Abstract
Lint percentage is a major yield component in cotton breeding programmes. To identify quantitative trait loci (QTLs) and candidate genes related to lint percentage, we used an intraspecific recombinant inbred line population of 137 lines derived from Gossypium hirsutum cv. CCRI36 and G. hirsutum acc. G2005 for QTL mapping of lint percentage. Based on a high-density genetic map and phenotype data collected in four growing environments, we identified a total of 28 QTLs for lint percentage. Three stable QTLs (qLP-At5-2, qLP-Dt7-1 and qLP-Dt7-2) were detected in at least two environments. Two genes (Gh_A05G1584 and Gh_A05G1689) containing nonsynonymous single nucleotide polymorphisms (SNPs) were identified by association analysis using published data. The quantitative real-time PCR results showed that the expression levels of Gh_A05G1584 were higher in cv. CCRI36 than in acc. G2005 during all fibre development stages; Gh_A05G1689 was mainly expressed in 15 and 25 days post-anthesis fibres and its expression level was higher in cv. CCRI36. These results suggest candidate genes for lint percentage and provide molecular information for use in cotton breeding programmes aimed at improving yield.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cotton is one of the most important cash crops in the world, and cotton fibres account for the majority of natural fibres used in the textile industry. Among the four cultivated Gossypium species (G. herbaceum, G. arboreum, G. hirsutum and G. barbadense), upland cotton (G. hirsutum) is most important cultivated variety, accounting for approximately 95% of the world’s cotton production (Chen et al. 2007). Yield is the main factor that determines cotton planting benefits, and improving cotton yield is the primary goal of cotton breeders.
Lint percentage (LP) is an important component of cotton lint yield and an index for the evaluation of lint yield improvement in cotton varieties (Culp and Harrell 1975; Su et al. 2016). LP has been greatly improved in the past several decades through conventional breeding methods. However, the genetic and molecular mechanisms responsible for LP in cotton are not well understood. With the rapid development of molecular marker technology, many quantitative trait loci (QTLs) for LP have been identified and mapped on different chromosomes (Liu et al. 2015, 2017; Ning et al. 2014; Shi et al. 2015; Wang et al. 2015a, 2016). According to the Cotton QTL Database (http://www2.cottonqtldb.org:8081/index), a total of 327 QTLs, distributed on 26 chromosomes, have been identified by linkage and association analysis as being related to LP. Due to the limitations of sequencing technology and reference genomes, prior to 2016 many QTLs were identified by simple sequence repeat (SSR) markers. Ning et al. (2014) used an SSR-based genetic map containing 279 SSR loci to identify nine QTLs for LP on seven chromosomes in an upland cotton population. Liu et al. (2015) constructed a high-density genetic map containing 1675 SSR loci to identify eight stable QTLs for LP identified in at least two environments ). However, due to the low resolution and polymorphic rate of SSR markers, it is difficult to construct a saturated genetic map that covers the whole genome.
The rapid development of next-generation sequencing (NGS) technologies, together with reduced costs of such technologies, has enabled the development and implementation of single nucleotide polymorphism (SNP) markers that cover the whole genome. Restriction site-associated DNA sequencing (RAD-seq) (Jia et al. 2016; Wang et al. 2015b), genotyping-by-sequencing (GBS) (Diouf et al. 2018) and specific-locus amplified fragment sequencing (SLAF-seq) (Su et al. 2016; Zhang et al. 2016) have been successfully applied to the detection of SNP markers in cotton. Zhang et al. (2016) identified 344 candidate genes using a high-density genetic map containing 5521 SNPs that was constructed using SLAF-seq, with a total distance of 3259.37 cM and an average marker distance of 0.78 cM. Diouf et al. (2018) constructed a high-density genetic map of cotton using the GBS method, resulting in map linkage of 5178 markers with a total distance of 4768.10 cM. NGS technologies have also greatly promoted the development of genome-wide association studies (GWAS) of cotton yield traits (Su et al. 2016). Su et al. (2016) identified 81,675 SNPs in 355 upland cotton accessions using SLAF-seq and obtained 12 SNPs associated with LP via GWAS. These authors also reported the identification of a candidate gene, Gh_A02G1268, that might regulate seed development. A cell wall-associated receptor-like kinase 3 protein (GhWAKL3) highly correlated with increased LP was identified based on a combination of identity-by-descent detection, QTL and candidate association analyses (Ma et al. 2019). Song et al. (2019) used 276 upland cotton accessions and a CottonSNP63K array to obtain 10,660 SNPs, of which 23 SNPs were identified to be associated with LP; two candidate genes, Gh_D05G0313 and Gh_D05G1124, were defined as the most promising potential regulators of LP. In a previous study conducted in our laboratory, we developed a recombinant inbred line (RIL) population containing 137 individuals from an intraspecific cross between upland cotton cv. CCRI36 and acc. G2005 and then constructed a high-density genetic map containing 6295 SNPs and 139 SSRs based on this RIL population by RAD-seq. In the present study, we used phenotypic data from four environments to identify valuable QTLs related to LP in intraspecific upland cotton populations. The candidate intervals and genes correlated to cotton lint percentage were obtained for further fine mapping and map-based cloning.
