Abstract
Fusarium oxysporum f. sp. cubense tropical race 4 is a lethal pathogen, affecting banana production by causing vascular wilt. To date, the molecular basis of resistant cultivars is largely unknown. In this study, suppression subtractive hybridization (SSH) libraries were constructed for the roots of two iso-line cultivars—a Foc-susceptible cultivar and its tolerant mutant—using both uninfected controls and plants artificially challenged with pathogen conidiospores. Unigenes from SSH libraries were used to develop a cDNA array, and then microarray analysis was carried out to verify those genes exhibiting differential expression. A total of 257 unigenes was obtained from the SSH libraries and microarray detection confirmed that most of them were differentially expressed. Of these, 116 unigenes had matched proteins in gene ontology and were classified into 38 different functional groups. The transcript levels of up to 19 putative defense genes were followed for a whole infection period in four cultivars differing in resistance level. At the early stages of infection (within 48 h), nineteen genes varied by a factor of more than three, subsequently only four genes (Pectinesterase inhibitor, PEI; Peroxidase, POD; Pathogenesis-related protein 1, PR1; and Pathogenesis-related protein 3, PR3) maintained their up-regulation. This indicates that anti-oxidation, cell wall modification and synthesis of anti-fungal proteins are the most important aspects to study if we are to understand the Foc resistance mechanism in commercial banana plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bananas, including plantains, are the fourth most important crop in developing countries behind rice, wheat, and corn (Heslop-Harrison and Schwarzacher 2007). The fruit is a staple food in sub-Saharan Africa, South and Central America and much of Asia, while the leaves are used for shelter and wrapping food and the male bud can be eaten as a vegetable (Lescot et al. 2008). Fusarium wilt, also known as Panama disease, currently poses a big challenge to the banana production industry. This disease has destroyed countless hectares of banana in Asia, Africa, Australia and the tropical Americas over several decades (Hwang and Ko 2004). It is therefore ranked as one of the most devastating plant diseases in terms of crop destruction (Carefoot and Sprott 1969). The pathogen is the soil-borne ascomycete, Fusarium oxysporum f. sp. cubense (Foc) (Aoki et al. 2014), of which at least four races have been identified so far. Among these, tropical race 4 (Foc TR4) is highly virulent on almost all banana cultivars; its spores prefer to attach first to the banana root tip, before the growing mycelia directly penetrate across the epidermal cell wall, colonize and occlude the xylem vessels (Li et al. 2011a). The infected plants usually present a reddish-brown discoloration in the rhizome and pseudostem, and the leaves eventually become bright yellow, wilt and collapse around the pseudostem, finally resulting in the death of the entire plant (Hwang and Ko 2004). Because of the survival of the chlamydospore, Foc infection will result in contamination of the soil, which may prevent planting susceptible cultivars for around 30 years (Stover 1990).
In addition to developing diagnostic procedures to inform quarantine strategies (Dita et al. 2010), various efforts have been made to control this disease, including fungicides, soil amendment with urea and rotation with paddy rice, however, none of these have been utilized commercially (Su et al. 1986; Hwang and Ko 2004). In general, the mechanism of Foc resistance is largely unknown. Beckman et al. (1961) reported that functional metaxylem elements in banana roots have cross-walls with minute pores which allow particles less than 2 μm to pass through, but not spores of Foc, suggesting a physical limitation for Foc spread. Physiological studies have revealed that, when exposed to Foc TR4 infection, tolerant cultivars increased lignin deposition and phenolic synthesis in root tissue (De Ascensao and Dubery 2000, 2003; van den Berg et al. 2007). In contrast, the activity of pectin methylesterase (PME) activity was found to be higher in a susceptible cultivar than a tolerant one (Ma et al. 2013), implying that suppressing PME activity and reinforcing cell walls may contribute to resistance. By using the suppression subtractive hybridization (SSH) technique, van den Berg et al. (2007) and Swarupa et al. (2013) found dozens of defense-associated genes. Recently, the transcriptome profiling of tolerant and susceptible Cavendish banana following Foc TR4 inoculation has been investigated and a very large dataset is available (Li et al. 2012, 2013a; Bai et al. 2013). The genome sequence of banana was published (D’hont et al. 2012). These studies annotated several thousand defense-related genes and revealed hundreds of differential expression ones at the early stages of the Foc-banana interaction. Furthermore, dozens of protein with broad-spectrum of stress resistance have been reported to be positive or negative regulated at early stage of Foc TR4 infection (Li et al. 2013b; Lu et al. 2013).
