Abstract
Competition-driven feeding niche separation is assumed to be an important driver of the morphological divergence of co-occurring animal species. However, despite a strong theoretical background, empirical studies showing a direct link between competition, diet divergence and specific morphological adaptations are still scarce. Here we studied the early steps of competition-driven eco-morphological divergence in two closely related passerines: the common nightingale (Luscinia megarhynchos) and the thrush nightingale (Luscinia luscinia). Our aim was to test whether previously-observed divergence in bill morphology and habitat in sympatric populations of both species is associated with dietary niche divergence. We collected and analysed data on (1) diet, using both DNA metabarcoding and visual identification of prey items, (2) habitat use, and (3) bill morphology in sympatric populations of both nightingale species. We tested whether the species differ in diet composition and whether there are any associations among diet, bill morphology and habitat use. We found that the two nightingale species have partitioned their feeding niches, and showed that differences in diet may be partially associated with the divergence in bill length in sympatric populations. We also observed an association between bill length and habitat use, suggesting that competition-driven habitat segregation could be linked with dietary and bill size divergence. Our results suggest that interspecific competition is an important driver of species’ eco-morphological divergence after their secondary contact, and provide insight into the early steps of such divergence in two closely related passerine species. Such divergence may facilitate species coexistence and strengthen reproductive isolation between species, and thus help to complete the speciation process.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Competition-driven feeding niche separation is considered to be one of the most important drivers of the morphological differentiation of co-occurring animal species (Brown and Wilson 1956; Schluter 2000). This is indicated by the frequent divergence of sympatric species in morphological traits related to foraging, such as jaw and tooth morphology, bill shape or body size (Grant and Grant 2002; Davies et al. 2007; Winkelmann et al. 2014; Drury et al. 2018). According to the classical niche theory, shifts in diet preferences enable the reduction of costly interspecific competition for food resources in sympatric species, which decreases the probability of the local extinction of the inferior competitor and enables long-term species’ coexistence in the same geographic area (MacArthur and Levins 1967; Pfennig and Pfennig 2009). However, despite such a strong theoretical background, studies showing a direct link between competition, diet divergence and specific morphological adaptations are still scarce (Schluter and McPhail 1992; Adams and Rohlf 2000; Grant and Grant 2002; Pfennig et al. 2007; Stuart and Losos 2013; Winkelmann et al. 2014). One reason for such a paucity of empirical studies are the practical challenges to collecting field data containing simultaneous information on species’ diet, morphology and ecology, and at the same time demonstrating a clear link between the divergence and competition.
To provide support for the competition-driven feeding niche divergence hypothesis and its effect on species morphology, studying the ongoing differentiation of young species with recent secondary contact is especially useful. Unlike analyses of distantly related species, where the steps of divergence are inferred retrospectively long after the divergence occurred (Tobias et al. 2014; Mönkkönen et al. 2017; Navalón et al. 2018; Felice et al. 2019), analyses of young species allow for the identification of early steps of differentiation (e.g. Wolf et al. 2008). Such early differentiation steps may even contribute to the evolution of reproductive isolation between the species and thus be an integral part of the speciation process (Sottas et al. 2018). Moreover, a comparison of sympatric populations affected by interspecific competition with allopatric populations without such competition enables the establishment of a connection between eco-morphological divergence and competition (Pfennig and Pfennig 2009).
Here we studied the early steps of eco-morphological divergence caused by interspecific competition in two closely related insectivorous passerines: the common nightingale (Luscinia megarhynchos, Brehm) and the thrush nightingale (Luscinia luscinia, Linnaeus). These species diverged approximately 1.8 Mya during Pleistocene climatic oscillations (Storchová et al. 2010), and their breeding areas currently overlap in a secondary contact zone spanning across Europe, where they occasionally hybridize (Sorjonen 1986). Reproductive isolation between them is incomplete and gene flow still occurs between the species (Storchová et al. 2010; Mořkovský et al. 2018; Albrecht et al. 2019). Both nightingale species are of similar size and have very similar morphology and ecology. They occupy dense shrubby vegetation in lowlands, often close to water bodies (Cramp and Brooks 1992). Both species are predominantly ground foragers, searching for food by exploring the litter under trees and shrubs (Cramp and Brooks 1992). For this purpose, they use strong and relatively long legs to displace the upper ground layer and uncover their invertebrate food, and then they pick the moving prey with their thin bill (Cramp and Brooks 1992). Nightingales also sometimes glean foliage on trees and shrubs, and very occasionally catch flying insects (Cramp and Brooks 1992). Previous studies have provided experimental evidence for intense interference competition between the two species (Reif et al. 2015; Souriau et al. 2018). A comparison of species morphology between sympatric and allopatric populations showed an increased divergence in bill size in sympatry compared to allopatry (Reifová et al. 2011a). Interspecific competition has also resulted in partial habitat segregation between the species in sympatry (Reif et al. 2018) that was shown to be associated with the bill size divergence (Sottas et al. 2018). However, it remains unclear whether the differences in habitat and bill size in these two nightingale species are linked to diet resources – presumably a principal driver of eco-morphological differentiation in birds (Gill 2006; Olsen 2017).
We expand on previous work by analysing the diet of both nightingale species in sympatry using DNA metabarcoding (Taberlet et al. 2018; Alberdi et al. 2019), as well as the visual identification of prey items under stereomicroscope (Poulin et al. 1994a). We further relate the diet data to bill morphology of both species and the habitats in their territories. We tested whether the observed divergence in bill size may be explained by differences in diet between the two species driven by their use of different habitats. Our results could elucidate the early steps of competition-driven eco-morphological divergence in passerine birds.
Materials and methods
Study area and sampling
We carried out sampling in Central Poland (Fig. 1), where breeding ranges of both species overlap. Although the degree of species’ co-occurrence varies across their sympatric zone (Reif et al. 2018), in our study area they bred very close to each other and their territories were often adjacent or slightly overlapping (J. Reif, personal observation). Both species were sampled in May 2016 and 2017 during the breeding seasons, when territories are already established. All sampled individuals were males. In total, 37 common nightingale (Luscinia megarhynchos, Brehm, hereafter CN) and 22 thrush nightingale (Luscinia luscinia, Linnaeus, hereafter TN) males were trapped using a mist net with tape luring. The two species were sampled evenly in both years, to avoid possible effects of variation in food availability between years causing a false diet divergence signal between the species. The Julian date (i.e. the number of days that passed since the 1st of January of a given year) of sampling was also comparable between species; nonetheless, to check for its possible effect on diet variation, it was included as a covariate into the models (see below). Each individual was ringed, and its species identity was determined according to species-specific morphological characteristics (Cramp and Brooks 1992; Kverek et al. 2008). Then we measured the bill length (measured to the skull), bill width and bill depth (both measured at the frontal margin of nostrils); see Reifová et al. (2011a) for more details. All measurements were conducted by the same person (JR), using the same equipment and were done three times to check for repeatability (see below). A list of the sampled birds including their dates of sampling, GPS coordinates and bill measurements is provided in Supplementary Table S1.
