Abstract
Verticillium wilt (VW, caused by Verticillium dahliae Kleb) is a destructive fungal soil-borne disease in Upland cotton (Gossypium hirsutum L.). High levels of VW resistance can be transferred into Upland from Pima cotton (G. barbadense L.) through interspecific introgression breeding. In this greenhouse study, VW resistance was evaluated in a multi-parent advanced generation inter-cross (MAGIC) introgressed line (IL) population, derived from a random mated Barbadense Upland population with five generations of intermating (called RMBUP-C4) between three Upland cotton cultivars and 18 CS-B Upland lines each carrying a pair of G. barbadense chromosome or arm in the TM-1 background. The objectives of this study were to, (1) evaluate VW resistance of 530 MAGIC ILs in the greenhouse; and (2) to identify lines with VW resistance in the MAGIC population based on a total of three replicated greenhouse tests. Approximately 8 plants for each line in each replicate were grown and screened for VW resistance using three parameters i.e., disease leaf severity rating, percentage defoliated leaves, and percentage infected plants, with a total of ~ 25,190 plants evaluated. A correlation analysis indicated that the three parameters were significantly and positively correlated with one another in each test. The disease leaf severity rating was the best parameter to assess VW resistance due to its relatively low coefficient of variation and its higher resolution to differentiate resistant genotypes from susceptible ones. Of the 530 genotypes, 5 showed resistance to VW, namely, NMIL348, NMIL518, NMIL405, NMIL290, NMIL307 and had higher levels of resistance to VW with mean disease leaf severity ratings, percentage of defoliated leaves, and percentage of infected plants across three tests ranging from 0.58–1.46, 9.46–26.74, and 25–95%, respectively. These lines can be used as parental lines to improve VW resistance in cotton breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Currently, Gossypium spp. is the most important cultivated fiber crop, and its world production was 106.49 million bales (23.18 MMT) in 2017 (USDA-ERS 2017a). The United States produced 20.9 million bales (4.55 MMT) of cotton (USDA-NASS 2018) and is the world’s third-largest cotton producer after India and China and the leading cotton exporter. In 2017, these three countries accounted for 63% of global cotton production (USDA-ERS 2017b). There are four cultivated cotton species, namely, G. hirsutum L. (Upland cotton), G. barbadense L. (Pima cotton), G. arboreum L. and G. herbaceum L. In the US, only two species are cultivated commercially, Upland and Pima, with Upland providing more than 97% of annual production (USDA-ERS 2018).
Verticillium wilts (VW), mainly caused by Verticillium dahliae and V. albo-atrum result in billions of dollars in annual crop losses worldwide (Klosterman et al. 2009). VW in cotton, caused by V. dahliae Kleb, is a limiting factor in cotton production in much of the cotton growing countries around the world (Pegg and Brady 2002). Carpenter (1914) was the first to report VW in cotton in Virginia in the eastern US. Later, it was reported by Shapovalov and Rudolph (1930) in California in the west. VW is known to significantly reduce cotton seed and lint yield (Friebertshauser and DeVay 1982; Wheeler and Woodward 2016). Zhang et al. (2012) compared healthy and VW diseased plants of 23 cultivars in the field and found that VW significantly reduced cotton yield, lint percentage, 50% span length and micronaire, but not 2.5% span length and fiber strength. To date, there are a few reports on VW resistance in commercial cotton cultivars (Zhang et al. 2012; Zhou et al. 2014). In the past, strategies such as soil fumigation, soil solarization, crop rotation and arbuscular mycorrhizal and endophytic fungi as a protector have been tested or used to control the disease (Wilhelm and Ferguson 1953; Pullman et al. 1981; Liu 1995; Xiao et al. 1998; Wheeler et al. 2012). Although chemical fungicides seem to be effective, they are not environmentally friendly and therefore non-sustainable (Nannipieri et al. 1990).
