Abstract
The selection of better-evaluated genotypes for a target region depends on the characterization of the climate conditions of the environment. With the advancement of computer technology and daily available information about the weather, integrating such information in selection and interaction genotype × environment studies has become a challenge. This article presents the use of the technique of artificial neural networks associated with reaction norms for the processing of climate and georeferenced data for the study of genetic behaviors and the genotype × environment interaction of soybean genotypes. The technique of self-organizing maps (SOM) consists of competitive learning between two layers of neurons; one is the input, which transfers the data to the map, and the other is the output, where the topological structure formed by the competition generates weights, which represent the dissimilarity between the neural units. The methodologies used to classify these neurons and form the target populations of environments (TPE) were the discriminant analysis (DA) and the principal component analysis (PCA). To study soybean genetic behavior within these TPE, the random regression model was adopted to estimate the components of variance, and the reaction norms were adjusted through the Legendre polynomials. The SOM methodology allowed for an explanation of 99% of the variance of the climate data and the formation of well-structured TPE, with the membership probability of the regions within the TPE above 80%. The formation of these TPE allowed us to identify and quantify the response of the genotypes to sensitive changes in the environment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The climate change scenario challenges agricultural research to provide intelligent solutions in a fast and economical way (Tigchelaar et al. 2018). Characterizing the conditions of crop growth is crucial to achieving this purpose (Xu 2016), allowing a deeper understanding of how the environment shapes phenotypic variations (for example, (Costa-Neto et al. 2021a; de los Campos et al. 2020; Heinemann et al. 2019; Ramirez-Villegas et al. 2018). Since 1960, several researchers have suggested the use of environmental information to explain the differences caused in cultivars due to the genetic-environment interaction (GxE) (Perkins and Jinks 1968; Crossa et al. 1999; Vargas et al. 1999). The environmental information used in these models of genomic selection usually focuses on the use of the information such as temperature, rainfall, and solar radiation, defined as co-variables within the models (Jarquín et al. 2014).
For research in plant breeding, especially for the selection of better-evaluated soybean genotypes for a target region, this approach is proven to be advantageous to discriminate genetic and non-genetic sources of culture adaptation (Costa-Neto et al. 2021c). In this context, new technologies available such as the historical description of the environment (Enviromics) (Costa-Neto et al. 2021b, c; Resende et al. 2021; Rogers et al. 2021) are crucial to improving conventional models, but bring the challenges of changing the already established systems. The integration of this new technology allied to the already established models allows the selection of cultivars with high yields in the face of the environmental conditions caused by climate changes and the consequent increase of the occurrence of abiotic stresses (Crossa et al. 2021).
The indication of genotypes may vary according to the macro-environment, climate and soil changes, different latitudes and longitudes, and years (Bourret et al. 2015; Gray et al. 2016), and it may also vary with changes within a micro-environment (Resende et al. 2016; Soares et al. 2016). Thus, the concept of envirotyping emerges to establish the quality of a certain environment (Cooper et al. 2014; Xu 2016); it uses multiple techniques to collect, process and integrate environmental information in genetic and genomic studies (Costa-Neto et al. 2021b), in addition to fostering breeding strategies to understand and deal with future scenarios of climate changes (de los Campos et al. 2020; Gillberg et al. 2019).
This information can be affected by many factors, such as the great amount of data, because they have complex structures, they are non-linear and because of the presence of redundancies and outliers (Gianola et al. 2011). Thus, non-linear methodologies are preferable to deal with a set of complex environmental data (Calus et al. 2004; Gianola et al. 2011). The use of these non-linear methodologies associated with environmental data has become more and more popular in recent years (for example Friedel 2012; Liukkonen et al. 2013; Strebel et al. 2013). However, the use of new technologies such as the envirotype and the use of neural networks associated with georeferenced data are crucial to improve conventional models and selecting high-yielding soybean cultivars in the face of environmental changes caused by climate change and abiotic stresses.
This study presents the use of the technique of neural networks associated with georeferenced data to implement the processing of climate and soil data, to describe and categorize this information with basis on the dissimilarity caused by the environmental variables, and subsequently to apply this in models of reaction norms in order to study and quantify genetic behaviors and genotype × environment interactions in soybean genotypes.