Materials and methods
Mapping population and trait evaluation
Gossypium hirsutum cv. CCRI36 and G. hirsutum acc. G2005 were used as the mapping parents (Jia et al. 2016). F1 seeds were obtained in Anyang, China, in the summer of 2006. The F1 plants were planted and self-pollinated to produce the F2 generation during the winter in Hainan Province in 2006. A total of 137 F2 plants were randomly selected and self-pollinated to obtain F2:3 seeds at the farm in Anyang in 2007. F2:9 RIL populations were obtained using the single seed descendant method in 2010.
Each year from 2011 to 2015, the RIL population and two parents were planted at the farm of the Cotton Research Institute of the Chinese Academy of Agricultural Sciences at Anyang using a randomized complete block design with three replicates. The lines were planted in single-row plots that measured 5 m in length with an inter-row spacing of 0.8 m. A total of 50 naturally opened bolls from each line were manually harvested each year in September, referred to as E1 (2011), E2 (2013), E3 (2014) and E4 (2015). Combined analysis (E5) was performed based on the mean values of LP in four environments. The LP (%) of each line was measured as the ratio of lint weight to seed cotton weight.
Data analysis and QTL detection
The phenotypic data for the two parents were analysed using a t-test. The phenotype data of the RIL population were analysed using SPSS version 21.0 (SPSS IBM Corp., Armonk, NY, USA). The two-way analysis of variance (ANOVA) with interaction was performed using R software (R Foundation for Statistical Computing, Vienna, Austria). The broad-sense heritability of LP was calculated as H2 = VG/(VG + VGE/n + Ve/rn), where VG is the genetic variance, VGE is the interaction variance of genotype × environment, Ve is the error variance, n represents the number of environments and r is the number of replications per environment (Knapp et al. 1985).
The genetic map for QTL mapping used in this present study was reported previously (Jia et al. 2016). The high-density genetic map contained 6295 SNPs and 139 SSR loci identified by RAD-seq. QTLs were identified by composite interval mapping using Windows QTL Cartographer version 2.5 (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm). The parameters of window size, walk speed and background markers were set as 5 cM, 1 cM and 10, respectively. In addition, 1000 permutation tests were used to detect statistical significance of the limit of detection (LOD) value. LOD score ≥ 2.5 was used to detect QTLs. QTLs identified in more than one environment were regarded as ‘stable’, and QTLs explaining > 10% of the phenotypic variance (PV) were regarded as ‘major’. The QTL nomenclature was adapted according to the method described in a previous study (McCouch et al. 1997).
Gene ontology analyses of the candidate genes
Gene ontology (GO) enrichment analysis of the candidate genes related to LP was performed using the AgriGO v2.0 programme (http://systemsbiology.cau.edu.cn/agriGOv2/), with the P value set at < 0.05 (Tian et al. 2017).
Association analysis of candidate genes with LP trait
The data obtained from genotyping the QTL regions and LP traits (9 environments) of 258 diverse accessions reported in the previous study were used for the association analysis (Fang et al. 2017). A total of 258 accessions were planted each year for 3 years (2007–2009) in three replicates in Anyang (AY), Nanjing (NJ) and Kuche (KC), respectively. The best linear unbiased prediction (BLUP) of the phenotype data was estimated using the R package lme4 (R Foundation for Statistical Computing). The kinship coefficients (K) and principal component analysis were evaluated using R package GAPIT (Lipka et al. 2012). Association analysis was performed using the mixed linear model in the R package GAPIT (Lipka et al. 2012). The non-synonymous SNPs were identified in at least four environments and BLUP data with P < 0.01.
Gene expression pattern, RNA extraction and quantitative real-time PCR analysis
The expression levels of the genes in various tissues were obtained from previously reported transcriptome data (Zhang et al. 2015). Cotton bolls of cv. CCRI36 and acc. G2005 were harvested at 0, 3, 5, 10, 15, 20 and 25 days post-anthesis (DPA). The fibres at 5, 10, 15, 20 and 25 DPA were separated from ovules in liquid nitrogen. Three biological replicates for each sample were harvested and stored at − 80 °C. Total RNA was isolated using an RNAprep Pure Plant kit (TIANGEN, Beijing, China). Quantitative real-time PCR (qRT-PCR) experiments were carried out on an ABI Prism 7500 system (Applied Biosystems, Foster City, CA, USA). The Gossypium hirsutum Actin (GhActin) gene was used as the reference gene. The relative expression levels were calculated using three biological replicates according to the 2−△△Ct method (Livak and Schmittgen 2001). The primers used in this study are listed in Electronic Supplementary Material (ESM) Table S1.