Although hundreds of genes were classified as defense-related, and there is an overview on banana defense response against Fusarium (Swarupa et al. 2014), the defense basis has yet not been elucidated in detail, i.e., which are the key genes and how they contribute to Fusarium wilt resistance remain unknown. In Arabidopsis, tomato and melon, resistance to different Fusarium oxysporum races has been proved to be an oligogenic trait. The best studied examples are tomato-F. oxysporum f. sp. lycopersici (Fol) interaction, in which the nucleotide binding site-leucine-rich repeat (NB-LRR) proteins encoding genes (I, I-2, I-3) recognize corresponding Avr proteins (SIX proteins) from different specific Fol races to activate the defense response (Takken and Rep 2010). In contrast, the resistance to Foc race 1 and race 4 in banana was suggested to be controlled respectively by a single dominant gene and a single recessive gene (Vikili 1965; Ssali et al. 2013), however, those genes have not been isolated, and the downstream factors triggered by R genes are also largely unknown. In addition, unlike self-pollinated and also model species rice whose quantitative trait loci (QTL) for bacterial blight disease resistance has been mapped (Han et al. 2014), the development of QTLs for commercial banana are quite difficulty due to their vegetative propagation and so far no QTL is available for Fusarium wilt resistance in banana cultivars. In a word, we still lack an understanding of the key defense-related factors, such as molecular messengers, enzymes, molecular markers and genes involved in Foc-resistance in banana. Alternatively, genetic closely related genotypes, such as iso-genetic lines, and resistant mutant and its susceptible wild type, could be used to screen those key candidate factors.
In this study, by using PCR-Select cDNA Subtraction, microarray and Real-time PCR technologies, a set of differential expression genes was identified from tolerant and susceptible banana nearly isogenic lines (NILs); in addition, dozens of key defense-related genes were followed for their expression pattern during a whole Foc TR4 infection period in four cultivars with different levels of resistance. Specifically, our goals were (i) to identify the candidate genes significantly involved in the early stages of the banana–Foc TR4 interaction; (ii) to explore several indicator genes (integrators) referring to disease resistance which were consecutively up-regulated in a whole period of Foc TR4 infection. This study would allow progression from simply describing the abundant differential expression of genes to identifying the critical factors.
Materials and methods
Materials
The plant materials used were Musa acuminata cv. Brazilian (AAA), cv. Williams-8818 (AAA), cv. Williams-8818-1 (AAA) and Musa acuminata x Musa balbisiana cv. Goldfinger (AAAB): the former two are popular commercial cultivars in China but highly susceptible to Foc TR4, while the latter two are highly tolerant. Williams-8818-1 is an NIL of Williams-8818, bred by ethyl methane sulfonate (EMS)-mediated somaclonal variation (Li et al. 2011b). The pathogen was Fusarium oxysporum f. sp. cubense tropical race 4, collected from an infested field in Xuwen city, Guangdong province, China (GPS: N20°18′6.2″, E101°18′55.3″). To confirm that the pathogen is a Foc TR4, 28S–18S ribosomal RNA intergenic spacer (IGS) was PCR amplified with specific primer set Foc TR4-F/Foc TR4-R (Dita et al. 2010) and sequenced for BLAST and phylogenic analysis.
Treatment
The propagation and inoculation of plantlets were conducted as previously described (Li et al. 2011b). To evaluate the level of disease resistance, plantlets were examined 20–35 days post inoculation (dpi), depending on when they died, but no later than 35 days. For gene expression analysis, roots were sampled separately at 0, 6, 24, 48, 72, 96, 120 144 h, and 8, 12, 16, 20 days post inoculation (hpi/dpi), immediately frozen in liquid nitrogen, and then kept at −80 °C until use, with each sample consisting of eight plantlets.