Materials and methods
Environmental data collection
This study used climate and soil information of 32 municipalities located within the Brazilian macro-region of soybean culture called MRS 3, in the state of Goiás. The municipalities chosen are part of the network of trials of value for cultivation and use (VCU) of the GDM Genética do Brasil S.A. company (GDM). The daily meteorological information that was given to this work is part of the collection of the Agrymet company. All data was kindly provided by GDM.
In order to characterize the sites being analyzed, a historical series of climate characteristics were used (Table 1), evaluated between the years of 2018 and 2020, from November to February. This time series was defined to capture all the climate variations throughout the whole development of the soybean culture in the region.
Soybean data
This study makes use of a great set of yield data formed by VCU trials of soybean varieties; the data set of this study was kindly provided by GDM Genética do Brasil S.A. company. The phenotypic data were the reports on grain yield (kg ha−1). This set of trials was carried out in multi-environmental conditions (MET) from 2018 to 2020 and standardized by the GDM company, in which each trial was composed of 17 genotypes. The trial was formed by four randomized blocks with three replications. Each plot is formed by a line of 4 m, with 19 seeds per meter.
Definition of the target population of environments (TPE)
The methodology of self-organized maps (SOM) of Kohonen, according to (Kohonen 2013), was used to characterize the patterns of the spatial distribution of the environmental variables. SOM is formed by two layers; one is the input, which transfers the data to the map, and the second one corresponds to the process of competitive learning of neurons, forming, this way, a topological structure (Chen et al. 2019). During the learning process of the network, the climate variables were informed as the input vector. According to the learning process, each input vector is attributed to an output neuron, attributing a weight associated with the input information. Based on these weights, the distance between neurons was calculated. The Euclidean mean distance standardized with a number of 1000 interactions was used for the processing. The construction of networks used the package “Kohonen” (Wehrens and Kruisselbrink 2018).
After the SOM learning process, the classification of the target population of environments (TPE) was carried out by using the procedures of discriminant analysis (DA) and the principal component analysis (PCA). Successive K-means were used for an interval of K-neurons, and the values of Bayesian Information Criterion (BIC) of the corresponding models and the coefficients of variation were calculated until the ideal number of clusters was found. The DA and PCA functions were implemented by using packages “ade4” (Dray and Dufour 2007) and “MASS” (Ripley et al. 2018). All the analyses were carried out on Software R version 4.2.1 (R Development Core Team 2022).
Components of variance
The components of variance were estimated according to the residual maximum likelihood method (Patterson and Thompson 1971), and the genetic values were predicted through the best linear unbiased predictor (Henderson 1975), according to (Gilmour et al. 2015). Random regression models were adjusted through the Legendre polynomials, considering all the possible levels of adjustment for each random effect, by using the following model:
where \({Y}_{ijk}\) is the ith individual (i = 1, 2,…, n) in the jth cluster (j = 1, 2,…, 7) in the kth replication (k = 1, 2,…,10); \({R}_{k}\) is the fixed effect of the replication; \({b}_{M}\) is the fixed coefficient of regression adjusted through the sixth degree of the Legendre polynomial for the common average trajectory of genotypes. The random effect, \({g}_{ikm}\) is the regression coefficient for the Legendre polynomial of degree m for the genetic value. ϕijm is the mth Legendre polynomial for the jth cluster of the ith individual; m is the adjustment of the degree of the Legendre polynomial, varying from 0 to 6, for the genetic and environmental effects, respectively; and \({\epsilon }_{ijk}\) is the residual random effect associated with \({Y}_{ijk}\).
In the matrix notation, the model above is described as follows:
where y is the vector of phenotypic observations; \(\varvec{\tau }\) is the vector of the effects of repetition (assumed as fixed); g is the vector of genetic effects (assumed as random); e is the error vector (random). X, Z refers to the incidence matrices for these effects.
In this model, g ~ N (0, Kg\(\otimes\)I) and e ~ N (0, R); where Kg is the matrix of co-variance for genetic effects; \(\otimes\) denotes the Kronecker product; I is an identity matrix with an appropriate order for the respective random effect; and R refers to the matrix of residual co-variances. Different structures of residual co-variance (homogeneous, diagonal and unstructured) were tested.
The polynomial order in models of random regression was selected by using the Akaike information criterion (AIC) (Schwarz 1978), as follows:
where LogL is the logarithm of the maximum value of the likelihood function (L), and p is the number of estimated parameters.