The diet samples were collected in two forms – faecal samples and stomach regurgitates—to obtain a more complex overview of the nightingale diet. To collect faecal samples, the bird was placed into a clean paper bag immediately after its capture and kept there until defecated (no longer than 5 min). To obtain stomach regurgitates, each bird was given 1 mL of 1.5% emetic solution (antimony potassium tartrate). The emetic was administered orally through a flexible tube attached to a syringe (Poulin et al. 1994b). The bird was then placed into a dark box for 10 min to regurgitate and then released. Both faecal samples and stomach regurgitates were stored in 99% ethanol. In total, we obtained 44 faecal samples (27 from CN and 17 from TN) and 42 regurgitate samples (29 from CN and 13 from TN).
Diet determination
Visual identification of prey items
We examined stomach regurgitates from individual birds under a dissecting scope. Most of the arthropods were fragmented, and thus their identification was based on the least digestible and most characteristic parts. We used published information (Tatner 1983; Arlettaz et al. 2017) and our own guide (available online https://multitrophicinteractions.blog/downloads) to identify the prey to the level of genus, and if this was not possible to the family, order or class level. We also measured the length of each arthropod individual or its body part to the nearest 0.1 mm. We estimated the body length of each prey according to the published order-specific equations using the lengths of different body parts (Calvemr and Woolledd 1982; Hódar 1997) (Supplementary Table S2). The visual identification of prey items resulted in 683 identified arthropod individuals belonging to 35 different genera and 11 orders (Supplementary Table S2).
DNA metabarcoding
DNA metabarcoding was used to identify prey arthropod taxa in both faecal samples and stomach regurgitates. Metagenomic DNA from both types of samples was extracted by a PowerSoil DNA isolation kit (MO BIO Laboratories Inc., USA). Amplicone libraries were prepared using cytochrome c oxidase I (COI)-specific primers, BF2 primer GCHCCHGAYATRGCHTTYCC and BR2 primer TCDGGRTGNCCRAARAAYCA, targeting a broad range of invertebrate taxa (Elbrecht and Leese 2017). To reduce issues associated with the formation of primer-dimers, sequencing libraries were prepared in three steps: (1) We performed PCR pre-amplification of the target COI region with gene-specific primers. To avoid over-representation of host’s COI in the amplicon libraries, a blocking primer exhibiting a perfect match to the host COI containing a C3 spacer modification on the 3′ end and partially overlapping the reverse universal primer (TCCGAAGAATCAGAAGAGGTGTTGGTAGAGKAC) was added to PCR reaction. (2) We performed PCR amplification with COI primers compatible with sequencing adaptors. (3) We used PCR-based ligation to attach the sequencing adaptors. See Appendix S1 for more details of all three PCR steps.
Technical PCR duplicates were prepared for all samples. Products from the 3rd PCR round were quantified by GenoSoft software (VWR International, Belgium) based on the band intensities after electrophoresis on a 1.5% agarose gel and mixed at equimolar concentrations. The final library was cleaned up using Agencourt AmpureXP beads (Beckman Coulter Life Sciences). Products of the desired size (450–700 bp) were extracted by PipinPrep (Sage Science Inc., USA) and sequenced on an Illumina Miseq (v3 kit, 300 bp paired-end reads) at the Central European Institute of Technology (CEITEC, Masaryk University, Brno, Czech Republic).
Skewer (Hongshan et al. 2014) was used for demultiplexing of sequencing data and for trimming of gene-specific primers. To determine reliable COI haplotypes (hereafter OTUs, i.e. Operational Taxonomic Units), we truncated all sequences after the 240th nucleotide, filtered low-quality sequences (containing more than two expected errors or any indeterminate nucleotide) and denoised the remaining high-quality sequences using dada2 (Callahan et al. 2016). Subsequently, an abundance matrix (representing read counts of each OTU in each sample) was constructed. To eliminate PCR or sequencing artefacts that were not corrected by dada2, we considered only these OTUs that were consistently present in both technical duplicates for a given sample. Consistency in OTU content among technical duplicates was high according to Procrustean analysis (r > 0.95, p < 0.0001); thus we merged them for the purpose of subsequent analyses. Next, naïve Bayesian RDP classifier (Wang et al. 2007) implemented in the dada2 pipeline was used for taxonomic assignment of OTUs. The reference dataset for the taxonomic assignment was constructed using COI sequences downloaded from the NCBI nt database (200 top blastn hits for each of our OTU, environmental samples were excluded).
Based on the existing knowledge of nightingales’ diet during the breeding season (Cramp and Brooks 1992), we considered OTUs assigned as non-invertebrates (e.g. fungi, algae or plants), representing 22% of reads after quality filtering, as non-target taxa for diet analyses and excluded them from the dataset. The rest of the identified OTUs was represented by 247 arthropod genera belonging to 27 orders. The number of genera identified in the stomach regurgitates was slightly higher (184 genera; Supplementary Fig. S1) than in faecal samples (160 genera; Supplementary Fig. S1). However, we did not find a significant difference in prey composition between the two forms of diet samples (Supplementary Table S3). We thus merged both datasets when they were available for the same individual. Additionally, we removed 4 nightingale individuals (3 CN and 1 TN) due to a low number of reads (less than 1000 reads per individual).
A matrix of read counts for individual COI haplotypes in each sample, along with sample metadata, taxonomic annotations and haplotype sequences, was merged into a joint database using the PHYLOSEQ package (McMurdie and Holmes 2013). All subsequent analyses focused on the relative abundance variation of arthropod (1) orders, which provides information about major differences in diet preferences, and (2) genera, which gives more subtle information on differences in diet.
As DNA metabarcoding provided more detailed information on diet than visual identification of prey items (247 vs. 35 identified genera, respectively, see above), we used the diet data obtained by DNA metabarcoding for further statistical analyses. Nevertheless, to check whether the datasets from visual identification and DNA metabarcoding provided consistent results, we compared the relative abundances of particular prey taxa in both datasets. Additionally, visual data were used for testing differences in prey size between the species.
Habitat description
To describe the habitat in nightingale territories, we delimited a radius of 50 m around the capture location of a given individual. This territory definition is a reasonable approximation of the space used by small territorial passerines including nightingales (Naguib et al. 2004; Wood et al. 2016). The habitat within each territory was described from two perspectives, (1) the vegetation composition and (2) the vegetation density, as both can affect the composition of arthropod communities (Schaffers et al. 2008; Kadlec et al. 2018) and thus the nightingale diet.
The vegetation composition was characterized by the presence of various tree and shrub species as well as the predominant type of ground layer. We distinguished three ground layer types composed of: (a) Urtica spp., (b) other herbs, and (c) no vegetation. We discriminated Urtica spp. from other herbs due to its large and dense stands 1–2 m in height, which creates a visually different microhabitat compared to all other common herb taxa in nightingale territories. We assumed that such a different physiognomy may also be linked to differences in food supply for nightingales. Moreover, nightingales often place their nests in Urtica spp. stands, possibly due to its protective function against predators (Becker 1995), which makes this plant of special importance for nightingale species. In the tree and shrub layer, we identified a total of 24 tree and shrub species. For further analyses, we removed tree and shrub species present in less than five nightingale territories (across both species combined), as their differential representation between species would be difficult to assess. The final dataset consisted of 19 tree and shrub species.
