey
To consider if the distinct transition in vegetation communities (i.e., forest type) influenced ecological processes that might affect lace monitor dispersal across the study area, we measured lace monitor abundance, sex ratio, and preferred prey (i.e., possums and gliders) resources in the two ecotonal forest types. The methods for estimating these measures are defined below:
Estimates of habitat-specific lace monitor population density estimates
Six repeated count surveys of lace monitors were conducted in the summer (December to January) of 2008/2009 at 76 sites using detections tallied daily from four concurrent sampling methods; box traps, baited sand pads, and visual count methods (Anson et al. 2013, 2014; Jessop et al. 2016). Each site was 500 m long and spaced 2 km apart along management tracks; this interval ensured spatial independence as it greatly exceeded the maximum home range area of lace monitors (Weavers 1993). Aluminium box traps (2 × 0.3 × 0.3 m) were positioned randomly along each transect and left in-situ for six days per sampling occasion, baited with beef infused with tuna emulsion oil. Visual surveys of tracks and adjacent forest were conducted by two observers driving at 10 km/h during the hottest hours of the day. Mixed searches recorded lace monitors opportunistically whilst observers were at each site conducting other activities (e.g., walking to check traps and sand pads) or driving through them (40 km/h). Climatic conditions can potentially influence the activity of these poikilothermic animals. Therefore surveys were restricted to days with reduced cloud cover and mean daytime temperatures above 26 °C(Jessop et al. 2013).
We used the Royle-Nichols abundance induced heterogeneity model to estimate lace monitor abundance at each site in PRESENCE v12.1 (Hines 2006). The model estimates the parameters λ and r, representing average abundance per sampling station and innate species detectability, respectively (Royle and Nichols 2003). These models were fitted with a Poisson distribution. We compared four models where λ was modelled with and without the effects of forest type or where detection rate was considered invariant across surveys or as survey-specific (i.e., complete identity) in all models (i.e. λ. and r). A null model was also compared to ensure that the top model produced a better fit to the data than chance alone. We ranked models using Akaike Information Criterion adjusted for small sample sizes (AICc) and Akaike weights to assess the relative support for each model (Burnham and Anderson 2003). Models with ∆AIC < 2 from the top-ranked model were considered substantially supported, provided that the null model did not receive similar support.
Estimates of habitat specific lace monitor sex ratio
From using data produced by using the genetic sexing techniques to assign sex to each individual captured, we assessed for significant sex ratio bias. We first used a one-sample binomial test to test if the sex ratio differed from parity. Next, we used a generalised linear model with a binomial distribution and logit link to test the effect of habitat differences on sex ratios.
Estimates of habitat specific prey population abundance
To estimate the relative abundance of common prey of lace monitor, we counted seven marsupial possums and glider species across the forest ecotone using 76 transects overlaid onto lace monitor sampling sites (Jessop et al. 2010; Anson et al. 2014). Each transect was 500 m long and spaced 2 km apart along management tracks; this interval ensured spatial independence as it greatly exceeds these marsupials’ maximum home range areas. We performed six repeated nocturnal (7 pm- 12 am) counts of possums and gliders along each transect over two years between January 2008 and December 2009.
We again used a Royle-Nichols abundance induced heterogeneity model to estimate arboreal possums and glider abundance (λ) at each site. We compared four models where λ was modelled with and without the effects of forest type or where detection rate was considered invariant across surveys or as survey-specific (i.e., full identity) in all models (i.e., λ. and r). A null model was also compared to ensure that the top model produced a better fit to the data than chance alone.