The soil habitat of VW, the ability of its survival structures to persist for years, and the fact that it can no longer be controlled by any means once it has entered the vascular bundle is what makes it a chronic economic problem in cotton production (Klosterman et al. 2009). The most feasible and sustainable way of controlling VW in cotton is breeding through detection and utilization of resistant cultivars or germplasm. Understanding the genetic events at the molecular level in the disease interaction between cotton plants and VW will increase our ability to exploit the existing resistance in cotton germplasm to reduce these losses through conventional breeding (Bolek et al. 2005). Acala cultivars developed in New Mexico and California have shown good fiber quality, yields and resistance to VW (Oakley 1998; Zhang et al. 2005). Resistance in Acala cotton is thought to be derived from G. barbadense. However, there has been no report to indicate the introgression of Pima germplasm to Acala cotton and the origin of the resistance (Zhang et al. 2005).
Introgressive hybridization or introgression is the genetic modification of one species by another through hybridization and repeated backcrossing (Anamthawat-Jónsson 2001). In plant breeding, it serves to transfer desirable traits such as resistance genes and important agronomic traits from one species to another (Anamthawat-Jónsson 2001). Upland cotton is known for its high yield potential, wide adaptation, fuzzed seed, and high lint percentage and accounts for 95% of world cotton production (Zhang et al. 2014a). Pima or Egyptian cotton has superior fiber quality, naked seed, lower lint percentage, and lower yield potential, but is only grown in semi-arid and arid areas in the world (Zhang et al. 2014a).
Cotton species and genotypes differ in their resistance to VW. Pima is a cultivated tetraploid species highly tolerant to VW (Wilhelm et al. 1974). Zhang et al. (2012) also found that Pima possessed higher levels of VW resistance than Upland cotton, but the performance was reversed when the root system was wounded after inoculation. However, the resistant traits in Pima have not been transferred into commercial Upland, except in breeding lines (Zhang et al. 2012). The major obstacle for successful introgression breeding has been hybrid breakdown, instability, and selective elimination of desirable genes during selfing (Zhang et al. 2014a). There is what is known as a reproductive barrier between the two species, whenever interspecific hybrids are carried out; hybrid breakdown is seen in the F2 and advanced generations (Zhang et al. 2014a).
Traditional experimental populations combine the genomes of two parents with contrasting phenotypes to identify regions of the genome affecting the trait. However, these populations tend to have a narrow genetic base. Multi-parent advanced generation inter-crosses (MAGIC) inter-mate multiple inbred founders for several generations prior to creating inbred lines. The result is a diverse population whose genomes are fine-scale mosaics of contributions from all founders (Huang et al. 2015). The development of MAGIC populations and chromosome substitution lines is a novel approach to traditional cotton breeding for disease resistance. Advances or lack thereof in VW resistance from these populations, however, remain to be seen. The objective of this study was to evaluate VW resitance under greenhouse conditions in an introgression line (IL) population of 530 MAGIC genotypes, derived from a random mated Barbadense Upland population with five generations of intermating (called RMBUP-C4, Jenkins et al. 2013) between ‘Sure-Grow 747’ (PVP 9800118), ‘PSC 355’, and ‘FM 966’ (PVP 200100209) and 18 CS-B lines (Stelly et al. 2005).
Materials and methods
Plant materials
A total of 530 MAGIC introgression lines (ILs) were used in this study. This population is a random mated population with G. barbadense alleles introgressed into Upland germplasm. The 530 MAGIC ILs were developed from an intermating population between 3 Upland cultivars, namely, ‘Sure-Grow 747’, ‘PSC 355’, and ‘FM 966’ and 18 chromosome substitution lines (CSLs) from G. barbadense (CS-B) (Stelly et al. 2005). These CS-B lines were CS-B01, CS-B02, CS-B04, CS-B06, CS-B07, CS-B10, CS-B16, CS-B17, CS-B18, CS-B25, CS-B05sh, CS-B11sh, CS-B12sh, CS-B14sh, CS-B15sh, CSB22sh, CS-B22Lo, and CS-B26Lo. This population involved five cycles of random mating beginning with 53 top-crossed F1 lines (Jenkins et al. 2013). After each cycle of random mating using a bulked pollen method (Jenkins et al. 2013), F1 lines were combined based on the original 18 CS-B lines as parents, thus producing 18 individual populations. These were planted as 18 individual populations and randomly mated among populations each additional cycle (Jenkins et al. 2013). The intermated population after one generation of self pollination with bulked seed was called RMBUP-C4 and publicly released in 2012 and registered in Crop Science Society of America (CSSA) by Jenkins et al. (2013).