Evaluation of the level of disease resistance
Fusarium wilt severity was evaluated and scored based on external and internal symptoms. Externally, 0 = no symptoms; 1 = initial yellowing, mainly in the lower leaves; 2 = yellowing of all the lower leaves with some discoloration of younger leaves; 3 = all leaves with intense yellowing; 4 = plant dead. Internally, 0 = 0 %; 1 = 1–15 %; 2 = 16–30 %; 3 = 31–45 %; 4 = 46–60 %; 5 = over 60 % of the rhizome discolored, estimated visually in longitudinal sections of the corm. Thirty plantlets were evaluated for each cultivar and the final score for external and internal symptoms was expressed as the sum of individual scores/30.
Construction of SSH libraries
The NILs of cv. Williams-8818 and cv. Williams-8818-1 were used to construct the libraries. Four groups of root samples (consisted by eight plantlets for each sample) were used: group 1, Williams-8818 harvested at 0 hpi; group 2, Williams-8818-1 harvested at 0 hpi; group 3, a mixture of equal volumes of plant tissue of Williams-8818 harvested at 6, 24 and 48 hpi; and group 4, a mixture of equal amounts of Williams-8818-1 harvested at 6, 24 and 48 hpi. From this material, a PCR-Select™ cDNA Subtraction Kit (Clontech) was used, following the manufacturer’s instructions, to construct four SSH libraries: SSH library 1, constructed with group 2 as the tester and group 1 as the driver, likewise; library 2, with group 1 as the tester and group 2 as the driver; library 3, with group 4 as the tester and group 3 as the driver; and library 4, with group 3 as the tester and group 4 as the driver. The fragments enriched in each library were inserted into pMD-18 Vector (Takara, Japan) and transformed into Escherichia coli DH5a cells.
Screening, sequencing and annotation analysis of clones in SSH libraries
In order to select those clones harboring a single recombinant plasmid with an insertion cDNA >190 bp (including primers), around 1 500 clones were randomly selected for each library and confirmed by PCR using nested PCR primers provided in the Kit. The appropriate clones (insertion >190 bp) were subjected to plasmid extraction, followed by sequencing of the inserted cDNA fragments using an ABI PRISM™ 3730 DNA Sequencer (Perkin Elmer Applied Biosystems, Foster City, CA, USA). Alignments were performed using software DNAStar Lasergene 7.1 (DNASTAR, Madison, USA) to remove the redundant sequences and identify the unigenes. To annotate, classify, and functionally map those unigenes, we used BLASTx to match the sequences using a cut-off E-value of 10−6, including the non-redundant protein database (NR, NCBI), and gene ontology (GO) database (http://geneontology.org/).
Preparation of cDNA microarray slides
In total, 257 unigenes were confirmed. Their cDNA inserts were amplified by PCR using nested PCR primers provided in the PCR Select-cDNA Subtraction Kit. The PCR products were precipitated once more with the addition of 100 μl of anhydrous ethanol and resuspended in 15 μl of 50 % dimethylsulphoxide (DMSO) at a final concentration of 0.1–0.5 μg μl−1 and then spotted onto amino silaned glass slides (CapitalBio. Corp, Beijing, China) with a SmartArrayer™ microarrayer (CapitalBio Corp.). Each clone was printed in triplicate. Four housekeeping genes (GenBank accession numbers AB022041, AF285176, AY904067 and DQ885479) from banana were used as internal controls. Eight sequences derived from intergenic regions in the yeast genome, showing no significant homology to any of the existing sequences in GenBank, were spotted multiple times onto the microarray to act as exogenous controls. Total banana RNA was spiked with a mixture of these exogenous control RNAs to validate the semi-quantitative microarray result.