The vegetation density was characterized along 4 transects (each 50 m in length) that were drawn from the bird capture location in each cardinal direction (N-S-W-E) (Supplementary Fig. S2). Along each transect, 5 locations (one spot every 10 m) were defined. At these locations, the density of vegetation in three vertical strata (up to 1 m, between 1 and 2 m, and above 2 m) was determined. The density of vegetation was categorized as: 0—no vegetation; 1—sparse vegetation with the next spot clearly visible; 2—dense vegetation, but the next spot was still visible; 3—very dense vegetation with very little or no visibility to the next spot. The most common category across the locations was assigned for each transect, and then the most common category across the four transects was assigned to each individual for each stratum.
To estimate the distances as precisely as possible, we used a 2 m high pole (marked every 10 cm) for small distances (i.e. to delineate the vertical strata), and a 30 m long rope or a laser range finder for longer distances.
Statistical analyses
All statistical analyses were done using packages running under R Statistical Software version 3.4.3 (R Core Team 2015). We first analysed between-species differences in bill morphology, diet and habitat use. In the next step, we tested for possible relationships among bill morphology, diet and habitat use. Relationships tested on spatially explicit data may be affected by spatial autocorrelation (sites being closer to each other may be more similar than more distant sites; Diniz-Filho et al. 2003). To check for the presence of spatial autocorrelations in our data, we performed a mantel test (Legendre et al. 2015) on each Euclidian geographical distance matrix calculated for the respective response variables used in the analyses. We used the mantel.test function from the R package ‘ade4’.
Nightingale diet composition and interspecific differences in diet
To assess the diet of both nightingale species, we evaluated (1) the dietary diversity for each nightingale species by measuring the Shannon’s diversity index (Hsh) and (2) the diet similarity between the two nightingale species by using Pianka’s index (O). The Shannon’s diversity index (Hsh) was calculated as:
where pi is the proportion of prey items belonging to the genus i (Spellerberg and Fedor 2003).
Pianka’s index (O) was calculated as:
where pi is the proportion of samples containing the prey of the genus i in the diet of CN and TN when n is the total number of prey genera (Pianka 1973). The index varies between 0 (total separation) and 1 (complete overlap).
The association between diet composition and species identity was firstly assessed by distance-based redundancy analyses (db-RDAs, Legendre and Andersson 1999) with nightingale species identity and Julian date included as explanatory variables. The diet composition was entered as a Bray–Curtis distance matrix for the relative abundances of insect (1) orders or (2) genera as the response variable. The relative abundance is the proportion of a prey item at a given order or genus level within all prey items in one sample. The significances of explanatory variables in db-RDAs were assessed by a permutation-based ANOVA.
Secondly, we used generalized linear models with a negative binomial distribution for community data (R package mvabund; Wang et al. 2012) to test which particular prey taxa were differentially represented in the diet of the two nightingale species. The response variable was the read counts for prey (1) orders or (2) genera, and nightingale species identity (i.e. CN or TN) was the explanatory variable. Log-transformed total number of reads per sample was specified as the model offset. The model was run for each taxon, therefore the false discovery rate method (Benjamini and Hochberg 1995) was subsequently used to account for false discoveries due to multiple testing.
Interspecific differences in bill morphology
We calculated the repeatability of the three measurements for each bill dimension (length, depth and width) using the rptR package (Stoffel et al. 2017) with 95% confidence intervals (CI) based on linear mixed models method and 1000 bootstrap samples. The repeatability estimates were 0.85 (CI = 0.77–0.90) for bill length, 0.75 (CI = 0.64–0.82) for bill depth, and 0.64 (CI = 0.51–0.74) for bill width, suggesting a low technical variability in bill measurements. We thus used the mean of the three values for each bill dimensions for further analyses. The three bill dimensions were reduced using a covariance Principal Component Analysis (PCA) into two non-independent composite variables (principal components, PCs). In the next step, we used a linear model where the two principal components were the respective response variables and nightingale species identity (CN or TN) was the explanatory variable.
In contrast to our previous studies (Reifová et al. 2011a; Sottas et al. 2018) expressing bill size relative to body size, we analysed here the absolute (not the relative) measures of bill size, as they should be directly linked to diet resources (Gill 2006; Olsen 2017). We thus also checked whether the absolute bill size also showed increased divergence in sympatry compared to allopatry by reanalysing morphological data from Reifová et al. (2011a) (see Appendix S2).
Interspecific differences in habitat use
We performed a Multiple Correspondence Analysis (MCA, Abdi and Valentin 2007) on the (1) vegetation composition data and (2) vegetation density data in nightingale territories. MCA is a data reduction technique particularly suitable for categorical data such as the presence/absence of plant taxa in our data set. The resulting uncorrelated MCA axes were used as composite descriptors of vegetation composition and vegetation density for further analyses as follows. To assess whether the species differ in habitat use, linear models were used where the respective response variables were vegetation composition or vegetation density and nightingale species identity was the explanatory variable.
Associations among bill morphology, diet and habitat
The associations between (1) diet and bill morphology, (2) diet and habitat, and (3) bill morphology and habitat were assessed using db-RDAs (Table 1). For this purpose, we used only morphological and habitat variables that showed significant differences between the species in the interspecific comparisons described above, as we were primarily interested whether differences in these traits could be associated with diet differentiation. The diet composition was entered as a Bray–Curtis distance matrix based on the relative abundance at (1) order or (2) genus levels. When a model showed a significant association, we added nightingale species identity as a predictor into the model to test for the effect of species on the tested association (all performed models are specified in Table 1). In all models, Julian date was included as a covariate to control for the possible effects of seasonality on the diet composition. The significance of the explanatory variables was assessed by permutation-based ANOVA.