The estimates of the components of variance (\({\sigma }_{g}^{2}\)) and the predicted genetic values (\({ \tilde{g}}_{{ij}}\)), in the original scale, were obtained through the following expressions (Kirkpatrick et al. 1990):
´
The genetic correlations (\({\rho }_{g}\)) between each pair of environmental clusters were obtained through the following expression:
where \({\widehat{\varvec{\sigma }}}_{\varvec{g}\left(\varvec{i}\varvec{j}\right)}\) is the genetic co-variance between the genotypes for the pair of environmental clusters i and j; \({\widehat{\varvec{\sigma }}}_{\varvec{g}\left(\varvec{i}\right)}^{2}\) and \({\widehat{\varvec{\sigma }}}_{\varvec{g}\left(\varvec{j}\right)}^{2}\) are the genetic variances between the genotype and environmental clusters i and j, respectively. The statistical analyses were carried out by using the software ASReml 4.1 (Gilmour et al. 2015) and R (R Development Core Team 2022).
Results
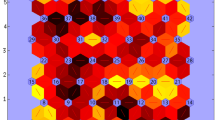
The topological formation of the SOM is represented in Fig. 1A. The scale of colors represents the synaptic weights of each variable in the 90 neurons of the map; this scale varies from blue colors, with lower weight values, to yellow colors, with greater synaptic weights. When evaluating weight distribution in the network, the effect of the variables on the different neurons is seen, as well as the similarity among them. In this stage, the neurons have not been divided into environmental clusters yet. In short, the network training was efficient, since it brought those neurons that presented similar weights closer, despite the use of different climate variables with different behaviors. It can be observed that, at first, solar radiation presented a greater differentiation among neurons, and that with variables altitude and latitude the first distributions of well-structured clusters are formed, since they presented the greatest synaptic weight values in the network. Variables of rainfall, wind speed, and relative humidity had the same behavior in the distribution of the neural network, just like variables temperature and AWC.
A Graphic representation of the variation of the synaptic weights in the 90 neurons formed by the methodology of self-organized maps of Kohonen for each climate and soil variable. B Mean coefficient of variation (CV) of the mean Euclidean distance and Bayesian Information Criterion (BIC) estimated for a growing number of TPE.
Figure 1B shows the BIC values and the coefficients of variation of the distances among the clusters for the growing values of k TPE. A clear decrease of BIC is seen up to value k = 5, after which the BIC value increases, clearly indicating that the best number of clusters is equal to five. The same can be observed in the trajectory of the CV values, which, when reaching the values of five clusters, shows no significant reduction of the coefficient of variation of the distances among the clusters with the increase of the k TPE value.
Figure 2A is the geographic representation of the classification of the municipalities in the state of Goiás, in which the municipalities used in the analysis are represented by different colors that form each cluster. For the principal component analysis, with the basis of the estimates of the mean Euclidean distance among the 90 neurons, only one discriminant function was enough to explain 99% of the variance, separating them into five TPEs (Fig. 2B). Among all the climate and soil variables, Altitude was the one that presented the greatest linear dependence (LD%) in the formation of the TPEs (97.43%), while the lowest as Rain (0.41%), SR (0.40%), and RH (1.70%) (Table 2). When observing Fig. 2C, it is possible to visualize the membership probability of each evaluated municipality in the three years in their respective clusters. In general, all the evaluated municipalities had a membership probability above 80%, even though TPE 1, TPE 3, and TPE 4 are geographically close and presented little chance of belonging to another cluster. Only TPE 1 presented a lower mean of membership (70%), which was on average 30% similar to TPE 2.
A Geographic disposition of the 17 municipalities of Goiás (GO) belonging to the five clusters formed by the method of discriminant analysis of principal components. B Graphic dispersion of the density of the first discriminant function for the five clusters formed. C Graphic representation of the membership probability of the 17 municipalities in the 3 years of evaluation for the five clusters formed. Axis x represents the observations of the municipalities in the 3 years and axis y represents the membership probability in their respective clusters
The average temperatures of the TPEs were between 25.3 °C (TPE 2) and 26.4 °C (TPE 4) (Table 2), while relative humidity had the same behavior from 76.18% (TPE 2) to 77.5% (TPE 4). Rainfall was lower in TPE 3 (8,22 mm.day−1) and greater in TPE 5 (9.02 mm day−1). Solar radiation was between 18.4 W m−2, day−1 (TPE 5) and 19.0 W m−2, day−1 (TPE 2). Wind speed (WS) was not greater than 1.5 m s−1 in the five TPEs. Available water capacity (AWC) was well balanced among the TPEs, among which TPE 5 presented the lowest values (0, 82% day−1). Regarding Altitude and Latitude, the orders of the TPEs had a similar pattern (TPE 4 < TPE 3 < TPE 1 < TPE 2 < TPE 5).