Preparation of fluorescent dye-labeled cDNA and hybridization
The gene expression profiles in all four groups were investigated by microarray analysis. An aliquot of 5 μg total RNA was used to produce Cy5/Cy3-labeled cDNA employing an RNA amplification combined with Klenow enzyme labeling strategy according to Guo’s protocol (Guo et al. 2005). Cy5/Cy3-labelled cDNA was hybridized with the microarray at 42 °C overnight. Following hybridization, the arrays were washed with 0.2 % SDS, 2× SSC at 42 °C for 5 min, and 0.2 % SSC for 5 min at room temperature.
Microarray data analysis
Arrays were scanned with a confocal laser scanner, LuxScan 3.0 (CapitalBio Corp.), and the resulting images were analyzed with SpotData Pro 2.0 software (CapitalBio Corp.). Spots with fewer than 50 % of the signal pixels exceeding the local background value for both channels (Cy3 and Cy5) plus two standard deviations of the local background were removed. A spatial and intensity-dependent (LOWESS) normalization method was employed to normalize the ratio values (Yang et al. 2002). Normalized ratio data were then log transformed. cDNA spots with less than two out of a total of three data points in each replicated hybridization were removed. Differentially expressed genes were identified using a t test and multiple test corrections were performed using false discovery rate (FDR) (Benjamini and Hochberg, 1995). Genes with FDR <0.05 and a change ≥1.5-fold were considered to be differentially expressed genes.
Quantitative real-time PCR analysis
To confirm the result of microarray assay and to gain an insight into the details of expression patterns exhibited by defense-related genes at a large scale, the expression level of 16 key-genes with putative defense-related features (based on the results of BLAST and GO analysis) and three more well-known genes (Pathogenesis-related protein 1, PR-1; Pathogenesis-related protein 3, PR-3 and a peroxidase, PIR7A) were confirmed by real-time PCR. β-Actin (Genbank accession AB022041) was used as internal reference. Primer sequences and putative function of each gene are listed in Table 1. Root samples from four genotypes were examined from inoculation day to the death of the susceptible genotype, i.e., samples collected at 0, 6, 24, 48, 72, 96, 120 144 hpi and 8, 12, 16, 20 dpi. Equal amount of root tissues from three plantlets of the same genotype collected at the same time point were pooled together and used as a repeat. Three repeats made a treatment. cDNA was synthesized using the SMART PCR cDNA Synthesis Kit (Clontech, San Francisco, USA). The PCR reaction was performed in three technical repeats with SYBR Green PCR Master Mix (TAKARA, Tokyo, Japan) using an iQ5 multicolor real-time PCR Detection System (BioRad, California, USA). Efficiency of each pair of primers was determined based on its standard curve obtained from a series of 4-fold diluted template cDNAs. The difference between the cycle threshold (Ct) of target gene and the Ct of β-Actin (△Ct = Ct target gene-Ct β-Actin) was used to obtain the normalized expression of target gene, which corresponds to \(2^{{ - \Delta {\text{C}}_{t} }}\). This value was finally expressed as fold change relative to a ‘calibrator’, which is the expression level for the particular transcript in control Williams-8818 roots at 0 h post innoculation (hpi).
Results
Disease symptoms exhibited by different cultivars
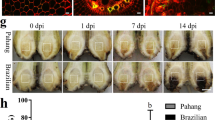
To gain an insight into the disease resistance levels of each genotype, the symptoms of the four cultivars responding to Foc TR4 colonization were described and compared. The well-known tolerant and susceptible commercial bananas, cv. Goldfinger and cv. Brazilian, were used as standard cultivars to monitor inoculation effects. The IGS nucleotide sequence amplified from the pathogen (GenBank accession KM108312) was nearly identical (>99.5 %) to that of Foc TR4 strain NRRL36114, and both BLAST and phylogeny analysis suggested that it is a Foc TR4 (data not shown). This pathogen could efficiently induce typical symptoms of Fusarium wilt in this study. Externally, Goldfinger had only a few yellowed lower leaves at 35 dpi which was similar to the tolerant line, Williams-8818-1. In contrast, all the leaves of the susceptible line, Brazilian and Williams-8818, were extensively yellowed at around 20 dpi and subsequently the whole plantlet died within 35 dpi. Internally, rhizomes of all four cultivars showed severe discoloration, with tissues in a longitudinal section mostly dead and blackened within 35 dpi; the order of severity was Brazilian, Williams-8818, Williams-8818-1 and Goldfinger (Table 2; Fig. 1).