Results
Nightingale diet composition and interspecific differences in diet
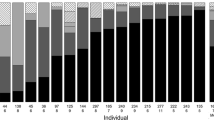
Sequencing of metagenomic DNA from the faecal samples and stomach regurgitates resulted in 1,307,666 reads across all samples. The mean sequencing depth per individual was 26,584 (range = 1181–70,626) in CN and 19,228 (range = 1027–120,262) in TN. In total, 247 invertebrate genera belonging to 27 orders were identified and the average number of identified genera per individual was 11.76 (range = 1–26). The diet of nightingales consisted predominantly of insects (84.45% of relative abundance); however, a few other arthropod classes were represented in the diet as well: Diplopoda (7.34%), Malacostraca (3.22%), Arachnida (2.74%), Collembola (1.14%) and Protura (0.73%). The most frequently occurring orders in the nightingale diet (all insects) were Coleoptera (45.22%), Diptera (12.84%), Hymenoptera (12.90%) and Lepidoptera (8.46%) (Fig. 2a, Supplementary Table S4).
a Overall diet composition for both nightingale species presented as the relative abundance of particular prey taxa from the total number of prey items identified at the order level in the diet of both nightingales (the common nightingale and thrush nightingale were considered jointly). Rare taxa (represented by < 0.5%) are labelled as ‘others’. b Differences in diet (relative abundance of each taxon) between the common nightingale (red bars) and the thrush nightingale (blue bars)
The two species had similar dietary diversity measured by Shannon’s diversity index (Hsh = 3.03 in TN; Hsh = 3.53 in CN) and showed moderate niche overlap at the genus level expressed by Pianka’s index (O = 0.54). Specifically, 145 genera were consumed only by CN (34 individuals), 42 only by TN (21 individuals) and 60 genera were consumed by both species (Supplementary Table S4). To test for interspecific differences in diet, we excluded rare orders or genera (occurring in less than 5 individuals) depending on which level the analysis was conducted. In total, we excluded 14 orders and 216 genera as their differential abundance between the species would be difficult to assess. The permutation-based ANOVA running on db-RDA revealed: (1) a significant effect of species identity (F1, 52 = 4.333, p = 0.003) but no effect of the Julian date (F1,52 = 1.119, p = 0.295) on the diet composition at the order level (model adjusted-R2 = 0.078); (2) a significant effect of species identity (F1, 52 = 1.876, p = 0.017) and Julian date (F1, 52 = 1.855, p = 0.013) on the diet composition at the genus level (model adjusted-R2 = 0.038).
According to the negative binomial generalized linear models, three orders – Coleoptera and Diptera (both belonging to class Insecta) and Protura (belonging to class Protura) were significantly differentially represented in the diet of the two the nightingale species after correction for multiple testing (Table 2a, Fig. 2b). Coleoptera represented a larger proportion of the diet in CN (58.39% of relative abundance) than in TN (27.80% of relative abundance). On the contrary, the orders Diptera (CN: 5.10% and TN: 26.57% of relative abundance) and Protura (CN: 0.03% and TN: 1.98% of relative abundance) were more common in the diet of TN than CN (Table 2a, Fig. 2b).
Nine genera were significantly differentially represented in the diet of the two nightingale species after correction for multiple testing. The genera Aphodius, Brachysomus, Dalopius, Heterotarsus, Notoxus Othiorhynchus (all belonging to the order Coleoptera), Lygus (order Hemiptera) and Formica (order Hymenoptera) were significantly more common in the CN diet, while unclassified Diptera genera were more common in the TN than CN diet (Table 2b).
Visual identification of prey items provided much less data and the representation of particular taxa was quite different from DNA metabarcoding dataset. It was thus difficult to compare the results from both datasets. Nevertheless, Coleoptera, which were highly represented in the visual data, were also more common in the CN than in the TN diet (Supplementary Table S5). This difference, however, was not statistically significant (Supplementary Table S5). Additionally, we used the data from visual identification to estimate the body length of prey items (Supplementary Table S2). We found that the size of prey items did not significantly differ between CN (mean = 8.31 mm, sd = 6.13) and TN (mean = 8.78 mm, sd = 4.76) (Wilcox test: p = 0.69).
Interspecific differences in bill morphology
We used a PCA to reduce the three bill dimensions into two uncorrelated PC axes. The first axis (PC1) explained 45% of the total variation in bill morphology and was strongly positively correlated with the bill width and depth (Supplementary Fig. S3 and Supplementary Table S6). The second axis (PC2) explained 33% of the total variability, and was strongly negatively correlated with the bill length (Supplementary Fig. S3 and Supplementary Table S6). While PC1 did not differ between the species (F1, 53 = 1.502, p = 0.226), PC2 showed marked differences (F1, 53 = 22.590, p < 0.001), with the species identity explaining 29% of the total variability in this model. We thus used the PC2 (hereafter referred as ‘bill length’) for subsequent analyses testing the association between bill length and diet. Using raw measurements of bill length (Supplementary Table S1), CN also showed significantly (t test: t = 4.930, df = 38.38, p < 0.001) longer bills (mean = 17.315 mm, sd = 0.580) than TN (mean = 16.452 mm, sd = 0.659). The larger bill length in CN cannot be explained by the body size, as CN is significantly smaller than TN (Reifová et al. 2011a).
The reanalysis of data on absolute bill size in sympatric and allopatric populations from Reifová et al. (2011a) (Appendix S2) confirmed that there is significantly increased divergence in bill length in sympatry compared to allopatry, which suggests that interspecific competition drives the bill length divergence between the species (Appendix S2).
Interspecific differences in habitat use
According to the MCA on variables describing vegetation composition, the first ordination axis (hereafter called vegetation composition 1) explained 15% of the variability in plant taxa records in the nightingale territories. It expresses a gradient from wet habitats, characterized by willow, reed and the presence of Urtica spp. on the ground, to dry habitats, characterized by the presence of oak, blackthorn, and bare or herbaceous ground layers (Supplementary Fig. S4a). The second axis (vegetation composition 2) explained 11% of the variability in plant taxa records in the nightingale territories. Plants occurring in forests or at forest edges, such as European ash or poplar, were linked with the negative part of the axis, whereas open habitats characterized by the presence of species such as rose or blackthorn represented the positive part of this axis (Supplementary Fig. S4a).
The MCA on the variables describing vegetation density revealed the first axis (vegetation density 1), explaining 18% of the variability, as a gradient from no vegetation in intermediate and upper strata to sparse vegetation in low strata (Supplementary Fig. S4b). The second axis (vegetation density 2) explained 16% of the variability and expressed a gradient from sparse vegetation in high/intermediate and upper strata to no vegetation in low strata (Supplementary Fig. S4b).
The results of the linear models revealed that vegetation composition 1 differs between the nightingale species (F1, 53 = 8.423, p < 0.001, adjusted-R2 = 0.121), with CN occurring more frequently in drier habitats while TN in wetter habitats. No differences between species were found in respect to the vegetation composition 2 (F1, 53 = 1.583, p = 0.214, adjusted-R2 = 0.011), vegetation density 1 (F1, 53 = 0.023, p = 0.288, adjusted-R2 = − 0.018) or vegetation density 2 (F1, 53 = 0.165, p = 0.686, adjusted-R2 = − 0.016).
Associations among bill morphology, diet and habitat use
We found a significant association between bill length and diet composition after controlling for the effect of Julian date at the genus level (p = 0.011, adjusted-R2 = 0.039), but not at the order level (Table 1). Longer bills were associated with the presence of genera Brachysomus and Aphodius (Coleoptera) and Operophtera (Lepidoptera) in the diet (Fig. 3d). Conversely, shorter bills were associated with the genera Julus (Julida, Diplopoda) and Strongylosoma (Polydesmida, Diplopoda), and Tipula (Diptera) (Fig. 3d). The association between the bill length and diet became insignificant when the nightingale species identity was included into the model (p = 0.259), suggesting that this association is to a large degree driven by between-species differences in bill length and diet in sympatry (Table 1). This was supported by very similar performances of the model where species identity was the only predictor of the diet (adjusted-R2 = 0.038) and the model where both species identity and bill length were included as explanatory variables (adjusted-R2 = 0.043) (Table 1); the variation explained by these two models did not differ significantly (F1, 51 = 1.777, p = 0.238). Variation partitioning analysis (Varpart analysis, Peres-Neto et al. 2006) showed that 1.6% of variation in diet composition was explained solely by the effect of bill length, 2% solely by the effect of species identity and 2.5% by correlated effect of bill length and species identity.