A subsample of the bulked seed with about 800 seed from the RMBUP-C4 population was grown and harvested individually in Leyendecker Plant Science Center, New Mexico State University (NMSU), Las Cruces, NM, in 2012. They were advanced to 750 progeny rows in 2013, which gave rise to the MAGIC population of 750 introgressed lines designated New Mexico Introgression Lines (NMILs), 530 of which were subsequently used in this study. A total of 3 replicated experiments were conducted in the greenhouse at the Fabian Garcia Research Center, NMSU, Las Cruces, NM, in the years 2016 (Test 1) and 2017 (Test 2 and Test 3).
Test 1: The experiment was a randomized complete block design (RCBD) with 530 MAGIC NMILs and two replications. The seeds were planted in 10-cm plastic pots with 10 seed pot−1. Each pot consisted of potting soil (Scotts 450, Scotts Co., Marysville, OH, USA) infected with V. dahliae pathogen from a previous greenhouse experiment. Planting for Test 1 was done through 9–10th May 2016.
The pathogen, V. dahliae was isolated from the cotton plants stored under cold temperatures and was placed on petri plates containing Czapek-Dox agar. After 10–15 days, the pathogen was cultured in Czapek-Dox broth at 25 °C at 150 rpm on a rotary shaker for 20 days. The conidial suspension was then filtered using a double layered cheese cloth to separate the conidia from mycelia. The conidial suspension was then quantified using a hemocytometer. The seedlings were inoculated with the conidial suspension when they were at the 2nd–3rd true leaf stage. To minimize escapes, double inoculations were carried out. The inoculation involved pipetting the inoculum unto the soil surface with no root wounding in each pot. At the end of inoculation, the plants were gently watered to ensure infiltration of inoculum into the soil. The first inoculation was done on June 13th, 2016 (34 days after planting, DAP) and the second was carried out on June 17th, 2016 (38 DAP). In both inoculations, 10 mL of inoculum with a concentration of 4.2 × 106 conidia mL-1 was applied to each pot. Assessment of disease was done on August 10–13th, 2016 (58–61 DAI). Plant management followed normal practices, as established in the program, including daily irrigation and bi-weekly application of fertilizers.
Test 2: The second experiment involved the same 530 genotypes with the same RCBD and two replications as Test 1. Planting for Test 2 was done 3–4th July 2017 in the greenhouse at the NMSU Fabian Garcia Research Center. The potting soil used was pathogen-free Miracle-Gro Moisture Control Potting Mix 2 CF (Scotts Co., Marysville, OH). The first inoculation was done on July 28th, 2017 (24 DAP) using 10 mL of inoculum applied to each pot (concentration 4.2 × 106 conidia mL-1); and the second round of inoculation was done on August 1st, 2017 in that 7 mL of inoculum was applied to every pot (concentration 5.2 × 106 conidia mL-1). A third and final round of inoculation was done on August 9th, 2017, 7 mL of inoculum was applied to every pot (concentration 3.4 × 106). VW assessment was done between September 28th and October 4th, 2017 (60–65 DAI) using a VW screening protocol from Zhang et al. (2012). Similar plant management as Test 1 was followed. Koch’s postulate was performed after each test to verify that the plants showing symptoms of Verticillium wilt were indeed caused by V. dahliae.
Test 3: A total of 133 genotypes used in Test 3 were chosen by selecting some of the top most resistant lines and bottom most susceptible lines in Test 1. The seeds were planted on April 28th, 2017 using the same method as Test 1, based on a RCBD with three replications. The first inoculation was done on May 17th, 2017 (18 DAP); the second round of inoculation was done on May 19th, 2017 (21 DAP) and the third round of inoculation was done on May 26th, 2017 (28 DAP). VW assessment was carried out on July 18th, 2017 (i.e., 68 DAI).