The Legendre polynomial was chosen according to the Akaike information criterion (AIC), in which model 2 had the best result (lowest value) of 5469.9 (Table 3). This model presents a heterogeneous residual structure, that is, it estimates a component of the residual variance for each TPE. Thus, this model was adopted to estimate the components of variance and to predict the genetic values for the tested soybean cultivars.
The behavior of the 17 cultivars across the TPEs is described in Fig. 3. Figure 3 A describes the average behavior of the phenotypes across the TPEs. As a whole, the yield had its maximum peak at 5500 kilos in TPE 2 and a minimum of 3000 kilos in TPE 3. It is possible to denote that the distribution of the means of the phenotypes across the environments had the same variation, but the ranking of the cultivars changed over the TPEs. The trajectories of the genetic effects (Fig. 3B) show a linear relationship with the complex genetic × environmental interaction, in which TPE 1 had the greatest variety of genetic values and decreased until TPE 5. Among the 17 genotypes evaluated through the model of random regression, genotype G39 stood out in first place for all the TPEs (Fig. 3B). When comparing phenotype behavior, G39 and G44 behaved similarly, but G44 had a medium genetic effect across the TPEs. The phenotype behavior of G16 had the worst classification in only two TPEs, but the genetic value observed is the lowest in almost all TPEs, and only in TPE 5 it was not classified as the lowest genetic effect. Thus, the genotype ranking changed in the environmental gradient very differently from the effect of the phenotypes.
Curves of the behaviors of the cultivars in the different TPEs. A Average behavior of phenotypes in the TPEs. B Reaction norms of the model of random regression of the 17 cultivars in the different TPEs. The colors highlight the cultivars that had the greatest genetic effect (G39) in blue, medium (G44) in yellow, and the smallest one (G16) in red, in the TPEs.
Along the environments, trait heritability varied between 0.25 (TPE 1) and 0.02 (TPE 4) (Fig. 4A), having a descending behavior from the first TPE to the fourth one, and a growing behavior until the fifth TPE. The genetic variances followed the behavior of heritability, where the greatest value was in TPE 1 (51,204) and the smallest in TPE 4 (4476). The greatest value of phenotypic variance was in TPE 3 (372,639) and the smallest in TPE 1 (208,303). The same distribution was found in the residual variance, with TPE 3 (361,759) and TPE 1 (157,099).
The greatest genetic correlations (\({\rho }_{g}\)) were between TPE 1 and TPE 2, with a value of 0.99, suggesting a low reordering between the genotypes on these sites (Fig. 4B). The smallest genetic correlation occurred between the extreme environments, TPE 1 and TPE 5 (\({\rho }_{g}\)= − 0.39). The greatest correlations were between TPE 1, TPE 2, and TPE 3, which were the ones with the greatest genetic yield potential, and the smallest among these TPEs with TPE 5, indicating a reordering of the classification of the genotypes in this TPE. Also, TPE 4 presented a medium genetic correlation among the TPEs, ranging from 0.48 for TPE 1, and 0.77 for TPE 3, being, thus, a TPE of transition between TPEs (1, 2, and 3) with TPE 5.
Discussion
Learning about the climate and soil conditions of a region is of major importance for the soybean breeding since certain genotypes are more stable in different environments; these materials are selected because they do not present undesirable changes in yield and are more resilient to local climate changes (Eberhart and Russell 1966). In addition, some genotypes are more adaptable, responding positively to the improvement in environmental conditions (Brawner et al. 2014).