Distance-based redundancy analyses ordination of the diet composition in nightingales. The dissimilarity matrix based on the relative abundance on diet samples at the order (a and c) or genus level (b and d) was the response variable, while vegetation composition 1 (a and b) or bill length (c and d), and Julian date (julian) were explanatory variables. Variation along the first two constrained axes is shown. Colour lozenges represent the prey taxa found in the diet at the order or genus level. Dots represent nightingale individuals (common nightingale in red, thrush nightingale in blue)
The diet was not related to the vegetation composition 1 at the order level (p = 0.819, adjusted-R2 = 0.003) or at the genus level (p = 0.369, adjusted-R2 = 0.023) (Table 1; Fig. 3a, b).
We found a significant association between the bill length and vegetation composition 1 (p = 0.009, adjusted-R2 = 0.085). Nightingales had longer bills in dry than in wet habitats (Fig. 3c, d). However, the association between bill length and vegetation composition 1 became insignificant (p = 0.143) if the nightingale species identity was included in the model, suggesting that this association is largely driven by interspecific differences (Table 1). Indeed, the model where bill length was related only to species identity explained only slightly less variation (adjusted-R2 = 0.310) than the model where both species identity and vegetation composition were included (adjusted-R2 = 0.325; Table 1) and this difference was not significant (F1, 51 = 2.147, p = 0.152). Variation partitioning analysis showed that 9% of variation in bill length was explained solely by the effect of vegetation, 28% solely by the effect of species identity and 29% by correlated effect of vegetation and species identity.
Spatial autocorrelation
We observed no linear correlation between space and (1) diet expressed as the read counts at the genus level (Mantel’s test, r = 0.039, p = 0.289), (2) vegetation composition 1 (Mantel’s test, r = − 0.081, p = 0.934) or (3) bill length (Mantel’s test, r = 0.069, p = 0.172).
Discussion
In this study, we explored the early steps of competition-driven eco-morphological divergence in two closely related insectivorous passerines, the common nightingale and the thrush nightingale. In accordance with previous studies (Reifová et al. 2011a; Reif et al. 2018; Sottas et al. 2018), we found that the two species diverged in bill morphology as well as in habitat use in sympatry. Such differences were not observed in the adjacent allopatric populations (Reifová et al. 2011a; Reif et al. 2018), even though possible geographical gradients were taken into account, suggesting that they arose in response to interspecific competition. The common nightingale in sympatry occurred more frequently in dry habitats and showed a longer bill, while the thrush nightingale was more abundant in wet habitats and had a shorter bill. Although the diet of both species largely overlapped in sympatric populations, we observed some significant differences in diet composition between the species, particularly for Coleoptera, which were more frequent in the common nightingale diet, and Diptera and Protura, which were more common in the thrush nightingale diet. Importantly, we found a significant relationship between bill length and diet. Individuals with a short bill fed more often on genera from the Diptera, Julida and Polydesmida orders, while individuals with a longer bill on genera from the Coleoptera and Lepidoptera orders. We also revealed a significant association between bill length and habitat, with bill length increasing towards drier habitats. Our results provide new insights into the early stages of competition-driven eco-morphological divergence in two closely related passerine birds. Below, we discuss details of this eco-morphological divergence as well as its implications for the long-term co-existence of the species and the evolution of reproductive isolation between them.
Shifts in feeding niches between competing species are crucial for long-term species coexistence in the same geographic area (MacArthur and Levins 1967), and are a keystone of many adaptive radiations in birds (Grant 1981; Freed et al. 2016) as well as in other taxa (Seehausen 2006; Losos 2011). Feeding niche divergence may be achieved directly by changes in diet preferences (Renner et al. 2012) as well as indirectly through foraging in different habitats or at different times (Pianka 1973; Schoener 1974). Our results, showing that the two nightingale species have differentiated in sympatry both in habitat composition and diet, suggest that they may have differentiated their feeding niches as a result of habitat segregation. Given that the composition of arthropod communities is often closely linked to vegetation (Schaffers et al. 2008; Kadlec et al. 2018), this possibility seems likely. On the other hand, we did not find a significant association between habitat composition and nightingale diet. This may be a result of our relatively low sample size and the fact that our diet data only reflected a snapshot of the diet at a given time and place. In any case, based on our data, we cannot exclude the alternative possibility that the species have differentiated their diet independently of habitat. Data on diet from allopatric populations could help to elucidate this issue.
Different diets can in turn drive a divergence in bill size. Our results showing a significant association between bill length and diet support this scenario. Moreover, the significant association between bill length and habitat gives some support to the hypothesis of feeding niche divergence due to competition-driven habitat segregation. Nevertheless, it should be noted that associations between bill length and diet as well as between bill length and habitat were to a large degree driven by species identity. However, our previous study demonstrated that at least the association between bill size and habitat cannot be explained entirely by species identity, and clearly demonstrated that bill size in sympatry is more divergent between species in different habitats than in the same habitats (Sottas et al. 2018). It is also worth noting that species identity explained less than 30% of the variation in bill length, suggesting that some other selective pressures are involved in shaping bill morphology in nightingales in addition to the mechanisms driving interspecific divergence. Together, these results indicate that nightingales could have differentiated their feeding niches as a result of habitat segregation, although more data is needed to confirm this conclusion.
Competition-driven habitat segregation resulting in changes in feeding niches has been described in a few other closely related species (e.g. Schluter and McPhail 1992; Fossog et al. 2014; Winkelmann et al. 2014). In a study on Ficedula flycatchers, the more dominant collared flycatcher displaced the submissive pied flycatcher from its preferred breeding habitat, which was associated with reduced access to the preferred food resources (Rybinski et al. 2016). Competition also played a role in the habitat segregation of two distinct eco-morphs of the cichlid fish Telmatochromis temporalis (Winkelmann et al. 2014), with the larger eco-morph outcompeting the smaller one on a favoured rock substrate. In both cases, habitat isolation promoted assortative mating within the same species or eco-morph, which strengthened the reproductive isolation between the incipient species. Habitat segregation resulting from interspecific competition may be thus a common driver of eco-morphological divergence in sympatric species at the early stages of their divergence. In cases of still not yet completely reproductively isolated species, as in the two nightingale species studied here (Storchová et al. 2010; Reifová et al. 2011b; Mořkovský et al. 2018), it may also contribute to the increase in reproductive isolation between the species by strengthening prezygotic isolation.