Assessment of Verticillium wilt resistance
The response of the cotton seedlings to VW based on leaf severity rating was evaluated at 58–68 DAI (i.e., 84–92 DAP) using a protocol (with a 0–5 rating scale) developed by Zhang et al. (2012), as follows:
- 0:
-
No symptom
- 1:
-
< 25% chlorotic/necrotic leaves, no leaf or cotyledon abscission.
- 2:
-
25–50% chlorotic/necrotic leaves, less than 2 leaves including cotyledons abscised.
- 3:
-
50–75% chlorotic/necrotic leaves, 2–3 leaves including cotyledons abscised.
- 4:
-
> 75% chlorotic/necrotic leaves, more than 3 leaves including cotyledons abscised.
- 5:
-
Complete defoliation or plant death.
In addition to leaf severity ratings, data were also collected on an individual plant basis for the following traits: the total number of leaves and the number of defoliated leaves. Percentage of defoliated leaves, percentage of infected plants, and average leaf severity rating for each genotype in each replication were calculated.
Statistical analyses
The data were subjected to an analysis of variance (ANOVA) using SAS 9.3 (SAS Institute Inc., Cary, NC, USA). PROC MIXED statistical procedure was used to determine the statistical significance of various sources of variation. Genotype was treated as a fixed effect and replication as a random effect. Significant interaction between genotype and test run was found; therefore, the analysis was conducted by test. A Pearson correlation analysis was also performed between VW resistance traits using the genotypic means from each test.
Results
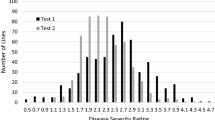
In Test 1, genotypic differences for average leaf severity rating and percentage of defoliated leaves at 58–60 DAI were significant (P < 0.05); however, there were no significant differences among the 530 lines in the percentage of infected plants, i.e., disease incidence (Table 1). The leaf severity ratings for different genotypes ranged from 0.48 to 3.68 with a mean of 1.68 in this test. The 10 lines with the lowest leaf severity ratings ranged from 0.48 to 0.64, as compared to 2.65–3.68 for the 10 lines with the highest ratings. The percentage of defoliated leaves ranged from 8.0 to 67.19% with a mean of 26.91%. The 10 lines with the lowest defoliated leaves ranged from 8.0 to 10.57%, as compared to 41.04–67.19% for the 10 genotypes with the highest values. In this test, the percentages of infected plants by genotypes ranged from 46.25 to 100% with a mean of 73.67%. In fact, more than 50 lines had 100% infected plants. However, there were still 26.33% of the plants escaped from the VW infection or did not show apparent VW symptoms.
In Test 2, genotypic differences in average leaf severity rating and percentage of infected plants at 60–63 DAI were significant (P < 0.05), while the percentage of defoliated leaves was not significantly different among the 530 lines (Table 1). The percentage of infected plants ranged from 65.72 to 100% with a mean of 99.3% in this test, indicating a high level of infection. In this test, only 5% of the lines tested did not receive 100% plant infection. The 10 lines with the lowest leaf severity ratings ranged from 1.09 to 1.66, as compared to 3.12–3.42 for the 10 genotypes with the highest ratings. Although no significant genotypic difference in the percentage of defoliated leaves was detected in this test, 10 lines had the lowest percentage of defoliated leaves ranging from 12.17 to 19.55%, as compared to 46.40–52.31% for 10 lines with the highest values.
To test if genotype × test interactions existed, a combined ANOVA was performed for the two greenhouse tests (Test 1 and 2), and significant genotypic variations were again detected for disease severity rating (F = 1.24, P < 0.01) and percent defoliated leaves (F = 1.16, P < 0.05). However, no significant genotype difference in the percentage of infected plants and no genotype × test interaction for any of the three traits were detected.