Effects of habitat type on lace monitor gene flow
To evaluate the effect of habitat variation on sex-biased dispersal, we assigned individuals to their respective habitat types recorded at capture. We used the mean corrected assignment index and spatial autocorrelation analyses to test for habitat effects on sex-biased dispersal. Next, we used BayesAss (ver. 3.04; Wilson and Rannala 2003) to estimate recent migration rates (i.e., within the last one to three generations) among and within banksia woodland and eucalyptus forest. This program allows the estimation of asymmetrical gene flow using a Bayesian clustering algorithm (Markov chain Monte Carlo; MCMC) to make inferences about levels of migration (i.e., the proportion of migrants between 0 and 1) and population inbreeding using diploid neutral genetic markers. We used the following settings: number of iterations, 10,000,000; sampling frequency, 100; length of burn-in, 1,000,000. The delta (D) values were set to 0.50, 0.30 and 0.80 for allele frequency, migration rate and inbreeding, respectively, to ensure good mixing and adequate sampling from the posterior distribution. We constructed 95% confidence intervals around the mean recent migration rates as mean ± 1.96 * standard deviation. Some BayesAss runs can have poor MCMC sampling, where values approach the bounds of the priors (Faubet et al. 2007, Meirmans 2014). Hence, following the protocol Faubet et al. (2007) suggested, we ran BayesAss 10 times under identical conditions but with different random number seeds. We then calculated the Bayesian deviance information criterion (DIC) value for each run and reported the estimated dispersal rates using the run with the lowest DIC value (Faubet et al. 2007, Meirmans 2014).
Results
Measures of sex-biased dispersal
We used genotypes from 111 lace monitors genetically sexed (i.e., using PCR) as 35 females and 76 males. Our first analysis of sex-biased dispersal estimated that male lace monitors had a mean negative mAIc value (-0.260 ± 0.193) that was significantly different (Mann–Whitney U test: Z = 2.735, P = 0.006) from the mean positive mAIc (0.594 ± 0.246) value estimated for females (Fig. 2A). The Hs scores also significantly differed (P = 0.007) between males (0.68) and females (0.64). These results indicated that males comprised a higher proportion of rarer genotypes indicative of a greater number of male migrants in the study area. In contrast, females were estimated to have an overall mean positive mAIc value, indicating that females on average showed a greater similarity in genotypes and evidence of greater philopatry. However, it is important to note that some females also had negative AIc values (i.e., 9 of 35 individuals) suggesting that some females were possible immigrants into the population (Fig. 2A). The vAIc score was not significantly different (P > 0.05) between males (14.95) and females (9.52).
The mean assignment index values (± SEM) (blue dots) were estimated for male and female lace monitors across the entire study area (A) and within banksia woodland and eucalyptus forest (B). Individual lace monitor assignment values are presented along with the sex specific distribution of these values. Sample sizes (N) for each group are reported below each figure
Spatial autocorrelation analyses, pending the distance class (i.e., 0.25, 0.5, 1, 2, 5 and 10 km), indicated significant spatial heterogeneity in the genetic structure between and within male and female lace monitors (Table 1). Generally, females exhibited greater spatial heterogeneity in genetic structure than males. However, there was no evidence of sex-biased dispersal, given the lack of significant positive relatedness within the first distance interval estimated at any distance class (Fig. 3; Fig. S9). There was no evidence of significant associations between geographic distance and pairwise genetic relatedness in male (LMM: β = 0.001 ± -0.001 – 0.012 [95% confidence intervals]; marginal R2 = 0.001) and female (LMM: β = -0.051 ± -0.161 – 0.064; marginal R2 = 0.001) lace monitors.
Examples of spatial correlogram plots reporting the genetic correlation coefficient (r) as a function of distance for female (A-B) and male lace monitors (C-D) at two different distance classes (1 and 5 km). The bootstrapped 95% confidence interval error bars are shown. Where both the upper and lower confidence intervals do not cross zero within a specific distance interval, individuals can be considered more significantly related (error bars for r > 0) and hence philopatric; or less significantly related and comprise migrants (error bars for r < 0) than the mean relatedness of individuals (r = 0). The pairwise sample size for each distance interval is reported by the number at the top of each error bar
Effect of forest type on lace monitor abundance, sex ratio, and preferred prey
There was evidence that eucalypt forest and banksia woodland were associated with significant differences in two of the three common processes influencing animal dispersal. Model ranking of Royle-Nichols models indicated a strong forest type effect (untransformed B = 0.66 ± 0.24; wi = 0.86) compared to the null model (ΔAICc = 3.79; wi = 0.06) on lace monitor site abundance (Table 2a). This index of population density was estimated to be nearly two-fold higher in banksia woodland (5.41 ± 0.99 individuals/site) compared to sites sampled in eucalypt forest (2.79 ± 0.79 individuals/site) (Fig. 4A). Lace monitors demonstrated a significant male bias (1.9 M: 1 F) within the study population (One sample binomial test: -3.091, P < 0.002). However there was no evidence of a significant forest type-related effect (B = -0.308, 95% CI = -1.35—-0.29) on sex ratio (GLM: χ2 = 0.68, P = 0.41, Fig. 4B).