This study sought to classify and analyze, through the methodology of self-organized maps, a time series of data under the scenario of a dynamic change of the climate in the Brazilian macro-region 3 of the soybean culture. The use of artificial neural networks (ANNs) proved to be highly efficient to interpret the climate dynamics in the region, where, after the formation of the TPEs, the discriminant analysis was able to explain 99% of the variation of the synaptic weights of the network. The model of self-organized maps is efficient to analyze climate and soil data since the way the information is dealt with by the network creates the possibility of a better performance if compared to conventional models (Bustos-Korts et al. 2022). In contrast with conventional approaches, this study sought a sensitive approach to the dynamic environment. Therefore, in the interpretation of environmental data, the information on topography, such as altitude and latitude, and the information on solar radiation are important for an initial interpretation of the network, since they presented greater synaptic weights (Fig. 1A), while the most sensitive changes in the network are caused by the dynamics of continuous climate variables (temperature, relative humidity, AWC, wind speed, and rainfall) over time.
Although there are different approaches in the study of the G × E interaction, there are still a few studies in the literature that describe a recommendation according to continuous environmental change in soybean culture. Environmental variables are usually attributed as discrete phenomena, generating clusters with similar environmental traits, so that the environments are treated as levels of categorical variables (Alexandre Bryan Heinemann et al. 2022). The modeling of the spatial variation and of temporal dynamics is a challenge for studies of interaction in the soybean culture. Here, the use of ANN as an environmental descriptor guaranteed that the quality in the formation of the environmental clusters was balanced in the face of the complexity of climate information. Given this, this study represents an important contribution to the better understanding of the G × E interaction in soybean crops, allowing a more accurate recommendation of cultivars according to continuous environmental changes. Furthermore, the approach used in this study, using artificial neural networks as an environmental descriptor, can be applied in other crops and a climate change scenario, providing valuable information for the selection of more adapted and resistant genotypes.
In soybean breeding, random regression is very useful, since it allows the prediction of genetic values of individuals evaluated in different years, sites, and common environments, with an effect of ordering and selection (Schaeffer 2004). The functions of co-variance can express, in a more realistic way, the phenomena associated with longitudinal data, being superior to models of repeatability and multi-traits (Meyer 1998). In addition, Legendre polynomials have been used to model curves of the behavior of perennial plants (Li et al. 2017).
The genetic trajectories of the reaction norms reinforce the presence of the genotype x environment interaction since their trajectories are non-linear and cross with each other, which implies a different classification for each environment. Besides that, the trajectories can also be interpreted as genetic variability. The more distant trajectories are from each other, the more genetically distinct the genotypes (Gomulkiewicz and Kirkpatrick 1992). The advantage of this strategy is that the response of selection can be predicted, not only in the expression of the genotype submitted to any environment, but also in the quantification of the environmental sensitivity through the genetic trajectories, that is, based on the capacity of response to the changes of the environment (Alves et al. 2020).
In addition, reaction norms describe the genetic values of each cultivar across the environmental gradient. The model of random regression can predict the genetic value for any cultivar of any environmental cluster (between the first and the last TPE). The trajectories demonstrated that the cultivars had similar performances from TP1 to TPE 3 (Fig. 3B), which reveals that the recommendation of cultivars for these regions can be similar. Genetic correlations reinforce the efficiency of the recommendation (Fig. 4B). Although these three environmental clusters have a high environmental genetic correlation, only TPE 1 presented a greater value of heritability, and it is a more propitious environment in the practice of selection of cultivars. The high correlation among these TPEs can support the idea of grouping them in the same region as done by RESENDE et al. (2021); however, it was seen that even if there is no difference in the ranking of the genotypes for these environments, the genetic variance was greater in TPE 1 (Fig. 3B), corroborating with the idea that the practice of selection in this environment will lead to greater genetic gains.
Even though the number of sites in this study does not provide full coverage of the Brazilian macro-region M3 of the soybean culture, the study allowed the identification of well-defined TPEs. The results of this study indicate that although altitude is the main descriptive variable, the climate dynamics caused by continuous variables play an important role in the formation of environmental clusters. When the focus is selecting genotypes for specific environments, this model can benefit by predicting genotype performance for the site, taking into consideration the behavior of an average environment, as long as there is enough climate information for the categorization, as seen by Chenu et al. (2013). This approach can also be used in a scenario of climate change, in which the frequency of hot and dry climates is expected to increase in the future (Rattis et al. 2021).
Conclusion
The use of artificial neural networks (ANNs) proved to be highly efficient to interpret the climate dynamics in the region, where it was possible to discriminate and classify these environments into well-defined TPEs by using dynamic information on the climate. With the classification of the TPEs, it was possible to study the GxE interaction and visualize what the soybean genetic behavior is like for this macro-region, in the form of reaction norms. The genetic trajectories reinforce the presence of the GxE interaction and allowed us to quantify the response of the genotypes to changes in climate. This methodology can be useful to optimize time and resources in soybean breeding programs since the choice of the most adequate genotypes can made based on sensitive changes in the environment.