In Test 3, genotypic differences in average leaf severity rating and the percentage of infected plants at 60–63 DAI were significant (P < 0.05), while the percentage of defoliated leaves was not significantly different among the 133 lines selected based on Test 1 and 2 (Table 1). Similar to Test 1, the percentage of infected plants ranged from 33.97 to 100% with a mean of 74.46% in this test. The severity ratings ranged from 0.60 to 3.05 with a mean of 1.80. The 10 lines with the lowest ratings ranged from 0.60 to 1.13, as compared to 2.48–3.05 for the 10 lines with the highest ratings. Although the percentages of defoliated leaves did not significantly differ among the genotypes tested, 10 lines had lowest values ranging from 9.25 to 14.85%, as compared to 10 lines with the highest values at 36.28–43.35%.
In addition, a combined ANOVA on the common lines tested in the three tests also showed significant genotypic variation (F = 1.50–2.00, P < 0.001) and genotype × test interactions (F = 1.23–1.62, P < 0.01) for the three traits including the disease severity rating. However, Based on Tests 1, 2 and 3, five lines were determined to be the most consistently resistant to VW. These lines are NMIL348, NMIL518, NMIL307, NMIL290 and NMIL405 with average leaf severity ratings ranging from 0.83 to 1.17 and percentage of defoliated leaves ranging from 8.0 to 23.8% (Table 2). These selected lines were in the top 30 lines performing consistently in all the three tests based on leaf severity rating. There were 11 lines found to be highly susceptible to VW, namely, NMIL698, NMIL227, NMIL706, NMIL237, NMIL258, NMIL259, NMIL654, NMIL360, NMIL023, NMIL528 and NMIL 399, with average leaf severity ratings ranging from 2.42 to 2.98 and percentage of defoliated leaves ranging from 29.0 to 40.3% (Table 2). These selected lines were in the bottom 40 lines performing consistently in all the three tests based on leaf severity rating.
Discussion
In this study, three greenhouse tests were conducted to screen a MAGIC cotton population with G. barbadense alleles introgressed into Upland germplasm for VW resistance. Of the 530 lines tested, we identified 5 lines that showed relatively higher levels of resistance (with leaf severity rating < 1.18 averaged from three replicated tests) against VW, accounting for 0.75% of the tested genotypes.
As shown in this study, genotypic variation in VW resistance can be detected in greenhouse screening based on the analysis of variance (ANOVA) on replicated tests. Selected resistant genotypes were also shown to be resistant when retested in this study. However, genotypes with low severity ratings should be retested in order to confirm if they are indeed resistant and not mere escapes due to low inoculum or a criterion of the disease symptoms not being met. Susceptible genotypes, however, don’t necessarily need to be retested, and some may be used as susceptible checks to measure the success of the field or greenhouse screening.
Field and greenhouse tests are used to evaluate VW resistance in cotton. Greenhouse tests are more favorable because they tend to minimize experimental errors with uniform and a high level of inoculum. Although field tests do reflect the natural order of infection and can detect resistance on a mature plant basis in the field, environmental conditions tend to influence the results greatly. Field tests also require a nearly 6-month cotton growing season, and uniform inoculation and infection is very difficult to accomplish in field tests. Greenhouse tests tend to moderate some of these errors by allowing the experimenter to have more control over some of the factors that can cause variation in the response of inoculated plants. Greenhouse tests allow for more uniformity and environmental control which are key factors to successful screening for VW resistance (Zhou et al. 2014; Zhang et al. 2014b). However, resistant genotypes identified in the greenhouse should be always tested and confirmed for their responses to VW in field conditions before their use in production or breeding.
There have been many methods used to inoculate cotton plants with V. dahliae; these include root inoculation, pot immersion, bare-root dipping and stem injection/puncture (Zhang et al. 2014b). In cotton, the most popular method is root inoculation. Bolek et al. (2005) found that stem inoculation achieved more consistent results than root inoculation in a comparison using similar lines in a field study. However, the root inoculation method is preferred because the stem puncture method fails to account for resistance that may be conferred by the plant root system. Success in using the root inoculation method has been reported by Zhang et al. (2012), Fang et al. (2013) and Zhou et al. (2014) in which replicated tests were conducted in the greenhouse and the results were consistent between tests. In the present study, root inoculation was achieved by pipetting the inoculum directly onto the soil surface in a 10 × 10 cm pot containing on average ~ 8 plants, with a total of 56 plants evaluated for each genotype in three replicated tests. Additionally, the utilization of the double or triple inoculation method minimized escapes and ensured plant infection by the pathogen.