Model ranking of Royle-Nichols models again indicated a strong effect of forest type (untransformed B = 0.78 ± 0.14; wi = 1) compared to the null model (ΔAICc = 91.12; wi = 0.00) on possum and glider abundance (Table 2b). With possum and glider abundance being greater in banksia woodland (6.59 individuals/transect, CI = 4.96—8.77) compared to transects surveyed in eucalypt forest (3.01 individuals/transect, CI = 2.29 -3.97; Fig. 4C).
Effects of forest type on sex-biased dispersal and migration rate
Population-level assignment analyses estimated a significant difference in mAIc value between male and female lace monitors in banksia woodland (Mann–Whitney U test: Z = 2.133, P = 0.033). However, there was no difference in genetic structure between male and female mAIc values sampled in the eucalypt forest (Mann–Whitney U test: Z = 0.097, P = 0.922) (Fig. 2B). Spatial autocorrelation analyses indicated no evidence of a habitat by sex effect on spatial genetic heterogeneity nor evidence that habitat type influenced sex-biased dispersal, given the lack of significant positive relatedness within the first distance interval (Fig. 3E). BayesAss analyses estimated that non-migrant lace monitors comprised the majority of individuals sampled within each forest type (~ 75% of individuals) (Fig. 5). However, there was strong evidence for near equal bi-directional migrant exchange (25%–26% migrant movement) between both forest types. These results indicated that habitat type did not affect migration across the study area.
Discussion
Studies that evaluate if informed, unlike random, dispersal is present within a population are essential to document if its individuals have the adaptive capacity to alter movement and gene flow tendencies to improve fitness (Clobert et al. 2009). Sex-biased dispersal is a common example of adaptive dispersal as it allows individuals of the dispersing sex to avoid fitness costs associated with spatial philopatry (Greenwood 1980; Trochet et al. 2016). We found method-specific evidence of sex-biased and forest type influenced dispersal in lace monitors. The mean assignment index (mAIc) method indicated evidence of a male sex-biased dispersal in lace monitors. Here increased dispersal was inferred by male lace monitors having a more negative mean assignment value than females to indicate that males are more likely than average to be recent immigrants into the local population (Mossman and Waser 1999; Goudet et al. 2002).
Male biased dispersal in lace monitors is consistent with findings from other reptiles that also report males being the more dispersive sex (Keogh et al. 2006; Ujvari et al. 2008, François et al. 2021). The operational sex ratio of this lace monitor population appeared strongly male-biased, and this attribute may provide an important basis for intrasexual competition and hence selection for male-biased dispersal in this species (King and Green 1999, Trochet et al. 2016). If so, male-biased dispersal, as advocated for other species, could reduce competition or inbreeding among male lace monitors (Greenwood 1980; Goudet et al. 2002). Conversely, genetic evidence of increased philopatry for female lace monitors may provide a basis for increased fitness by allowing individuals to exploit local habitat resources of known quality, including nesting sites, rather than dispersing into habitats of unknown and possibly inferior quality (Hendry et al. 2004; Jessop et al. 2018). Additionally, phenotypic differences at the time of dispersal, such as body size, could further impose sex-specific dispersal costs during dispersal and add to the adaptive basis of male-biased dispersal in lace monitors (Bonte et al. 2012).