References
Alves RS, de Resende MDV, Azevedo CF, Silva FF, Rocha JRASC, Nunes ACP, Carneiro APS, dos Santos GA (2020) Optimization of Eucalyptus breeding through random regression models allowing for reaction norms in response to environmental gradients. Tree Genet Genomes. https://doi.org/10.1007/s11295-020-01431-5
Bourret A, Bélisle M, Pelletier F, Garant D (2015) Multidimensional environmental influences on timing of breeding in a tree swallow population facing climate change. Evol Appl. https://doi.org/10.1111/eva.12315
Brawner JT, Hodge GR, Meder R, Dvorak WS (2014) Visualising the environmental preferences of Pinus tecunumanii populations. Tree Genet Genomes. https://doi.org/10.1007/s11295-014-0747-8
Bustos-Korts D, Boer MP, Layton J, Gehringer A, Tang T, Wehrens R, Messina C, de la Vega AJ, van Eeuwijk FA (2022) Identification of environment types and adaptation zones with self-organizing maps; applications to sunflower multi-environment data in Europe. Theor Appl Genet 135(6):2059–2082. https://doi.org/10.1007/S00122-022-04098-9/FIGURES/10
Calus MPL, Bijma P, Veerkamp RF (2004) Effects of data structure on the estimation of covariance functions to describe genotype by environment interactions in a reaction norm model. Genet Sel Evol. https://doi.org/10.1051/gse:2004013
Chen N, Chen L, Ma Y, Chen A (2019) Regional disaster risk assessment of China based on self-organizing map: clustering, visualization and ranking. Int J Disaster Risk Reduct 33:196–206. https://doi.org/10.1016/J.IJDRR.2018.10.005
Chenu K, Deihimfard R, Chapman SC (2013) Large-scale characterization of drought pattern: a continent-wide modelling approach applied to the Australian wheatbelt–spatial and temporal trends. New Phytol 198(3):801–820. https://doi.org/10.1111/NPH.12192
Cooper M, Messina CD, Podlich D, Totir LR, Baumgarten A, Hausmann NJ, Wright D, Graham G (2014) Predicting the future of plant breeding: complementing empirical evaluation with genetic prediction. Crop Pasture Sci 65(4):311–336. https://doi.org/10.1071/CP14007
Costa-Neto G, Crossa J, Fritsche-Neto R, Batán E, de México E, de Posgraduado C (2021a) Enviromic assembly increases accuracy and reduces costs of the genomic prediction for yield plasticity in maize. Front Plant Sci 12:717552
Costa-Neto G, Fritsche-Neto R, Crossa J (2021b) Nonlinear kernels, dominance, and envirotyping data increase the accuracy of genome-based prediction in multi-environment trials. Heredity. https://doi.org/10.1038/s41437-020-00353-1
Costa-Neto G, Galli G, Carvalho HF, Crossa J, Fritsche-Neto R (2021c) EnvRtype: a software to interplay enviromics and quantitative genomics in agriculture. G3 Genes Genomes Genet. https://doi.org/10.1093/g3journal/jkab040
Crossa J, Vargas M, Van Eeuwijk FA, Jiang C, Edmeades GO, Hoisington D (1999) Interpreting genotype x environment interaction in tropical maize using linked molecular markers and environmental covariables. Theor Appl Genet. https://doi.org/10.1007/s001220051276
Crossa J, Fritsche-Neto R, Montesinos-Lopez OA, Costa-Neto G, Dreisigacker S, Montesinos-Lopez A, Bentley AR (2021) The modern plant breeding triangle: optimizing the use of genomics, phenomics, and enviromics data. Front Plant Sci. https://doi.org/10.3389/fpls.2021.651480
de los Campos G, Pérez-Rodríguez P, Bogard M, Gouache D, Crossa J (2020) A data-driven simulation platform to predict cultivars’ performances under uncertain weather conditions. Nat Commun 11(1):4876. https://doi.org/10.1038/s41467-020-18480-y
Dray S, Dufour AB (2007) The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22(4):1–20. https://doi.org/10.18637/JSS.V022.I04
Eberhart SA, Russell WA (1966) Stability parameters for comparing varieties. Crop Sci. https://doi.org/10.2135/cropsci1966.0011183X000600010011x
Friedel MJ (2012) Data-driven modeling of surface temperature anomaly and solar activity trends. Environ Model Softw. https://doi.org/10.1016/j.envsoft.2012.04.016