To demonstrate an association between diet, bill size and habitat, it is necessary to understand the functional link between these variables. It is possible that the longer bill in the common nightingale, occurring more often in dry habitats, might be caused by larger prey sizes. Our analysis of prey size in both nightingale species, however, did not support this hypothesis. The bill morphology may also be affected by the feeding strategy (e.g. Miller et al. 2017). The representatives of Coleoptera found more frequently in the common nightingale diet (e.g. Brachysomus, Othiorhynchus, Notoxus) are beetle genera occurring on the ground. In fact, most of these Coleoptera genera occurred almost exclusively in the common nightingale diet and were absent in the thrush nightingale diet (Table 2b). We also found underground Coleoptera larvae (e.g. Tenebrionidae) in the common nightingale but not in the thrush nightingale diet. By contrast, Diptera, which occurred more often in the thrush nightingale diet, are normally found on vegetation, and thrush nightingales might capture them by gleaning or while flying. This may be related to the fact that thrush nightingale habitats often occur in river floodplains in sympatry (Reif et al. 2018) where ground foraging is not possible during spring when these areas are flooded. The divergence in bill morphology between the two nightingale species may thus result from their different feeding strategies in different habitats. Indeed, a study on Acrocephalus warblers found that short bills were associated with gleaning feeding techniques, whereas long bills were typical for species that pursue hidden and difficult-to-access prey (Leisler and Winkler 2015). Some other studies on passerine birds have suggested that longer bills increase the dietary niche breadth (Brandl et al. 1994). The dietary diversity measured by Shannon index was, however, comparable in our two nightingale species.
Based on our results, we cannot exclude other explanations for the bill size divergence between the two nightingale species in sympatry. First, bill size may be affected by temperature. It has been shown that bill size increases with temperature as it plays an important role in thermoregulation and heat lost (Greenberg et al. 2012; Luther and Greenberg 2014; Friedman et al. 2017; Tattersall et al. 2017). This could explain why the common nightingale, occurring in drier and potentially hotter habitats in sympatry (Reif et al. 2018), has a longer bill. However, since the habitat driven climatic variability is likely very small within our study area compared to other sources of climate variation (e.g. seasonality), we consider this explanation unconvincing. Second, bill size may also be shaped by song (Derryberry et al. 2012). It has been shown that bird song can be affected by vegetation structure (‘acoustic adaptation hypothesis’, Boncoraglio and Saino 2007). Nevertheless this explanation is unlikely in our system as no difference in vegetation density was observed between the species, and the species show song convergence rather than divergence in sympatry (Vokurková et al. 2013; Souriau et al. 2018).
In conclusion, our results, together with previously published studies (Reifová et al. 2011a; Reif et al. 2018; Sottas et al. 2018), suggest that interspecific competition between the two nightingale species in their secondary contact zone has led to partial habitat segregation, and this could have played a role in the feeding niche divergence we observed in this study. However, based on our data we cannot provide direct evidence for this scenario with a link between diet composition and habitat segregation. Nevertheless, our data indicate that the feeding niche divergence in the two nightingale species has in turn resulted in bill size divergence in sympatry. The observed habitat-partitioning and feeding niche divergence may together facilitate the coexistence of the two nightingale species in the same geographical area by reducing resource overlap. Since the two nightingale species are still not completely reproductively isolated (Storchová et al. 2010, Reifová et al. 2011b, Mořkovský et al. 2018), competition-driven eco-morphological divergence may also strengthen the degree of reproductive isolation and thus contribute to the speciation process.
Data availability
Sequencing data are available from the European Nucleotide Archive under the study accession number: PRJEB37727. Metadata to the individual sequence samples are provided in the Supplementary Table S7. All other data are attached as Supplementary Material.
References
Abdi H, Valentin D (2007) Multiple correspondence analysis. Mult Corresp Anal Soc Sci 2:651–666
Adams DC, Rohlf FJ (2000) Ecological character displacement in Plethodon: biomechanical differences found from a geometric morphometric study. Proc Natl Acad Sci 97:4106–4111. https://doi.org/10.1073/pnas.97.8.4106
Alberdi A, Aizpurua O, Bohmann K et al (2019) Promises and pitfalls of using high-throughput sequencing for diet analysis. Mol Ecol Resour 19:327–348. https://doi.org/10.1111/1755-0998.12960
Albrecht T, Opletalová K, Reif J et al (2019) Sperm divergence in a passerine contact zone: indication of reinforcement at the gametic level. Evolution 73:202–213. https://doi.org/10.1111/evo.13677
Arlettaz R, Christe P, Schaub M (2017) Food availability as a major driver in the evolution of life-history strategies of sibling species. Ecol Evol 7:4163–4172. https://doi.org/10.1002/ece3.2909
Becker J (1995) Sympatric occurrence and hybridization of the Thrush Nightingale (Luscinia luscinia) and the Nightingale (Luscinia megarhynchos) at Frankfurt (Oder), Brandenburg. Vogelwelt 116:109–118
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300. https://doi.org/10.1111/j.2517-6161.1995.tb02031.x
Boncoraglio G, Saino N (2007) Habitat structure and the evolution of bird song: a meta-analysis of the evidence for the acoustic adaptation hypothesis. Funct Ecol 21:134–142. https://doi.org/10.1111/j.1365-2435.2006.01207.x
Brandl R, Kristín A, Leisler B (1994) Dietary niche breadth in a local community of passerine birds: an analysis using phylogenetic contrasts. Oecologia 98:109–116. https://doi.org/10.1007/BF00326096
Brown WL, Wilson EO (1956) Character displacement. Syst Zool 5:49–64. https://doi.org/10.2307/2411924
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Calvemr MC, Woolledd RD (1982) A technique for assessing the taxa, length, dry weight and energy content of the arthropod prey of birds. Aust Wildl Res 9:293–301. https://doi.org/10.1071/WR9820293
Cramp S, Brooks DJ (1992) Handbook of the birds of Europe, the Middle East and North Africa. The birds of the western Palearctic, vol VI. Oxford University Press, Warblers
Davies TJ, Meiri S, Barraclough GT, Gittleman LJ (2007) Species co-existence and character divergence across carnivores. Ecol Lett 10:146–152. https://doi.org/10.1111/j.1461-0248.2006.01005.x