The forest type in which individuals were captured was also associated with differences in the mean assignment index of each sex. In particular, male lace monitors reported a strong negative mean assignment index value in banksia woodland compared to no differences in eucalypt forest. These results suggest that ecological or environmental processes within this forest type could influence male-biased dispersal across the population (Fraser et al. 2004). Our measures indicated that banksia woodland had an almost two-fold higher lace monitor and arboreal prey abundance than the adjacent eucalypt forest, which may provide two possible explanations for this difference. Animal dispersal is widely recognised as a density- and habitat quality-dependent process with higher dispersal rates generally predicted out of higher-density and poorer quality habitat patches (Clobert et al. 2004; Bowler and Benton 2005; Clobert 2012). The higher dispersal of male lace monitors in banksia woodland might reflect their responses to counter-selection from increased local population densities that could increase both resource or kin competition. The absence of sex-biased dispersal in the eucalypt forest is also intriguing. It may suggest that selection for sex-biased dispersal is weaker in this forest type, possibly as an advent of lower lace monitor population densities reducing intraspecific competition. Other forest type-dependent processes, including structural habitat resistance that impedes movement to alter dispersal costs, could be conceived as additional mechanisms that might attenuate sex-biased dispersal in this forest type. Similarly, we cannot discount that because of the smaller sample size obtained from eucalypt forest that we simply lacked statistical power to demonstrate that this habitat type was also associated with sex biased dispersal in lace monitors (Berry et al. 2004). Further experimental research is needed to tease apart which mechanisms may lead to sex and habitat-related differences in lace monitor genetic differentiation.
Importantly, other tests used in this study did not detect similar evidence of sex or habitat biased dispersal in lace monitors. First, we did not detect sex-related differences in the variance associated with assignment values (i.e., vAIc). While this test can provide a more robust inference to assess sex-biased dispersal than the mean value (Goudet et al. 2002). It also rapidly loses sensitivity to detect differences in dispersal between the sexes when there is some capacity for both sexes to disperse or because there is insufficient power to detect differences due to small sample sizes (Goudet et al. 2002). For example, other studies have indicated that non-significant differences in vAIc arise when both male and females can disperse (Mossman and Waser 1999; Goudet et al. 2002). Thus, we interpret these results to suggest that while males contain a greater proportion of rare migrant associated genotypes, females too contain sufficient migrants (i.e., negative AIc values) to limit statistical differences in the mean variance of assignment values between each sex.
Additionally spatial autocorrelation analyses, linear mixed effect models and Bayesian assignment tests provided no evidence of sex or habitat biased lace monitor dispersal. These results arose because either sex or habitat types produced non-significant trends between genetic and geographic distance or because different habitat types contained and high and similar levels of immigration. These results clearly point to the absence of spatial genetic structuring within the population. Fine scale genetic structure within populations can be weak or non-existent for several reasons (Smouse and Peakall 1999). For example, even if there is a difference in the proportion of dispersers within each sex, the less dispersive sex may maintain enough ongoing dispersal that sees gene flow prevent fine scale genetic structure from arising within a sex. Furthermore, given lace monitors, possess extensive daily movements, large home ranges, promiscuous mating and are dietary generalists (Guarino 2001, 2002; Jessop et al. 2012, 2018). These traits are expected to promote ongoing gene flow that negate genetic drift or the physical clustering of related genotypes and limit spatial genetic structuring within populations (Smith et al. 1996; Aguillon et al. 2017). Similarly, the high level of largely symmetrical migrant exchange (i.e. > 20%) between forest types suggests an impressive dispersal capacity of lace monitors across this complex landscape. Indeed, it is likely that the absence of fine scale genetic structuring at the landscape scale and relatively high rates of migration explain why this species can maintain current gene flow across vast areas of its extensive range distribution (~ 2 million km2), with inter-population genetic structuring only recorded at Eastern Australia’s most significant biogeographic barriers (Smissen et al., 2013).