Gianola D, Okut H, Weigel KA, Rosa GJM (2011) Predicting complex quantitative traits with bayesian neural networks: a case study with Jersey cows and wheat. BMC Genet. https://doi.org/10.1186/1471-2156-12-87
Gillberg J, Marttinen P, Mamitsuka H, Kaski S (2019) Modelling G×E with historical weather information improves genomic prediction in new environments. Bioinformatics 35(20):4045–4052. https://doi.org/10.1093/BIOINFORMATICS/BTZ197
Gilmour aR, Gogel BJ, Cullis BR, Welham SJ, Thompson R (2015) ASReml user guide release 4.1 structural specification. VSN International Ltd. Hemel Hempstead
Gomulkiewicz R, Kirkpatrick M (1992) Quantitative genetics and the evolution of reaction norms. Evolution. https://doi.org/10.1111/j.1558-5646.1992.tb02047.x
Gray LK, Rweyongeza D, Hamann A, John S, Thomas BR (2016) Developing management strategies for tree improvement programs under climate change: insights gained from long-term field trials with lodgepole pine. For Ecol Manag. https://doi.org/10.1016/j.foreco.2016.06.041
Heinemann AB, Ramirez-Villegas J, Rebolledo MC, Neto C, Castro AP (2019) Upland rice breeding led to increased drought sensitivity in Brazil. Field Crop Res. https://doi.org/10.1016/j.fcr.2018.11.009
Heinemann A, Bryan, Costa-Neto G, Fritsche-Neto R, da Matta DH, Fernandes IK (2022) Enviromic prediction is useful to define the limits of climate adaptation: a case study of common bean in Brazil. Field Crop Res 286:108628. https://doi.org/10.1016/J.FCR.2022.108628
Henderson CR (1975) Best linear unbiased estimation and prediction under a selection model. Biometrics. https://doi.org/10.2307/2529430
Jarquín D, Crossa J, Lacaze X, Du Cheyron P, Daucourt J, Lorgeou J, Piraux F, Guerreiro L, Pérez P, Calus M, Burgueño J, de los Campos G (2014) A reaction norm model for genomic selection using high-dimensional genomic and environmental data. Theor Appl Genet. https://doi.org/10.1007/s00122-013-2243-1
Kirkpatrick M, Lofsvold D, Bulmer M (1990) Analysis of the inheritance, selection and evolution of growth trajectories. Genetics. https://doi.org/10.1093/genetics/124.4.979
Kohonen T (2013) Essentials of the self-organizing map. Neural Netw. https://doi.org/10.1016/j.neunet.2012.09.018
Li Y, Suontama M, Burdon RD, Dungey HS (2017) Genotype by environment interactions in forest tree breeding: review of methodology and perspectives on research and application. Tree Genet Genom. https://doi.org/10.1007/s11295-017-1144-x
Liukkonen M, Laakso I, Hiltunen Y (2013) Advanced monitoring platform for industrial wastewater treatment: multivariable approach using the self-organizing map. Environ Model Softw. https://doi.org/10.1016/j.envsoft.2013.07.005
Meyer K (1998) Estimating covariance functions for longitudinal data using a random regression model. Genet Sel Evol. https://doi.org/10.1051/gse:19980302
Patterson HD, Thompson R (1971) Recovery of inter-block information when block sizes are unequal. Biometrika. https://doi.org/10.1093/biomet/58.3.545
Perkins JM, Jinks JL (1968) Environmental and genotype-environmental components of variability III. Multiple lines and crosses. Heredity. https://doi.org/10.1038/hdy.1968.48
R Development Core Team R (2022) R: a language and environment for statistical computing. In: R foundation for statistical computing. https://doi.org/10.1007/978-3-540-74686-7
Ramirez-Villegas J, Heinemann AB, Pereira de Castro A, Breseghello F, Navarro-Racines C, Li T, Rebolledo MC, Challinor AJ (2018) Breeding implications of drought stress under future climate for upland rice in Brazil. Glob Change Biol. https://doi.org/10.1111/gcb.14071
Rattis L, Brando PM, Macedo MN, Spera SA, Castanho ADA, Marques EQ, Costa NQ, Silverio DV, Coe MT (2021) Climatic limit for agriculture in Brazil. Nat Clim Change 11(12):1098–1104. https://doi.org/10.1038/s41558-021-01214-3