Results
Phenotype characteristics of lint percentage under four environments
The phenotypic data for LP of the two parents and RIL population are shown in Table 1. The LP of cv. CCRI36 was significantly higher than that of acc. G2005 in the four environments. The RIL population underwent transgressive segregation according to normal distributions in the four environments. The ANOVA results revealed that LP was significantly influenced by genotype, environment and genotype × environment (P < 0.001) (Table 2). The broad-sense heritability of LP was 91.47%, suggesting that the LP trait was stable under significant genotype × environment effects.
QTL mapping for LP
A total of 28 QTLs for LP were identified on 13 chromosomes based on the previously published high-density genetic map (Jia et al. 2016). These QTLs explained 4.18–17.83% of the PV, with LOD scores ranging from 2.53 to 9.24 (see Table 3). Eight of these 28 QTLs and three novel QTLs were identified by combined analysis (E5). Fourteen and 14 QTLs were mapped on the At and Dt subgenomes, respectively. Gossypium hirsutum cv. CCRI36 conferred positive additive alleles for 9 QTLs, and G. hirsutum acc. G2005 conferred positive additive alleles for 19 QTLs. QTL qLP-At5-2 was identified in three environments (E2, E3 and E4) as well as in the combined analysis (E5), explaining 5.15–8.99% of the PV, with LOD scores ranging from 2.96 to 5.03. QTL qLP-Dt7-1 was identified in two environments (E1 and E2) and in the combined analysis (E5), explaining 5.10–6.74% of the PV, with LOD scores ranging from 3.06 to 3.91. QTL qLP-Dt7-2 was identified in three environments (E1, E2 and E3) and in the combined analysis (E5), explaining 4.29–7.95% of the PV, with LOD scores ranging from 2.54 to 4.67. QTL qLP-At4-2 was identified in one environment (E3) and in the combined analysis (E5), and QTLs qLP-At5-1, qLP-Dt1-2, qLP-Dt10-1 and qLP-Dt10-2 were identified in one environment (E4) and in the combined analysis (E5).
Identification of favourable alleles of QTLs for LP
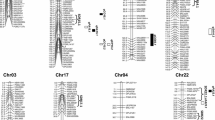
Three stable QTLs (qLP-At5-2, qLP-Dt7-1 and qLP-Dt7-2) and six major QTLs (qLP-At4-1, qLP-At5-1, qLP-Dt1-1, qLP-Dt10-1, qLP-Dt10-2 and qLP-Dt10-3) were used for further analysis to identify the favourable alleles of QTLs for LP. qLP-At5-1, qLP-At5-2 and qLP-Dt1-1 showed positive additive effects originating from cv. CCRI36, and qLP-At4-1, qLP-Dt7-1, qLP-Dt7-2, qLP-Dt10-1, qLP-Dt10-2 and qLP-Dt10-3 showed positive additive effects originating from acc. G2005 (Table 3). The population was divided into two groups (group 1 and group 2) based on the favourable alleles of nine QTLs (Fig. 1). The favourable alleles of group 1 and group 2 were conferred by CCRI36 and G2005, respectively. The phenotypic values of group 1 in the qLP-At5-1, qLP-At5-2 and qLP-Dt1-1 regions were higher than those of group 2 (Fig. 1). The phenotypic values of group 2 in the qLP-At4-1, qLP-Dt7-1, qLP-Dt7-2, qLP-Dt10-1, qLP-Dt10-2 and qLP-Dt10-3 regions were higher than those of group 1 (Fig. 1). The results indicate that these QTLs can provide available information to develop functional markers for molecular marker-assisted selection (MAS) in cotton.
Boxplot of phenotypic characteristics of the two groups classified based on favourable alleles in the recombinant inbred line population. Group 1 comprises favourable alleles conferred by Gossypium hirsutum cv. CCRI36; group 2 indicates favourable alleles conferred by G. hirsutum acc. G2005. The environments are given along the x-axis: E1 2011 in Anyang, E2 2013 in Anyang, E3 2014 in Anyang, E4 2015 in Anyang. Asterisks indicate a significant difference at *P = 0.05 and **P = 0.01. LP Lint percentage
GO analysis of genes in three stable QTL regions
According to the reference genome (Zhang et al. 2015), 455, 117 and 55 genes were identified in the regions of the three stable QTLs qLP-At5-2, qLP-Dt7-1 and qLP-Dt7-2, respectively. Lint fibre production is a key index for cotton LP, which is closely associated with fibre development (Ma et al. 2019). Based on the transcriptome data of upland cotton TM-1 (Zhang et al. 2015), the expression patterns of these genes in 11 tissues (root, stem, leaf, petal, stamen, pistil, 0 DPA ovule and fibres at four developmental stages) were obtained. In the qLP-At5-2, qLP-Dt7-1 and qLP-Dt7-2 QTL regions, 339, 81 and 46 genes, respectively, with FPKM ≥ 1 in at least one of the five investigated tissues (0 DPA ovule, 5, 10, 20 and 25 fibres) were used in subsequent analyses.