We also recognize that the capacity of multilocus spatial autocorrelation analyses to detect sex-biased dispersal can be strongly influenced by sample size and, to a lesser extent, the number of loci evaluated (Berry et al. 2004; Banks and Peakall 2012). Again, it is possible our study lacked adequate and predominantly female sample sizes to apply these three methods successfully. Adding additional samples and loci are now needed to discount if lace monitors definitively lack sex or habitat informed gene flow across the different statistical methods used in this study.
In conclusion, adaptive dispersal is widely regarded as a critical trait that can influence fitness and metapopulation function in species responses to environmental and ecological change (Clobert et al., 2009). However, for any animal to alter its dispersal, it inevitably requires the sensory capacity to register changes in potentially multiple selections and counter-selection processes operating within their environment. Our study shows method dependent evidence of sex and habitat-related effects on dispersal in lace monitors, suggesting the possibility of this species adaptively responding to demographic or ecological variation at the landscape scale. Moreover, evidence of high dispersal capacity suggests that this species can attenuate the effects of ecological or environmental processes that could otherwise lead to genetic structuring within the population at the landscape scale.
Availability of data and material
Will be provided on request to the lead author.
Code availability
Not applicable.
References
Aguillon SM, Fitzpatrick JW, Bowman R, Schoech SJ, Clark AG, Coop G, Chen N (2017) Deconstructing isolation-by-distance: The genomic consequences of limited dispersal. PLoS Genet 13:e1006911
Anson JR, Dickman CR, Boonstra R, Jessop TS (2013) Stress triangle: do introduced predators exert indirect costs on native predators and prey? PLoS ONE 8:e60916
Anson JR, Dickman CR, Handasyde K, Jessop TS (2014) Effects of multiple disturbance processes on arboreal vertebrates in eastern Australia: implications for management. Ecography 37:357–366
Baguette M (2003) Long distance dispersal and landscape occupancy in a metapopulation of the cranberry fritillary butterfly. Ecography 26:153–160
Banks SC, Peakall R (2012) Genetic spatial autocorrelation can readily detect sex-biased dispersal. Mol Ecol 21:2092–2105
Barton, K., and M. K. Barton. 2019. Package ‘MuMIn’. R package version 1.
Bates D, Mächler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67(1). https://doi.org/10.18637/jss.v067.i01
Berry O, Tocher MD, Sarre SD (2004) Can assignment tests measure dispersal? Mol Ecol 13:551–561
Bohonak AJ (1999) Dispersal, gene flow, and population structure. Q R Biol 74:21–45
Bonte D, Van Dyck H, Bullock JM, Coulon A, Delgado M, Gibbs M, Lehouck V, Matthysen E, Mustin K, Saastamoinen M (2012) Costs of dispersal. Biol Rev 87:290–312
Bowler DE, Benton TG (2005) Causes and consequences of animal dispersal strategies: Relating individual behaviour to spatial dynamics. Biol Rev Camb Philos Soc 80:205–225
Burnham KP, Anderson DR (2003) Model selection and multimodel inference: a practical information-theoretic approach. Springer Science & Business Media, London
Cayuela H, Rougemont Q, Prunier JG, Moore JS, Clobert J, Besnard A, Bernatchez L (2018) Demographic and genetic approaches to study dispersal in wild animal populations: A methodological review. Mol Ecol 27:3976–4010
Ciofi C, Tzika AC, Natali C, Watts PC, Sulandari S, Zein MS, Milinkovitch MC (2011) Development of a multiplex PCR assay for fine-scale population genetic analysis of the Komodo monitor Varanus komodoensis based on 18 polymorphic microsatellite loci. Mol Ecol Resour 11:550–556