Resende RT, Marcatti GE, Pinto DS, Takahashi EK, Cruz CD, Resende MDV (2016) Intra-genotypic competition of Eucalyptus clones generated by environmental heterogeneity can optimize productivity in forest stands. For Ecol Manag. https://doi.org/10.1016/j.foreco.2016.08.041
Resende RT, Piepho HP, Rosa GJM, Silva-Junior OB, Silva FF, de Resende MDV, Grattapaglia D (2021) Enviromics in breeding: applications and perspectives on envirotypic-assisted selection. Theor Appl Genet. https://doi.org/10.1007/s00122-020-03684-z
Ripley B, Venables B, Bates DM, Firth D, Hornik K, Gebhardt A (2018) Support functions and datasets for venables and ripley’s MASS. 169. http://www.stats.ox.ac.uk/pub/MASS4/
Rogers AR, Dunne JC, Romay C, Bohn M, Buckler ES, Ciampitti IA, Edwards J, Ertl D, Flint-Garcia S, Gore MA, Graham C, Hirsch CN, Hood E, Hooker DC, Knoll J, Lee EC, Lorenz A, Lynch JP, McKay J, Holland JB (2021) The importance of dominance and genotype-by-environment interactions on grain yield variation in a large-scale public cooperative maize experiment. G3 Genes Genomes Genet. https://doi.org/10.1093/g3journal/jkaa050
Schaeffer LR (2004) Application of random regression models in animal breeding. Livest Prod Sci. https://doi.org/10.1016/S0301-6226(03)00151-9
Schwarz G (1978) Estimating the dimension of a model. Ann Stat. https://doi.org/10.1214/aos/1176344136
Soares AAV, Leite HG, Souza AL, Silva SR, Lourenço HM, Forrester DI (2016) Increasing stand structural heterogeneity reduces productivity in Brazilian eucalyptus monoclonal stands. For Ecol Manag. https://doi.org/10.1016/j.foreco.2016.04.035
Strebel K, Espinosa G, Giralt F, Kindler A, Rallo R, Richter M, Schlink U (2013) Modeling airborne benzene in space and time with self-organizing maps and bayesian techniques. Environ Model Softw. https://doi.org/10.1016/j.envsoft.2012.12.001
Tigchelaar M, Battisti DS, Naylor RL, Ray DK (2018) Future warming increases probability of globally synchronized maize production shocks. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.1718031115
Vargas M, Crossa J, Van Eeuwijk FA, Ramírez ME, Sayre K (1999) Using partial least squares regression, factorial regression, and AMMI models for interpreting genotype x environment interaction. Crop Sci. https://doi.org/10.2135/cropsci1999.0011183X003900040002x
Wehrens R, Kruisselbrink J (2018) Flexible self-organizing maps in Kohonen 3.0. J Stat Softw 87(7):1–18. https://doi.org/10.18637/JSS.V087.I07
Xu Y (2016) Envirotyping for deciphering environmental impacts on crop plants. Theor Appl Genet. https://doi.org/10.1007/s00122-016-2691-5
Acknowledgements
The authors would like to thank the GDM Genética do Brasil S.A. for providing the environmental and soybean data. This study was supported the the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001, Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). In addition to our gratitude to the organizations that supported this study, we would like to express our sincere appreciation to the late Professor Fabyano Fonseca e Silva for his invaluable contributions to our research.
Author information
Authors and Affiliations
Contributions
BGL: Conceptualization, Methodology, Software, Formal analysis and Supervision. LFS: Conceptualization and Writing-Orinal Draft. MAP: Validation, Software and Data Curation. LAP: Writing—Review and Editing, Formal analysis. FLS: Writing—Review and Editing, Project administration.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Leichtweis, B.G., de Faria Silva, L., Peixoto, M.A. et al. Envirotype approach for soybean genotype selection through the integration of georeferenced climate and genetic data using artificial neural networks. Euphytica 220, 8 (2024). https://doi.org/10.1007/s10681-023-03267-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-023-03267-1