All 466 genes were used in the GO analysis to identify potential biological functions, which were classified into three main GO categories (cellular component, molecular function and biological process) and eight GO terms (Fig. 2; ESM Table S2). Microtubule-associated complex (5 genes) was the main subcategory in the cellular component category (Fig. 2). In the molecular function category, tubulin binding (6 genes), microtubule binding (6 genes), cytoskeletal protein binding (6 genes), motor activity (5 genes) and microtubule motor activity (5 genes) were the principal subcategories (Fig. 2). In the biological process category, movement of cell or subcellular component (5 genes) and microtubule-based movement (5 genes) were the main subcategories (Fig. 2).
Prediction of candidate genes by association analysis
To identify the candidate genes related to LP, the genotyping data in three QTL regions (qLP-At5-2, qLP-Dt7-1 and qLP-Dt7-2) of 258 diverse accessions and the LP phenotype data reported in a previous study were used for association analysis to detect the significant site (Fang et al. 2017). Two non-synonymous SNPs (A05_16215549 and A05_17576051) were identified in the qLP-At5-2 regions (Table 4). The GG haplotype of the A05_16215549 (A/G) SNP locus had positive phenotypic effects on LP (Fig. 3a). The AA haplotype of the A05_17576051 (A/G) SNP locus had positive phenotypic effects on LP (Fig. 3b). Thus, two genes (Gh_A05G1584 and Gh_A05G1689) were shown to contain non-synonymous SNPs (Table 4).
Phenotypic characteristics of two single nucleotide polymorphism (SNP) loci and expression levels of two candidate genes during fibre development stages. a Phenotypic characteristics for LP based on SNP A05_16215549, 07AY (2007 in Anyang), 07KC (2007 in Kuche), 07NJ (2007 in Nanjing), 08AY (2008 in Anyang), 08KC (2008 in Kuche), 08NJ (2008 in Nanjing), 09AY (2009 in Anyang), 09KC (2009 in Kuche), 09NJ (2009 in Nanjing), BLUP (BLUP analysis based on 9 environments). b Phenotypic characteristics of LP based on SNP A05_17576051. c Expression levels of Gh_A05G1584 during 7 fibre development stages in cv. CCRI36 and acc. G2005. d Expression levels of Gh_A05G1689 during 7 fibre development stages in cv. CCRI36 and acc. G2005. Asterisks indicate a significant difference at *P = 0.05 and **P = 0.01
RT-PCR analysis was used to identify the differences in the expression levels of these two genes at various fibre development stages between the two parents cv. CCRI36 and acc. G2005. The expression levels of Gh_A05G1584 in CCRI36 were higher than those in G2005 during all fibre development stages (Fig. 3c); Gh_A05G1689 was mainly expressed in 15 and 25 DPA fibres and its expression level was higher in CCRI36 (Fig. 3d).
Discussion
High yield has always been the primary goal of cotton breeding programmes. LP is a relative trait defined as the ratio of lint weight to boll weight. It is one of the important components of yield traits, and its improvement can help to increase lint yield. The broad-sense heritability of LP in the present study was 91.47% and higher than that of other traits in the same RIL population reported in previous studies (Jia et al. 2016, 2018). In other segregation populations (Diouf et al. 2018; Wang et al. 2015a), the broad-sense heritability of LP was also high. Wang et al. (2015a) reported that the broad-sense heritability of the LP was 61.21%. The broad-sense heritability of LP in an F2:3 population reported by Diouf et al. (2018) was as high as 82.65%. In the natural population, LP showed the same characteristics. The broad-sense heritability of the LP was 69.72% in 355 upland cotton accessions grown in four environments (Su et al. 2016). Sun et al. (2018) reported the broad-sense heritability of LP as high as 88.66% in 719 upland cotton accessions grown in seven environments. In the present study, the RIL population and the two parents were planted at the farm of the Cotton Research Institute of the Chinese Academy of Agricultural Sciences, Anyang City, Henan Province for 4 years. Data on daily average temperature and weather from May to September were collected for 4 years. Analysis of these data showed that the average temperature in May, August and September had significant environmental effects (P < 0.001) (ESM Table S3). The numbers of clear days, cloudy days and rainy days during these 5 months for each year during the 4-year study period were clearly different (ESM Table S4), suggesting that that the weather in Anyang City was different in these 4 years (ESM Tables S3 and S4). These results indicate that LP was significantly influenced by the environment, albeit it is a stably inherited trait in various environments. Therefore, we conclude that breeding for high yield through genetic selection of the LP trait is an efficient method.