Derryberry EP, Seddon N, Claramunt S et al (2012) Correlated evolution of beak morphology and song in the neotropical woodcreeper radiation. Evolution 66:2784–2797. https://doi.org/10.1111/j.1558-5646.2012.01642.x
Diniz-Filho JAF, Bini LM, Hawkins BA (2003) Spatial autocorrelation and red herrings in geographical ecology. Glob Ecol Biogeogr 12:53–64. https://doi.org/10.1046/j.1466-822X.2003.00322.x
Drury JP, Tobias JA, Burns KJ et al (2018) Contrasting impacts of competition on ecological and social trait evolution in songbirds. PLoS Biol 16:1–23. https://doi.org/10.1371/journal.pbio.2003563
Elbrecht V, Leese F (2017) Corrigendum: validation and development of COI metabarcoding primers for freshwater macroinvertebrate bioassessment. Front Environ Sci 5:1–11. https://doi.org/10.3389/fenvs.2017.00038
Felice RN, Tobias JA, Pigot AL, Goswami A (2019) Dietary niche and the evolution of cranial morphology in birds. Proc R Soc B Biol Sci 286:20182677. https://doi.org/10.1098/rspb.2018.2677
Fossog BT, Ayala D, Acevedo P et al (2014) Habitat segregation and ecological character displacement in cryptic African malaria mosquitoes. Evol Appl 8:326–345. https://doi.org/10.1111/eva.12242
Freed LA, Medeiros MCI, Cann RL (2016) Multiple reversals of bill length over 1.7 million years in a Hawaiian bird lineage. Am Nat 187:363–371. https://doi.org/10.1086/684787
Friedman NR, Harmáčková L, Economo EP, Remeš V (2017) Smaller beaks for colder winters: thermoregulation drives beak size evolution in Australasian songbirds. Evolution 71:2120–2129. https://doi.org/10.1111/evo.13274
Gill FB (2006) Ornithology, 3rd edn. Macmillan Publishers Limited, New York
Grant PR (1981) Speciation radiation and the adaptive of Darwin’s finches. Am Sci 69:653–663
Grant PR, Grant RB (2002) Unpredictable evolution in a 30 year study of Darwin’s finches. Science 296:707–711. https://doi.org/10.1126/science.1070315
Greenberg R, Cadena V, Danner RM, Tattersall G (2012) Heat loss may explain bill size differences between birds occupying different habitats. PLoS ONE 7:1–9. https://doi.org/10.1371/journal.pone.0040933
Hódar JA (1997) The use of regression equations for the estimation of prey length and biomass in diet studies of insectivore vertebrates. Misc Zool 20:1–10
Hongshan J, Rong L, Shou-Wei D, Shuifang Z (2014) Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinform 15:1–12. https://doi.org/10.1186/1471-2105-15-182
Kadlec T, Štrobl M, Hanzelka J et al (2018) Differences in the community composition of nocturnal Lepidoptera between native and invaded forests are linked to the habitat structure. Biodivers Conserv 27:2661–2680. https://doi.org/10.1007/s10531-018-1560-8
Kverek P, Storchová R, Reif J, Nachman MW (2008) Occurrence of a hybrid between the Common Nightingale (Luscinia megarhynchos) and the Thrush Nightingale (Luscinia luscinia) in the Czech Republic confirmed by genetic analysis. Sylvia 44:17–26
Legendre P, Andersson MJ (1999) Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecol Monogr 69:1–24
Legendre P, Fortin M-J, Borcard D (2015) Should the Mantel test be used in spatial analysis? Methods Ecol Evol 6:1239–1247. https://doi.org/10.1111/2041-210X.12425
Leisler B, Winkler H (2015) Evolution of island warblers: beyond bills and masses. J Avian Biol 46:236–244. https://doi.org/10.1111/jav.00509
Losos JB (2011) Lizards in an evolutionary tree: ecology and adaptive radiation of anoles, vol 10. University of California Press, Berkeley
Luther D, Greenberg R (2014) Habitat type and ambient temperature contribute to bill morphology. Ecol Evol 4:699–705. https://doi.org/10.1002/ece3.911
MacArthur R, Levins R (1967) The limiting similarity, convergence, and divergence of coexisting species. Am Nat 101:377–385. https://doi.org/10.1086/282505
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Miller ET, Wagner SK, Harmon LJ, Ricklefs RE (2017) Radiating despite a lack of character: ecological divergence among closely related, morphologically similar honeyeaters (Aves: Meliphagidae) co-occurring in arid Australian environments. Am Nat 189:E14–E30. https://doi.org/10.1086/690008
Mönkkönen M, Devictor V, Forsman JT et al (2017) Linking species interactions with phylogenetic and functional distance in European bird assemblages at broad spatial scales. Glob Ecol Biogeogr 26:952–962. https://doi.org/10.1111/geb.12605
Mořkovský L, Janoušek V, Reif J et al (2018) Genomic islands of differentiation in two songbird species reveal candidate genes for hybrid female sterility. Mol Ecol 27:949–958. https://doi.org/10.1111/mec.14479
Naguib M, Amrhein V, Kunc HP (2004) Effects of territorial intrusions on eavesdropping neighbors: communication networks in nightingales. Behav Ecol 15:1011–1015. https://doi.org/10.1093/beheco/arh108
Navalón G, Bright JA, Marugán-Lobón J, Rayfield EJ (2018) The evolutionary relationship among beak shape, mechanical advantage, and feeding ecology in modern birds. Evolution 73:422–435. https://doi.org/10.1111/evo.13655
Olsen AM (2017) Feeding ecology is the primary driver of beak shape diversification in waterfowl. Funct Ecol 31:1985–1995. https://doi.org/10.1111/1365-2435.12890
Peres-Neto PR, Legendre P, Dray S, Bocard D (2006) Variation partitioning of species data matrices: estimation and comparison of fraction. Ecology 87:2614–2625
Pfennig KS, Pfennig DW (2009) Character displacement: ecological and reproductive responses to a common evolutionary problem. Q Rev Biol 84:253–276. https://doi.org/10.1086/605079
Pfennig DW, Rice AM, Martin RA (2007) Field and experimental evidence for competition’s role in phenotypic divergence. Evolution 61:257–271. https://doi.org/10.1111/j.1558-5646.2007.00034.x
Pianka ER (1973) The structure of lizard communities. Annu Rev Ecol Syst 4:53–74. https://doi.org/10.1146/annurev.es.04.110173.000413
Poulin B, Leeebvre G, Mcneil R (1994a) Diets of land birds from northeastern Venezuela. Condor 96:354–361. https://doi.org/10.2307/1369320
Poulin B, Lefebvre G, McNeil R (1994b) Effect and efficiency of tartar emetic in determining the diet of tropical land birds. Condor 96:98–104. https://doi.org/10.2307/1369067
Reif J, Jiran M, Reifová R et al (2015) Interspecific territoriality in two songbird species: potential role of song convergence in male aggressive interactions. Anim Behav 104:131–136. https://doi.org/10.1016/j.anbehav.2015.03.016
Reif J, Reifová R, Skoracka A, Kuczyński L (2018) Competition-driven niche segregation on a landscape scale: evidence for escaping from syntopy towards allotopy in two coexisting sibling passerine species. J Anim Ecol 87:774–786. https://doi.org/10.1111/1365-2656.12808
Reifová R, Kverek P, Reif J (2011a) The first record of a female hybrid between the Common Nightingale (Luscinia megarhynchos) and the Thrush Nightingale (Luscinia luscinia) in nature. J Ornithol 152:1063–1068. https://doi.org/10.1007/s10336-011-0700-7