Clarke RT, Rothery P, Raybould AF (2002) Confidence limits for regression relationships between distance matrices: estimating gene flow with distance. J Agric Biol Environ Stat 7:361–372
Clobert J (2012) Dispersal ecology and evolution. Oxford University Press, New Jersey
Clobert J, Galliard L, Cote J, Meylan S, Massot M (2009) Informed dispersal, heterogeneity in animal dispersal syndromes and the dynamics of spatially structured populations. Ecol Lett 12:197–209
Clobert J, Ims RA, Rousset F (2004) Causes, mechanisms and consequences of dispersal. Ecology, genetics and evolution of metapopulations. Elsevier, Netherlands, pp 307–335
Cure K, Thomas L, Hobbs J-PA, Fairclough DV, Kennington WJ (2017) Genomic signatures of local adaptation reveal source-sink dynamics in a high gene flow fish species. Sci Rep 7:1–10
Dobson FS (1982) Competition for mates and predominant juvenile male dispersal in mammals. Anim Behav 30:1183–1192
Dubey S, Brown G, Madsen T, Shine R (2008) Male-biased dispersal in a tropical Australian snake (Stegonotus cucullatus, Colubridae). Mol Ecol 17:3506–3514
Edelaar P, Jovani R, Gomez-Mestre I (2017) Should I change or should I go? Phenotypic plasticity and matching habitat choice in the adaptation to environmental heterogeneity. Am Nat 190:506–520
Faubet P, Waples RS, Gaggiotti OE (2007) Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol Ecol 16(6):1149–1166
Favre L, Balloux F, Goudet J, Perrin N (1997) Female-biased dispersal in the monogamous mammal Crocidura russula: evidence from field data and microsatellite patterns. Proc Royal Soc London Ser B: Biolog Sci 264:127–132
Fitch AJ, Goodman AE, Donnellan SC (2005) Isolation and characterisation of microsatellite markers for the Australian monitor lizard, Varanus acanthurus (Squamata: Varanidae) and their utility in other selected varanid species. Mol Ecol Notes 5:521–523
François D, Ursenbacher S, Boissinot A (2021) Isolation-by-distance and male-biased dispersal at a fine spatial scale: a study of the common European adder (Vipera berus) in a rural landscape. Conserv Genet 22:823–837. https://doi.org/10.1007/s10592-021-01365-y
Fraser DJ, Lippé C, Bernatchez L (2004) Consequences of unequal population size, asymmetric gene flow and sex-biased dispersal on population structure in brook charr (Salvelinus fontinalis). Mol Ecol 13:67–80
Gadgil M (1971) Dispersal: population consequences and evolution. Ecology 52:253–261
Garant D, Forde SE, Hendry AP (2007) The multifarious effects of dispersal and gene flow on contemporary adaptation. Funct Ecol 21:434–443
Goudet J, Perrin N, Waser P (2002) Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Mol Ecol 11:1103–1114
Goudet, J. 2001. FSTAT, a program to estimate and test gene diversities and fixation indices, version 2.9. 3.http://www2.unil.ch/popgen/softwares/fstat.htm
Greenwood PJ (1980) Mating systems, philopatry and dispersal in birds and mammals. Anim Behav 28:1140–1162
Guarino F (2001) Diet of a large carnivorous lizard, Varanus varius. Wildl Res 28:627–630
Guarino F (2002) Spatial ecology of a large carnivorous lizard, Varanus varius (Squamata: Varanidae). J Zool 258:449–457
Halverson J, L Spelman (2002) Sex determination and its role in management. In: Komodo dragons: Biology and Conservation. Smithsonian Institution Pr: Washington p:165–177
Hanski I (1999) Metapopulation ecology Oxford University Press, Oxford Google Scholar. Oxford University Press, New Jersey
Hendry AP, Castric V, Kinnison MT, Quinn TP, Hendry A, Stearns S (2004) The evolution of philopatry and dispersal. Evolution illuminated: salmon and their relatives. Elsiver, Nedharlond, pp 52–91
Hines, J. E. 2006. Program PRESENCE. See http://www.mbrpwrc.usgs.gov/software/doc/presence/presence.html
Iannucci A, Altmanová M, Ciofi C, Ferguson-Smith M, Milan M, Pereira JC, Pether J, Rehák I, Rovatsos M, Stanyon R (2019) Conserved sex chromosomes and karyotype evolution in monitor lizards (Varanidae). Heredity 123:215–227