A total of 28 QTLs related to LP were identified in the present study using a high-density genetic map under four environments (Table 3). These 28 QTLs were distributed on 13 chromosomes, among which Chr16, Chr05, Chr20 and Chr09 contained five, four, three and three QTLs, respectively. A previous meta-analysis of QTLs (Said et al. 2015) demonstrated that the hotspots of QTLs related to LP were mainly distributed on Chr03, Chr07, Chr11, Chr13, Chr16 and Chr24. Thus, most QTLs identified in the present study may be new. The positive additive alleles of nine and 19 QTLs were derived from cv. CCRI36 and acc. G2005, respectively. Low-value parent acc. G2005 provided a higher number of positive additive loci than the high-value parent cv. CCRI36. Similar results were obtained in previous studies on the mapping of QTLs related to LP (Li et al. 2016; Liu et al. 2018; Ning et al. 2014; Yu et al. 2013). Taken together, it appears that not all favourable alleles are derived from high-value parents (Xiao 1996), especially alleles related to the LP trait in cotton. These results suggest that it might be necessary to aggregate the favourable alleles from different parents (not only high-value parents) to improve the LP. Of the 28 QTLs identified, three stable QTLs (qLP-At5-2, qLP-Dt7-1 and qLP-Dt7-2) were detected under at least 2 environments, and six major QTLs (qLP-At4-1, qLP-At5-1, qLP-Dt1-1, qLP-Dt10-1, qLP-Dt10-2 and qLP-Dt10-3) explained > 10% of the PV (Table 3).The population can be clearly divided into two groups based on the favourable alleles of nine QTLs of the two parents (Fig. 1). These results show that these QTLs are reliable and are available for future MAS in breeding programmes for high-yield cotton.
The objective of QTL mapping was to obtain available genetic information for molecular breeding. To further confirm the commonality and reliability of the QTLs obtained in the present study, we compared the physical intervals of the QTLs associated with LP in previous studies with our results. Nine QTLs (qLP-At3-1, qLP-At5-2, qLP-At9-2, qLP-At10-2, qLP-At13-1, qLP-At13-2, qLP-Dt4-1, qLP-Dt5-1 and qLP-Dt7-2) were identified in previous studies by e-PCR (Huang et al. 2017; Jia et al. 2014; Ma et al. 2018; Mei et al. 2013; Qin et al. 2015; Shi et al. 2015). QTL qLP-At3-1 was mapped to an adjacent region of NAU862 that was shown to be associated with LP under two environments in an association analysis study (Qin et al. 2015). A pair of loci (HAU1185 and TMB1791) detected in a natural population were located in the interval of the stable QTL qLP-At5-2 (Jia et al. 2014). qLP-At9-2 and qLP-At13-2 were identified as qGhLP-c9 and qGhLP-c13, respectively, in a genome-wide association study (GWAS) (Huang et al. 2017). Ma et al. (2018) reported that SNP loci A10_96602651, A13_10477365 and D05_25310910 associated with LP were located in the region of the QTLs qLP-At10-2, qLP-At13-1 and qLP-Dt5-1, respectively (Ma et al. 2018). qLP-Dt4-1 was identified as qGhLP-c22-2 in a study of Huang et al. (2017), and D04_5572787 was also mapped to the qLP-Dt4-1 region (Ma et al. 2018). These common QTLs were identified in different populations and markers, confirming their reliability and potential use for future fine mapping and mapping-based cloning.
In the present study we identified 455, 117 and 55 genes in the regions of the three stable QTLs qLP-At5-2, qLP-Dt7-1 and qLP-Dt7-2, respectively. The genotypes and phenotypes of 258 accessions reported in a previous study (Fang et al. 2017) were used for additional analyses to investigate the candidate genes associated with LP in the three candidate intervals. Two non-synonymous SNPs (− log10P > 2) were identified in more than four environments, and two candidate genes (Gh_A05G1584 and Gh_A05G1689) were also identified (Table 4). The expression levels of the two candidate genes were significantly different between the two parents during the fibre development stages (Fig. 3). Gh_A05G1584 (GhFREE1) is homologous to AtFREE1, which is the key regulator of intracellular trafficking and vacuole biogenesis and plays an essential role in plant growth and development (Gao et al. 2014; Kolb et al. 2015). The osmotic regulation of vacuole enlargement is important for cotton fibre development and cell expansion (Wang et al. 2010, 2014; Wang and Ruan 2010). Gh_A05G1689 (GhTLP3) is a member of the thaumatin-like protein family and encodes a pathogenesis-related 5-like protein. GbTLP1 participates in secondary cell-wall synthesis (Tu et al. 2007) and enhances resistance against Verticillium dahliae in transgenic tobacco (Munis et al. 2010). GrTLP3 is considered a candidate gene related to fibre length and fibre strength (Islam et al. 2016). Li et al. (2017) identified a candidate gene (GhTLP) significantly correlated with fibre length by RNA-seq and QTL mapping (Li et al. 2017). These results provide candidate genes for future cloning of the genes related to LP, and understanding of the associated molecular mechanisms provides a foundation for cotton yield breeding.