The construction of SSH libraries and transcript profiles with Foc TR4 infection
Forward and reverse subtractions were conducted between lines Williams-8818-1 and cv. Williams-8818 using root material, i.e. roots harvested at 0 hpi and root mixtures from 6, 24, 48 hpi were used separately. Randomly, about 1500 clones were selected for PCR amplification of the insertions from each SSH library, of which only those longer than 190 bps were sequenced. For SSH libraries 1, 2, 3 and 4, 303, 287, 262 and 319 sequences were cloned, respectively and 153, 84, 106 and 118 unigenes were obtained comprising a total of 257 unigenes (GenBank accession numbers: JZ482667 to JZ482923) with an average of 218 bps, ranging from 78 to 492 bps. Of the 257 unigenes, 58 cDNAs were singletons, the other 199 cDNAs belonged to divergent clusters, and each cluster contained 2–8 sequences. This result suggested the number of key genes with differential expression may not be as large as we expected.
All 257 unigenes were used for blast searching using BLASTx (e-value < 0.000001) against the NCBI and banana non-redundant (nr) protein database. The former search indicated that 116 unigenes were similar to known proteins from Arabidopsis thaliana, rice, maize, poplar, and grape, with dozen of them involved in defense responses (Supplementary Table S1); while the latter matched 100 unigenes to many kinds of proteins (data not shown). Functional annotation by gene ontology conducted via Blast2GO resulted in 116 unigenes mapping to 431 GO terms (each mapping to 1–12), which were further grouped into 38 functional categories (Fig. 2 and Supplementary Table S1). Among them, genes assigned to the categories cells, cell parts, organelles, binding, catalytic, cellular processes and metabolic processes occupied the seven largest groups, suggesting the involvement of these key activities in a living cell. Three groups were defined as defense-related genes, i.e., two small groups of antioxidants (5 genes) and immune system process (6 genes), and a moderately large group of response to stimuli (39 genes), indicating the onset of many defense procedures following Foc TR4 infection.
In order to confirm the differential expression of each gene, all 257 unigenes were amplified and then used for microarray analysis. RNA samples were the same as those used to construct the SSH libraries, i.e., the RNA from group 1 (Williams-8818, 0 hpi), group 2 (Williams-8818-1, 0 hpi), group 3 (Williams-8818’ mixture, 6, 24 and 48 hpi), and group 4 (Williams-8818-1’ mixture, 6, 24 and 48 hpi) compared to group 1, respectively. The cDNAs with FDR <0.05 and a change ≥1.5-fold were considered to be differentially expressed genes. Totally, there were 74 unigenes exhibited differential expression, comprising 0, 10, 26, 23 up-regulated and 9, 24, 16, 18 down-regulated cDNAs from groups 1–4, respectively (Table 3). It should be pointed out that group 1 did not has differentially expressed cDNA when the proportional change was set to ≥2-fold (data not shown), suggesting that the microarray assay was of good quality, which in turn confirmed that the SSH libraries were robust. In addition, most of the genes have the same trends of up- or down-regulation in both susceptible and tolerant genotypes after Foc TR4 infection, implying that the perception of Foc TR4 by different banana genotypes is somewhat the same (Table 3).
Of the unigenes showed significant differential expression, about half members referred to the known proteins with the majority involved in stress response. The largest group belongs to antifungal and antitoxin genes, including homologues of PR proteins, disease resistance response protein, germin-like protein, non-symbiotic hemoglobin, nadp-dependent malic enzyme, alcohol dehydrogenase and so on. The second group was antioxidant-like genes, such as homologues of catalase, isoflavone reductase, metallothionein-like protein, glutathione s-transferase. Genes related to cell wall metabolism were also observed, i.e., peroxidase, pectinesterase inhibitor, lipoxygenase, etc. (Table 3). It was noteworthy that up to half of the differential expression uingenes were unknown proteins, thus the potential important roles of them should be further characterized in future.