Reifová R, Reif J, Antczak M, Nachman MW (2011b) Ecological character displacement in the face of gene flow: evidence from two species of nightingales. BMC Evol Biol 11:138. https://doi.org/10.1186/1471-2148-11-138
Renner SC, Baur S, Possler A et al (2012) Food preferences of winter bird communities in different forest types. PLoS ONE 7:1–10. https://doi.org/10.1371/journal.pone.0053121
Rybinski J, Sirkiïa PM, McFarlane SE et al (2016) Competition-driven build-up of habitat isolation and selection favoring modified dispersal patterns in a young avian hybrid zone. Evolution 70:2226–2238. https://doi.org/10.1111/evo.13019
Schaffers AP, Raemakers IP, Sykora KW, Braak C (2008) Arthropod assemblages are best predicted by plant. Ecology 89:782–794. https://doi.org/10.1890/07-0361.1
Schluter D (2000) Ecological character displacement in adaptive radiation. Am Nat 156:S4–S16. https://doi.org/10.1086/303412
Schluter D, McPhail JD (1992) Ecological character displacement and speciation in sticklebacks. Am Nat 140:85–108. https://doi.org/10.1086/285404
Schoener TW (1974) Resource partitioning in ecological communities. Science 185:27–39. https://doi.org/10.1126/science.185.4145.27
Seehausen O (2006) African cichlid fish: a model system in adaptive radiation research. Proc R Soc B Biol Sci 273:1987–1998. https://doi.org/10.1098/rspb.2006.3539
Sorjonen J (1986) Mixed singing and interspecific territoriality-consequences of secondary contact of two ecologically and morphologically similar nightingale species in Europe. Ornis Scand 17:53–67. https://doi.org/10.2307/3676753ER
Sottas C, Reif J, Kuczyński L, Reifová R (2018) Interspecific competition promotes habitat and morphological divergence in a secondary contact zone between two hybridizing songbirds. J Evol Biol 31:914–923. https://doi.org/10.1111/jeb.13275
Souriau A, Kohoutová H, Reif J et al (2018) Can mixed singing facilitate coexistence of closely related nightingale species? Behav Ecol 29:925–932. https://doi.org/10.1093/beheco/ary053
Spellerberg IF, Fedor PJ (2003) A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon-Wiener’ index. Glob Ecol Biogeogr 12:177–179. https://doi.org/10.1046/j.1466-822X.2003.00015.x
Stoffel MA, Nakagawa S, Schielzeth H (2017) rptR: repeatability estimation and variance decomposition by generalized linear mixed-effects models. Methods Ecol Evol 8:1639–1644. https://doi.org/10.1111/2041-210X.12797
Storchová R, Reif J, Nachman MW (2010) Female heterogamety and speciation: reduced introgression of the Z chromosome between two species of nightingales. Evolution 64:456–471. https://doi.org/10.1111/j.1558-5646.2009.00841.x
Stuart YE, Losos JB (2013) Ecological character displacement: glass half full or half empty? Trends Ecol Evol 28:402–408. https://doi.org/10.1016/j.tree.2013.02.014
Taberlet P, Bonin A, Zinger L, Coissac E (2018) Environmental DNA: for biodiversity research and monitoring. Oxford University Press, Oxford
Tatner P (1983) The diet of urban magpies Pica pica. Ibis 125:97–107. https://doi.org/10.1111/j.1474-919X.1983.tb03086.x
Tattersall GJ, Arnaout B, Symonds MRE (2017) The evolution of the avian bill as a thermoregulatory organ. Biol Rev 92:1630–1656. https://doi.org/10.1111/brv.12299
Tobias JA, Cornwallis CK, Derryberry EP et al (2014) Species coexistence and the dynamics of phenotypic evolution in adaptive radiation. Nature 506:359–363. https://doi.org/10.1038/nature12874
Vokurková J, Petrusková T, Reifová R et al (2013) The causes and evolutionary consequences of mixed singing in two hybridizing songbird species (Luscinia spp.). PLoS ONE 8:e60172. https://doi.org/10.1371/journal.pone.0060172
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
Wang Y, Naumann U, Wright ST, Warton DI (2012) Mvabund—an R package for model-based analysis of multivariate abundance data. Methods Ecol Evol 3:471–474. https://doi.org/10.1111/j.2041-210X.2012.00190.x
Winkelmann K, Genner MJ, Takahashi T, RuBer L (2014) Competition-driven speciation in cichlid fish. Nat Commun 5:1–8. https://doi.org/10.1038/ncomms4412
Wolf JBW, Harrod C, Brunner S et al (2008) Tracing early stages of species differentiation: ecological, morphological and genetic divergence of Galápagos sea lion populations. BMC Evol Biol 8:1–14. https://doi.org/10.1186/1471-2148-8-150
Wood EM, Swarthout SEB, Hochachka WM et al (2016) Intermediate habitat associations by hybrids may facilitate genetic introgression in a songbird. J Avian Biol. https://doi.org/10.1111/jav.00771
Acknowledgements
We would like to thank Abel Souriau and Lucie Baránková for the field assistance and Hanka Pinkasová for her work on DNA isolation. Petr Klimeš and Petr Kozel assisted with identification of ants and beetles, respectively. We acknowledge the CF Genomics of CEITEC supported by the NCMG research infrastructure (LM2015091 funded by MEYS CR) for their support with obtaining scientific data presented in this paper. Access to computing and storage facilities owned by parties and projects contributing to the National Grid Infrastructure MetaCentrum, provided under the programme ‘Projects of Large Infrastructure for Research, Development, and Innovations’ (LM2010005), is greatly appreciated. The study was supported by a student grant of the Grant Agency of Charles University (6462/2017) to C.S., the Czech Science Foundation (Grant No. 18–14325S) and the Charles University grant PRIMUS/19/SCI/008 to R.R. Work of K.S. was supported by European Research Council project BABE 805189.
Author information
Authors and Affiliations
Contributions
RR, JR, JK and CS conceived ideas and designed methodology; CS, JR and TO collected the data in the field; LS and KS performed laboratory analyses; CS, JK and JR analysed the data; RR, CS, JR and JK wrote the manuscript. All authors contributed critically to the drafts and gave final approval for publication.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Human and animal rights
This work was carried out in accordance with ethical animal research requirements of Poland according to Polish law (the Act On the Protection of Animals used for Scientific or Educational Purposes, 15.01.2015, item 266, implementing Directive 2010/63/EU of the European Parliament and of the European Council of 22.09.2010). Experiments on birds were approved by the Local Ethic Committee for Scientific Experiments on Animals in Poznań, Poland (permission No. 17/2015) and the Polish National Nature Conservation Authority (permission no. DZP-WG.6401.03.123.2017.dl.3). Ringing was performed under license No. 114225895 of the national ringing centre in Gdańsk (Ornithological Station of the Polish Academy of Sciences).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sottas, C., Reif, J., Kreisinger, J. et al. Tracing the early steps of competition-driven eco-morphological divergence in two sister species of passerines. Evol Ecol 34, 501–524 (2020). https://doi.org/10.1007/s10682-020-10050-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10682-020-10050-4