Jessop TS, Urlus J, Lockwood T, Gillespie G (2010) Preying possum: assessment of the diet of lace monitors (Varanus varius) from coastal forests in southeastern Victoria. Biawak 4:59–66
Jessop TS, Smissen P, Scheelings F, Dempster T (2012) Demographic and phenotypic effects of human mediated trophic subsidy on a large Australian lizard (Varanus varius): meal ticket or last supper? PLoS ONE 7:e34069
Jessop TS, Kearney MR, Moore JL, Lockwood T, Johnston M (2013) Evaluating and predicting risk to a large reptile (Varanus varius) from feral cat baiting protocols. Biol Invasions 15:1653–1663
Jessop TS, Anson JR, Narayan E, Lockwood T (2015) An introduced competitor elevates corticosterone responses of a native lizard (Varanus varius). Physiol Biochem Zool 88:237–245
Jessop TS, Ariefiandy A, Purwandana D, Ciofi C, Imansyah J, Benu YJ, Fordham DA, Forsyth DM, Mulder RA, Phillips BL (2018) Exploring mechanisms and origins of reduced dispersal in island Komodo dragons. Proc R Soc B 285:20181829
Jessop, T. S., G. Gillespie, and M. Letnic. 2016. Examining multi-scale effects of the invasive fox on a large varanid (Varanus varius White, 1790) mesopredator. Pages 221–236 in Interdisciplinary World Conference on Monitor Lizards’.(Ed. M. Cota.) .
Kark S (2013) Ecotones and ecological gradients. Ecological systems. Springer, New York, pp 147–160
Keogh JS, Webb JK, Shine R (2006) Spatial genetic analysis and long-term mark–recapture data demonstrate male-biased dispersal in a snake. Biol Let 3:33–35
Lane A, Shine R (2011) Intraspecific variation in the direction and degree of sex-biased dispersal among sea-snake populations. Mol Ecol 20:1870–1876
Lowe WH, Addis BR (2019) Matching habitat choice and plasticity contribute to phenotype–environment covariation in a stream salamander. Ecology 100:e02661
Lukoschek V, Waycott M, Keogh JS (2008) Relative information content of polymorphic microsatellites and mitochondrial DNA for inferring dispersal and population genetic structure in the olive sea snake, Aipysurus laevis. Mol Ecol 17:3062–3077
Massot M, Clobert J, Ferriere R (2008) Climate warming, dispersal inhibition and extinction risk. Glob Change Biol 14:461–469
McCurry MR, Mahony M, Clausen PD, Quayle MR, Walmsley CW, Jessop TS, Wroe S, Richards H, McHenry CR (2015) The relationship between cranial structure, biomechanical performance and ecological diversity in varanoid lizards. PLoS ONE 10:e0130625
McPeek MA, Holt RD (1992) The evolution of dispersal in spatially and temporally varying environments. Am Nat 140:1010–1027
Meirmans PG (2014) Nonconvergence in Bayesian estimation of migration rates. Mol Ecol Res 14(4):726–733
Mossman C, Waser P (1999) Genetic detection of sex-biased dispersal. Mol Ecol 8:1063–1067
Olsson M, Shine R (2003) Female-biased natal and breeding dispersal in an alpine lizard, Niveoscincus microlepidotus. Biol J Lin Soc 79:277–283
Paetkau D, Calvert W, Stirling I, Strobeck C (1995) Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol 4:347–354
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenALEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Peterman WE, Connette GM, Semlitsch RD, Eggert LS (2014) Ecological resistance surfaces predict fine-scale genetic differentiation in a terrestrial woodland salamander. Mol Ecol 23:2402–2413
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Prugnolle F, De Meeˆus T (2002) Inferring sex-biased dispersal from population genetic tools: a review. Heredity 88:161–165
Pusey AE (1987) Sex-biased dispersal and inbreeding avoidance in birds and mammals. Trends Ecol Evol 2:295–299
Raufaste N, Bonhomme F (2000) Properties of bias and variance of two multiallelic estimators of FST. Theor Popul Biol 57:285–296