However, 100% disease incidence is a difficult task to accomplish in all the plots (or pots) even with double or triple artificial inoculations in the greenhouse. This was illustrated from Test 1 and Test 3, where one quarters of the seedlings did not display apparent VW symptoms. It is known that successful VW disease infections as indicated by apparent foliar disease symptoms depend on cotton genotype, virulence of the pathogen, inoculation density, and soil temperature, moisture, nutrients and other microorganisms. Even though the three greenhouse studies were conducted using the same VW isolate under the same soil and environmental conditions, the effective inoculum level may be still too low in Test 1 and 3 to ensure 100% disease incidence. However, 99.3% of the seedlings were infected with apparent VW symptoms in Test 2, but the disease incidence still significantly differed among genotypes, indicating a non-random effect on this trait in the study. This was also the case for Test 3 when 74.46% overall disease incidence was observed. Therefore, among the 25% of the plants without apparent VW symptoms in Test 1 and 3, there may be some unknown resistance mechanism in the root that prevented the successful infection of the VW pathogen when inoculum level was not high enough. In our current (results not shown) and previous greenhouse studies (Zhou et al. 2014), the VW incidence is always significantly and positively correlated with the disease severity rating. So the disease incidence may be used as a measure for VW resistance, when the inoculum level is low and unknown, especially in the field conditions (Zhang et al. 2012). However, when 100% plants are infected with apparent symptoms in studies such as Test 2, incidence will be invalid to quantitatively assess germplasm for VW resistance. Although subjective, the disease severity rating takes into consideration disease incidence (from 0 to 100%) and leaf symptoms including defoliation (also from 0 to 100%), it has been and should be the most reliable measure for VW resistance under any conditions.
References
Anamthawat-Jónsson K (2001) Molecular cytogenetics of introgressive hybridization in plants. Methods Cell Sci 23(1–3):139–148
Bolek Y, El-Zik KM, Pepper AE, Bell AA, Magill CW, Thaxton PM, Reddy OUK (2005) Mapping of Verticillium wilt resistance genes in cotton. Plant Sci 168:1581–1590
Carpenter CW (1914) The Verticillium wilt problem. Phytopathology 4:393
Fang H, Zhou H, Sanogo S, Flynn R, Percy RG, Hughs SE (2013) Quantitative trait locus mapping for Verticillium wilt resistance in a backcross inbred line population of cotton (Gossypium hirsutum × Gossypium barbadense) based on RGA-AFLP analysis. Euphytica 194:79–91
Friebertshauser G, DeVay J (1982) Differential effects of the defoliating and nondefoliating pathotypes of Verticillium dahliae upon the growth and development of Gossypium hirsutum. Phytopathology 72:872–877
Huang EB, Verbyla KL, Verbyla A, Raghavan C, Singh V, Guar P, Cavanagh CR (2015) MAGIC populations in crops: current status and future prospects. Theor Appl Genet 128:999–1017
Jenkins JN, McCarty JC, Gutierrez OA, Hayes RW, Jones DC (2013) Registration of RMBUP-C4, a random-mated population with Gossypium barbadense L. alleles introgressed into Upland cotton germplasm. J Plant Regist 7:224–228
Klosterman SJ, Atallah ZK, Vallad GE, Subbarao KV (2009) Diversity, pathogenicity, and management of Verticillium species. Annu Rev Phytopathol 47:39–62
Liu RJ (1995) Effect of vesicular-arbuscular mycorrhizal fungi on Verticillium wilt of cotton. Mycorrhiza 4:293–297