Quantitative real-time PCR analysis of nineteen defense-related genes
To confirm the result of microarray assay and to gain a larger-scale insight into transcript profiling of defense-related genes in banana roots in a whole Foc TR4 infection period, sixteen key defense-related genes enriched in SSH libraries, as well as three already well-known genes, were tracked for their expression levels over a whole infection period, i.e., from inoculation to death of the susceptible genotype cv. Brazilian (20 days in this experiment). The real-time PCR results showed that most of the genes exhibited limited change at the early stages of infection, and a few members exhibited significant changes at the late stages. Compared to the expression level of cv. Williams-8818 at 0 hpi, there were just 12 genes that exhibited 3-fold up-regulation in each genotype at certain time points. When these 12 genes were further compared to 0 hpi for the same genotypes, only four of them (Pectinesterase inhibitor, PEI; Peroxidase, POD; PR1 and PR3) exceeded this threshold (Supplementary Table S2), with PEI, PR1 and PR3 kept generally in up-regulation during the whole infection period (Fig. 3). The upregulated expressions of PR10 and PEI were in lines with the results of microarray assay. The significant changes of transcripts levels of many well-known defense-related genes in certain time points during Foc infection period suggested that they were activated in commercial bananas under stress.
Discussion
In this study, SSH and microarray approaches were used to enrich and identify disease tolerance-related genes that have functional significance. Genes enriched in the SSH libraries belong to a total of 38 functional groups, indicating that the whole plant launched diverse responses to counter the infection (Fig. 2 and Supplementary Table S1). Among them were potentially defense-related genes including genes for antioxidant production, immune system process and responses to stimuli (Li et al. 2012; Bai et al. 2013). Seventy-four unique genes were identified to be significantly expressed changes by microarray analysis (Table 3). The expression profiles of dozens of key defense-related genes were further examined using real-time PCR analysis over the whole infection period (Supplementary Table S2).
The SSH approach and sampling times
The disease severity assessment showed that the external symptoms were quite different between tolerant cultivars and susceptible ones, but all genotypes were extensively death in rhizome (Table 2; Fig. 1), indicating that disease resistance in tolerant genotypes may mainly occur during the later stages of infection. However, Li et al. (2011a, 2012) and Li et al. (2013a) reported that Foc TR4 has exhibited more activities in susceptible cultivars relative to in resistance ones during early stages of infection (48–96 hpi), implying that those early responding genes may play more important roles in mediating resistance. To be in line with previous studies, we also sampled at early stages for SSH and microarray assays. As NILs, the genetic differences between cv. Williams-8818-1 and cv. Williams-8818 are limited and thus particularly suitable for SSH analysis. When the four libraries were merged together, there was a total of only 257 unigenes, suggesting that two NILs have many identical genes with similar expression levels or patterns, which in turn proved the robust of SSH for gene screening from NILs. Of the 16 genes from SSH libraries subjected to real-time PCR assay, fourteen showed expression change ≥3-fold at a certain time point during the whole infection period, while eight exhibited this change at early stages (within 48 hpi), suggesting that more than half genes with differential expression could be enriched from a nearly-isogenic background during the early infection stages by SSH (Supplementary Table S2). It should be note that this hypothesis cannot be tested using these data, because of only 16 genes were examined and the validity of a 3-fold difference threshold still needs to be confirmed.
Although only 116 unigenes could be annotated and 431 GO terms were matched, they included 38 functional categories. By contrast, transcriptome sequencing produced thousands of unigenes but just 45 and 35 GO categories were assigned in previous studies (Li et al. 2012; Wang et al. 2012). Our data suggested that the differentially expressed genes are relatively limited in number but diverse in functions. Many genes enriching in antioxidants, immune system process and response to stimuli indicated the onset of various defense processes following Foc TR4 infection. Li et al. (2012) and Bai et al. (2013) also postulated that high levels of defense-related transcripts contributed to Foc TR4 resistance.