References
Chen ZJ, Scheffler BE, Dennis E, Triplett BA, Zhang T, Guo W, Chen X, Stelly DM, Rabinowicz PD, Town CD, Arioli T, Brubaker C, Cantrell RG, Lacape JM, Ulloa M, Chee P, Gingle AR, Haigler CH, Percy R, Saha S, Wilkins T, Wright RJ, Van Deynze A, Zhu Y, Yu S, Abdurakhmonov I, Katageri I, Kumar PA, Mehboob Ur R, Zafar Y, Yu JZ, Kohel RJ, Wendel JF, Paterson AH (2007) Toward sequencing cotton (Gossypium) genomes. Plant Physiol 145:1303–1310
Culp TW, Harrell DC (1975) Influence of lint percentage, boll size, and seed size on lint yield of upland cotton with high fiber strength. Crop Sci 15:741–746
Diouf L, Magwanga RO, Gong W, He S, Pan Z, Jia Y, Kirungu JN, Du X (2018) QTL mapping of fiber quality and yield-related traits in an intra-specific Upland cotton using genotype by sequencing (GBS). Int J Mol Sci 19:441
Fang L, Wang Q, Hu Y, Jia Y, Chen J, Liu B, Zhang Z, Guan X, Chen S, Zhou B, Mei G, Sun J, Pan Z, He S, Xiao S, Shi W, Gong W, Liu J, Ma J, Cai C, Zhu X, Guo W, Du X, Zhang T (2017) Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat Genet 49:1089–1098
Gao C, Luo M, Zhao Q, Yang R, Cui Y, Zeng Y, Xia J, Jiang L (2014) A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr Biol 24:2556–2563
Gao Y, He C, Zhang D, Liu X, Xu Z, Tian Y, Liu X, Zang S, Pauly M, Zhou Y, Zhang B (2017) Two trichome birefringence-like proteins mediate xylan acetylation, which is essential for leaf blight resistance in rice. Plant Physiol 173:470–481
Gille S, Pauly M (2012) O-acetylation of plant cell wall polysaccharides. Front Plant Sci 3:12
Gille S, de Souza A, Xiong G, Benz M, Cheng K, Schultink A, Reca IB, Pauly M (2011) O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant cell 23:4041–4053
Huang C, Nie X, Shen C, You C, Li W, Zhao W, Zhang X, Lin Z (2017) Population structure and genetic basis of the agronomic traits of upland cotton in China revealed by a genome-wide association study using high-density SNPs. Plant Biotechnol J 15:1374–1386
Islam MS, Zeng L, Thyssen GN, Delhom CD, Kim HJ, Li P, Fang DD (2016) Mapping by sequencing in cotton (Gossypium hirsutum) line MD52ne identified candidate genes for fiber strength and its related quality attributes. Theor Appl Genet 129:1071–1086
Jia X, Pang C, Wei H, Wang H, Ma Q, Yang J, Cheng S, Su J, Fan S, Song M, Wusiman N, Yu S (2016) High-density linkage map construction and QTL analysis for earliness-related traits in Gossypium hirsutum L. BMC Genom 17:909
Jia X, Wang H, Pang C, Ma Q, Su J, Wei H, Song M, Fan S, Yu S (2018) QTL delineation for five fiber quality traits based on an intra-specific Gossypium hirsutum L. recombinant inbred line population. Mol Genet Genom 293:831–843
Jia Y, Sun X, Sun J, Pan Z, Wang X, He S, Xiao S, Shi W, Zhou Z, Pang B, Wang L, Liu J, Ma J, Du X, Zhu J (2014) Association mapping for epistasis and environmental interaction of yield traits in 323 cotton cultivars under 9 different environments. PLoS ONE 9:e95882
Knapp S, Stroup W, Ross W (1985) Exact confidence intervals for heritability on a progeny mean basis. Crop Sci 25:192–194
Kolb C, Nagel MK, Kalinowska K, Hagmann J, Ichikawa M, Anzenberger F, Alkofer A, Sato MH, Braun P, Isono E (2015) FYVE1 is essential for vacuole biogenesis and intracellular trafficking in Arabidopsis. Plant Physiol 167:1361–1373
Li C, Dong Y, Zhao T, Li L, Yu E, Mei L, Daud MK, He Q, Chen J, Zhu S (2016) Genome-wide SNP linkage mapping and QTL analysis for fiber quality and yield traits in the Upland cotton recombinant inbred lines population. Front Plant Sci 7:1356
Li X, Wu M, Liu G, Pei W, Zhai H, Yu J, Zhang J, Yu S (2017) Identification of candidate genes for fiber length quantitative trait loci through RNA-Seq and linkage and physical mapping in cotton. BMC Genom 18:427
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28:2397–2399