Nannipieri P, Grego S, Ceccanti B (1990) Ecological significance of the biological activity in soil. Soil Biochem 6:293–355
Oakley S (1998) Breeding for resistance to Verticillium wilt and root-knot nematode in California Acalas. In: Proceedings of the Beltwide Cotton Conferences, p 128
Pegg GF, Brady BL (2002) Verticillium wilts. CAB International, Wallingford, p 37
Pullman GS, DeVay JE, Garber RH, Weinhold AR (1981) Soil solarization: effects on Verticillium wilt of cotton and soilborne populations of Verticillium dahliae, Pythium spp., Rhizoctonia solani, and Thielaviopsis basicola. Phytopathology 9:954–959
Shapovalov M, Rudolph BA (1930) Verticillium hadromycosis (wilt) of Cotton in California. Plant Dis Rep 14:2
Stelly DM, Saha S, Raska DA, Jenkins JN, McCarty JC, Gutierrez OA (2005) Registration of 17 Upland (Gossypium hirsutum) cotton germplasm lines disomic for different G. barbadense chromosome or arm substitutions. Crop Sci 45:2663–2665
USDA-ERS (2017a) Cotton and Wool Outlook: August 2017. https://www.ers.usda.gov/publications/pub-details/?pubid=84691. Accessed on 25 July 2018
USDA-ERS (2017b) Cotton and Wool Outlook. http://usda.mannlib.cornell.edu/usda/ers/CWS//2010s/2017/CWS-09-14-2017.pdf. Accessed 25 July 2018
USDA-ERS (2018) Cotton Outlook. https://www.usda.gov/oce/forum/2018/commodities/Cotton.pdf. Accessed on 25 July 2018
USDA-NASS (2018) World Agricultural Production. https://www.nass.usda.gov/Statistics_by_Subject/result.php?B9D5B9F1-2AEC-3A16-B831-36725976844A§or=CROPS&group=FIELD%20CROPS&comm=COTTON. Accessed on 25 July 2018
Wheeler TA, Woodward JE (2016) Field assessment of commercial cotton cultivars for Verticillium wilt resistance and yield. Crop Prot 88:1–6
Wheeler TA, Bordovsky JP, Keeling JW, Mullinix BG Jr (2012) Effects of crop rotation, cultivar, and irrigation and nitrogen rate on Verticillium wilt in cotton. Plant Dis 7:985–989
Wilhelm S, Ferguson J (1953) Soil fumigation against Verticillium albo-atrum. Phytopathology 43:593–596
Wilhelm S, Sagen JE, Tietz H (1974) Resistance to Verticillium wilt in cotton: sources, techniques of identification, inheritance trends, and the resistance potential of multiline cultivars. Phytopathology 64:924–931
Xiao CL, Subbarao KV, Schulbach KF, Koike ST (1998) Effects of crop rotation and irrigation on Verticillium dahliae microsclerotia in soil and wilt in cauliflower. Phytopathology 10:1046–1055
Zhang J, Lu Y, Adragna H, Hughs E (2005) Genetic improvement of New Mexico Acala cotton germplasm and their genetic diversity. Crop Sci 45:2363–2373
Zhang J, Sanogo S, Flynn R, Baral JB, Bajaj S, Percy RG (2012) Germplasm evaluation and transfer of Verticillium wilt resistance from Pima (G. barbadense) to Upland cotton (G. hirsutum). Euphytica 187:147–160
Zhang J, Percy RG, McCarthy JC (2014a) Introgression genetics and breeding between Upland and Pima cotton: a review. Euphytica 198:1–12
Zhang J, Fang H, Zhou H, Sanogo S, Ma Z (2014b) Genetics, breeding, and marker-assisted selection for Verticillium wilt resistance in cotton. Crop Sci 4:1289–1303
Zhou H, Fang H, Sanogo S, Hughs S, Jones D, Zhang J (2014) Evaluation of Verticillium wilt resistance in commercial cultivars and advanced breeding lines of cotton. Euphytica 196:437–448
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Martinez, G., Abdelraheem, A., Darapuneni, M. et al. Evaluation of a multi-parent advanced generation inter-cross (MAGIC) introgressed line population for Verticillium wilt resistance in Upland cotton. Euphytica 214, 197 (2018). https://doi.org/10.1007/s10681-018-2278-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-018-2278-0