The role of ROS during the early stage of banana–Foc TR4 interaction
Fusarium oxysporum is a necrotrophic pathogen that can produce hydrolytic enzymes and induce the production of reactive oxygen species (ROS) as part of the typical events during the early stages of the pathogen–plant interaction (Berrocal-Lobo and Molina 2007). Fox elicitor treatment in Arabidopsis cell suspension cultures induced ROS production, and this depended on the activation of peroxidases and NADPH oxidases (Bae et al. 2006; Bindschedler et al. 2006; Davies et al. 2006). In the case of banana, Li et al. (2011b) revealed that rapid accumulation of O2·− and H2O2, and activation of SOD and CAT enzymes in roots occur during the early stages of Foc TR4 infection. Recently, large scale gene expression profiling has been undertaken for banana roots infected with Foc TR4, and it has been found that NADPH (Li et al. 2012), the respiratory burst oxidase homolog (Rboh) (Bai et al. 2013), peroxidase homologues (Wang et al. 2012) and genes encoding germin-like protein (Li et al. 2013a) are strongly induced. In this study, four catalase, several peroxidase and metallothionein 3-like protein homologues were enriched in the SSH libraries, as well as many antioxidant-related genes (Fig. 2 and Supplementary Table S1). Moreover, one catalase and two peroxidase unigenes tested by real-time PCR demonstrated significant changes in at least one of the cultivars tested (Supplementary Table S2). Therefore, it seems that ROS production was frequently observed in banana roots responding to Foc TR4 infection (Swarupa et al. 2014), but if and how ROS activation contributes to disease resistance requires further investigation.
Genes related to cell wall modification and stress response
Many studies report increases in cell wall-strengthening substances and enzyme activities, such as lignin, callous tissue, phenolic compounds, PAL, POD and PPO, following Foc TR4 inoculation (De Ascensao and Dubery 2000, 2003; van den Berg et al. 2007), suggesting that the reinforcement of the cell wall may provide additional physical barriers and thus delay and localize the advance of pathogens. In our study, genes related to cell wall metabolism and stress response were greatly enriched, including homologues of polyamine oxidase, isoflavone reductase, lipoxygenase, pectinesterase inhibitor, glutathione s-transferase, glyceraldehyde-3-phosphate dehydrogenase, NADP-dependent malic enzyme, non-symbiotic hemoglobin, alcohol dehydrogenase 1, elongation factor 1-alpha, PR3, PR1 and germin-like protein (Supplementary Table S1). Besides playing roles in ROS production, catalase and peroxidase, together with polyamine oxidase, isoflavone reductase, lipoxygenase and pectinesterase inhibitor, are involved in plant cell wall fortification (Brisson et al. 1994). Overexpression of PME inhibitor (PMEI) could control the methyl esterification status of polysaccharide Homogalacturonan (HG) by reducing the activity or transcripts of PME, resulting in promoting resistance of plants to pathogens (Lionetti et al. 2007; Pelloux et al. 2007; An et al. 2008; Wolf et al. 2009; Volpi et al. 2011). In this study, the level of PEI (a homolog of PMEI) transcripts varied greatly in each genotype during the whole infection period examined, implying a role of hinding cell wall degradation, which was in line with the results of Ma et al. (2013).
PR1, PR3 and PR10 identified in this study are defense response proteins. The synthesis of PR proteins is one prominent feature of plant defense responses to biotic stress. A large number of PR proteins have been characterized and classified into 17 functional families based on their primary structure, serological relationships and biological activities (van Loon et al. 2006). PR1, PR3 and PR10 are cell wall-degrading enzymes, which can digest the major components of chitin or glucan in fungi cell walls, and therefore inhibit fungal growth. Subramaniam et al. (2006) reported that overexpression of PR1 in transgenic banana conferred some Foc-1 resistance. We found, in line with this, that the PR1 transcript level increased in waves in each genotype as time passed.