Liu D, Liu F, Shan X, Zhang J, Tang S, Fang X, Liu X, Wang W, Tan Z, Teng Z, Zhang Z, Liu D (2015) Construction of a high-density genetic map and lint percentage and cottonseed nutrient trait QTL identification in upland cotton (Gossypium hirsutum L.). Mol Genet Genom 290:1683–1700
Liu R, Gong J, Xiao X, Zhang Z, Li J, Liu A, Lu Q, Shang H, Shi Y, Ge Q, Iqbal MS, Deng X, Li S, Pan J, Duan L, Zhang Q, Jiang X, Zou X, Hafeez A, Chen Q, Geng H, Gong W, Yuan Y (2018) GWAS analysis and QTL identification of fiber quality traits and yield components in Upland cotton using enriched high-density SNP markers. Front Plant Sci 9:1067
Liu X, Teng Z, Wang J, Wu T, Zhang Z, Deng X, Fang X, Tan Z, Ali I, Liu D, Zhang J, Liu F (2017) Enriching an intraspecific genetic map and identifying QTL for fiber quality and yield component traits across multiple environments in Upland cotton (Gossypium hirsutum L.). Mol Genet Genom 292(6):1281–1306
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△Ct method. Methods 25:402–408
Ma Z, He S, Wang X, Sun J, Zhang Y, Zhang G, Wu L, Li Z, Liu Z, Sun G, Yan Y, Jia Y, Yang J, Pan Z, Gu Q, Li X, Sun Z, Dai P, Gong W, Wu J, Wang M, Liu H, Feng K, Ke H, Wang J, Lan H, Wang G, Peng J, Wang N, Wang L, Pang B, Peng Z, Li R, Tian S, Du X (2018) Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat Genet 50:803–813
Ma X, Wang Z, Li W, Zhang Y, Zhou X, Liu Y, Ren Z, Pei X, Zhou K, Zhang W, He K, Zhang F, Liu J, Ma W, Xiao G, Yang D (2019) Resequencing core accessions of a pedigree identifies derivation of genomic segments and key agronomic trait loci during cotton improvement. Plant Biotechnol J 17:762–775
McCouch SR, Cho YG, Yano PE, Blinstrub M, Morishima H, Kinoshita T (1997) Report on QTL nomenclature. Rice Genet Newsl 14:11–131
Mei H, Zhu X, Zhang T (2013) Favorable QTL alleles for yield and its components identified by association mapping in Chinese Upland cotton cultivars. PLoS ONE 8:e82193
Munis MF, Tu L, Deng F, Tan J, Xu L, Xu S, Long L, Zhang X (2010) A thaumatin-like protein gene involved in cotton fiber secondary cell wall development enhances resistance against Verticillium dahliae and other stresses in transgenic tobacco. Biochem Biophys Res Commun 393:38–44
Ning Z, Chen H, Mei H, Zhang T (2014) Molecular tagging of QTLs for fiber quality and yield in the upland cotton cultivar Acala-Prema. Euphytica 195:143–156
Qin H, Chen M, Yi X, Bie S, Zhang C, Zhang Y, Lan J, Meng Y, Yuan Y, Jiao C (2015) Identification of associated SSR markers for yield component and fiber quality traits based on frame map and Upland cotton collections. PLoS ONE 10:e0118073
Said JI, Song M, Wang H, Lin Z, Zhang X, Fang DD, Zhang J (2015) A comparative meta-analysis of QTL between intraspecific Gossypium hirsutum and interspecific G. hirsutum x G. barbadense populations. Mol Genet Genom 290:1003–1025
Shi Y, Li W, Li A, Ge R, Zhang B, Li J, Liu G, Liu A, Shang H, Gong J, Gong W, Yang Z, Tang F, Liu Z, Zhu W, Jiang J, Yu X, Wang T, Wang W, Chen T, Wang K, Zhang Z, Yuan Y (2015) Constructing a high-density linkage map for Gossypium hirsutum x Gossypium barbadense and identifying QTLs for lint percentage. J Integr Plant Biol 57:450–467
Song C, Li W, Pei X, Liu Y, Ren Z, He K, Zhang F, Sun K, Zhou X, Ma X, Yang D (2019) Dissection of the genetic variation and candidate genes of lint percentage by a genome-wide association study in upland cotton. Theor Appl Genet 132:1991–2002
Stranne M, Ren Y, Fimognari L, Birdseye D, Yan J, Bardor M, Mollet JC, Komatsu T, Kikuchi J, Scheller HV, Sakuragi Y (2018) TBL10 is required for O-acetylation of pectic rhamnogalacturonan-I in Arabidopsis thaliana. Plant J 96:772–785
Su J, Fan S, Li L, Wei H, Wang C, Wang H, Song M, Zhang C, Gu L, Zhao S, Mao G, Pang C, Yu S (2016) Detection of favorable QTL alleles and candidate genes for lint percentage by GWAS in Chinese Upland cotton. Front Plant Sci 7:1576