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Ronce O (2007) How does it feel to be like a rolling stone? Ten questions about dispersal evolution. Annu Rev Ecol Evol Syst 38:231–253
Rousset F (2008) genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Royle JA, Nichols JD (2003) Estimating abundance from repeated presence–absence data or point counts. Ecology 84:777–790
Ryberg K, Olsson M, Wapstra E, Madsen T, Anderholm S, Ujvari B (2004) Offspring-driven local dispersal in female sand lizards (Lacerta agilis). J Evol Biol 17:1215–1220
Scheelings T, Jessop T (2011) Influence of capture method, habitat quality and individual traits on blood parameters of free-ranging lace monitors (Varanus varius). Aust Vet J 89:360–365
Slatkin M (1985) Gene flow in natural populations. Annu Rev Ecol Syst 16:393–430
Smissen PJ, Melville J, Sumner J, Jessop TS (2013) Mountain barriers and river conduits: phylogeographical structure in a large, mobile lizard (Varanidae: Varanus varius) from eastern Australia. J Biogeogr 40:1729–1740
Smith GT, Arnold GW, Sarre S, AbenspergTraun M, Steven DE (1996) The effects of habitat fragmentation and livestock-grazing on animal communities in remnants of gimlet Eucalyptus salubris woodland in the Western Australian wheatbelt.2. Lizards J Appl Ecol 33:1302–1310
Smouse PE, Peakall R (1999) Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82:561–573
Stevens VM, Whitmee S, Le Galliard JF, Clobert J, Böhning-Gaese K, Bonte D, Brändle M, Matthias Dehling D, Hof C, Trochet A (2014) A comparative analysis of dispersal syndromes in terrestrial and semi-terrestrial animals. Ecol Lett 17:1039–1052
Trochet A, Courtois EA, Stevens VM, Baguette M, Chaine A, Schmeller DS, Clobert J, Wiens JJ (2016) Evolution of sex-biased dispersal. Q Rev Biol 91:297–320
Tucker A, McCallum H, Limpus C, McDonald K (1998) Sex-biased dispersal in a long-lived polygynous reptile (Crocodylus johnstoni). Behav Ecol Sociobiol 44:85–90
Tucker JM, Allendorf FW, Truex RL, Schwartz MK (2017) Sex-biased dispersal and spatial heterogeneity affect landscape resistance to gene flow in fisher. Ecosphere 8:e01839
Ujvari B, Dowton M, Madsen T (2008) Population genetic structure, gene flow and sex-biased dispersal in frillneck lizards (Chlamydosaurus kingii). Mol Ecol 17:3557–3564
Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wang W, Li J, Wang H, Ran Y, Wu H (2020) Genetic evidence reveals male-biased dispersal of the Omei tree frog. J Zool 310:201–209
Weavers BW (1993) Home range of male lace monitors, Varanus varius (Reptilia: Varanidae), in south-eastern Australia. Wildl Res 20:303–313
Acknowledgements
We gratefully acknowledge the assistance of several volunteers, especially Tim Lockwood, for their help with fieldwork.
Funding
Zoos Victoria provided research funding for the “forest for life” project.
Author information
Authors and Affiliations
Contributions
Conceptualization, resources and funding acquisition: TSJ, JMS; Methodology: TSJ, PS, JA, CS; Data collection: TSJ, PS, JA; Analysis and the original draft: TSJ, PS, CS, JSM; Review and editing: TSJ, JSM.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethics approval
The research was carried out under research permits 10005634 from the Department of Sustainability and Environment, Victoria, and Animal Experimental Ethics Committee approval 0911328.2 (University of Melbourne).
Consent to participate
All authors agree to participate in the study.
Consent for publication
All authors consent to the publication of the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jessop, T.S., Smissen, P., Anson, J.R. et al. Do common dispersal influences inform a large lizard’s landscape-scale gene flow?. Evol Ecol 36, 987–1006 (2022). https://doi.org/10.1007/s10682-022-10208-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10682-022-10208-2