Sun Z, Wang X, Liu Z, Gu Q, Zhang Y, Li Z, Ke H, Yang J, Wu J, Wu L, Zhang G, Zhang C, Ma Z (2018) A genome-wide association study uncovers novel genomic regions and candidate genes of yield-related traits in upland cotton. Theor Appl Genet 131:2413–2425
Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45:W122–W129
Tu L, Zhang X, Liang S, Liu D, Zhu L, Zeng F, Nie Y, Guo X, Deng F, Tan J, Xu L (2007) Genes expression analyses of sea-island cotton (Gossypium barbadense L.) during fiber development. Plant Cell Rep 26:1309–1320
Wang L, Ruan Y (2010) Unraveling mechanisms of cell expansion linking solute transport, metabolism, plasmodesmtal gating and cell wall dynamics. Plant Signal Behav 5:1561–1564
Wang L, Li X, Lian H, Ni DA, He Y, Chen X, Ruan Y (2010) Evidence that high activity of vacuolar invertase is required for cotton fiber and Arabidopsis root elongation through osmotic dependent and independent pathways, respectively. Plant Physiol 154:744–756
Wang L, Cook A, Patrick JW, Chen XY, Ruan YL (2014) Silencing the vacuolar invertase gene GhVIN1 blocks cotton fiber initiation from the ovule epidermis, probably by suppressing a cohort of regulatory genes via sugar signaling. Plant J 78:686–696
Wang H, Huang C, Guo H, Li X, Zhao W, Dai B, Yan Z, Lin Z (2015a) QTL mapping for fiber and yield traits in Upland cotton under multiple environments. PLoS ONE 10:e0130742
Wang H, Jin X, Zhang B, Shen C, Lin Z (2015b) Enrichment of an intraspecific genetic map of upland cotton by developing markers using parental RAD sequencing. DNA Res 22:147–160
Wang H, Huang C, Zhao W, Dai B, Shen C, Zhang B, Li D, Lin Z (2016) Identification of QTL for fiber quality and yield traits using two immortalized backcross populations in Upland cotton. PLoS ONE 11:e0166970
Xiao J, Li J, Yuan L, Tanksley SD (1996) Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor AppI Genet 92:230–244
Yu J, Zhang K, Li S, Yu S, Zhai H, Wu M, Li X, Fan S, Song M, Yang D, Li Y, Zhang J (2013) Mapping quantitative trait loci for lint yield and fiber quality across environments in a Gossypium hirsutum x Gossypium barbadense backcross inbred line population. Theor Appl Genet 126:275–287
Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu B, Liu C, Wang S, Pan M, Wang Y, Wang D, Ye W, Chang L, Zhang W, Song Q, Kirkbride RC, Chen X, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu X, Zhang H, Wu H, Zhou L, Mei G, Chen S, Tian Y, Xiang D, Li X, Ding J, Zuo Q, Tao L, Liu Y, Li J, Lin Y, Hui Y, Cao Z, Cai C, Zhu X, Jiang Z, Zhou B, Guo W, Li R, Chen ZJ (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33:531–537
Zhang Z, Shang H, Shi Y, Huang L, Li J, Ge Q, Gong J, Liu A, Chen T, Wang D, Wang Y, Palanga KK, Muhammad J, Li W, Lu Q, Deng X, Tan Y, Song W, Cai J, Li P, Rashid HO, Gong W, Yuan Y (2016) Construction of a high-density genetic map by specific locus amplified fragment sequencing (SLAF-seq) and its application to Quantitative Trait Loci (QTL) analysis for boll weight in upland cotton (Gossypium hirsutum.). BMC Plant Biol 16:79
Zhong R, Cui D, Ye ZH (2017) Regiospecific acetylation of xylan is mediated by a group of DUF231-containing o-acetyltransferases. Plant Cell Physiol 58:2126–2138
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2017YFD0101603), the Chinese National Natural Science Foundation (31601346) and the China Agriculture Research System (CARS-15-06).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, H., Jia, X., Kang, M. et al. QTL mapping and candidate gene identification of lint percentage based on a recombinant inbred line population of upland cotton. Euphytica 217, 102 (2021). https://doi.org/10.1007/s10681-021-02823-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-021-02823-x