Conclusion
Compared with previous studies, two results are particularly noteworthy. The first is that most of the 19 defense-related genes exhibited their first significant change within 72 h of Foc TR 4 inoculation in all genotypes, regardless of their level of resistance. The second was that the expression of PEI, PR1 and PR3 generally remained up-regulated after inoculation, suggesting that the key times for the onset of banana–Foc TR4 interaction may be within 72 h, and cell wall-modified and anti-fungal genes play important roles in the whole process. In addition, the hundreds of differentially expressed genes identified by SSH screening, with dozens of them confirmed by microarray assay, are an important resource for the identification of key genes related to plant resistance in a non-model organism, namely commercial banana. This study advances the current understanding of a non-model organism’s resistance to F. oxysporum, highlights the characteristics of key defense-related genes in banana and should provide valuable guidance for disease-resistance breeding programs.
References
An SH, Sohn KH, Choi HW, Hwang IS, Lee SC, Hwang BK (2008) Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 228:61–78
Aoki T, O’Donnell K, Geiser DM (2014) Systematics of key phytopathogenic Fusarium species: current status and future challenges. J Gen Plant Pathol 80:189–201
Bae H, Kim MS, Sicher RC, Bae HJ, Bailey BA (2006) Necrosis- and ethylene-inducing peptide from Fusarium oxysporum induces a complex cascade of transcripts associated with signal transduction and cell death in Arabidopsis. Plant Physiol 141:1056–1067
Bai TT, Xie WB, Zhou PP, Wu ZL, Xiao WC, Zhou L, Sun J, Ruan XL, Li HP (2013) Transcriptome and expression profile analysis of highly resistant and susceptible banana roots challenged with Fusarium oxysporum f. sp. cubense tropical race 4. PLoS One 8:e73945. doi:10.1371/journal.pone.0073945
Beckman CH, Mace ME, Halmos S (1961) Physical barriers associated with resistance in Fusarium wilt of bananas. Phytopathology 51:507–515
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300
Berrocal-Lobo M, Molina A (2007) Arabidopsis defense response against Fusarium oxysporum. Trends Plant Sci 13:145–150
Bindschedler LV, Dewdney J, Blee KA, Stone JM, Asai T, Plotnikov J, Denoux C, Hayes T, Gerrish C, Davies DR, Ausubel FM, Bolwell GP (2006) Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J 47:851–863
Brisson LF, Tenhaken R, Lamb C (1994) Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell 6:1703–1712
Carefoot GL, Sprott ER (1969) Famine on the Wind. Angus and Robertson, London, p 222
Davies DR, Bindschedler LV, Strickland TS, Bolwell GP (2006) Production of reactive oxygen species in Arabidopsis thaliana cell suspension cultures in response to an elicitor from Fusarium oxysporum: implications for basal resistance. J Exp Bot 57:1817–1827
De Ascensao C, Dubery IA (2000) Panama disease: cell wall reinforcement in banana roots in response to elicitors from Fusarium oxysporum f. sp. cubense race four. Phytopathology 90:1173–1180
De Ascensao C, Dubery IA (2003) Soluble and wall-bound phenolics and phenolic polymers in Musa acuminata roots exposed to elicitors from Fusarium oxysporum f. sp. cubense. Phytochemistry 63:679–686
D’hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, Da Silva C, Jabbari K, Cardi C, Poulain J, Souquet M, Labadie K, Jourda C, Lengelle J, Rodier-Goud M, Alberti A, Bernard M, Correa M, Ayyampalayam S, McKain MR, Leebens-Mack J, Burgess D, Freeling M, Mbeguie A, Mbeguie D, Chabannes M, Wicker T, Panaud O, Barbosa J, Hribova E, Heslop-Harrison P, Habas R, Rivallan R, Francois P, Poiron C, Kilian A, Burthia D, Jenny C, Bakry F, Brown S, Guignon V, Kema G, Dita M, Waalwijk C, Joseph S, Dievart A, Jaillon O, Leclercq J, Argout X, Lyons E, Almeida A, Jeridi M, Dolezel J, Roux N, Risterucci AM, Weissenbach J, Ruiz M, Glaszmann JC, Quetier F, Yahiaoui N, Wincker P (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488:213–217
Dita MA, Waalwijk C, Buddenhagen IW, Souza MT Jr, Kemab GHJ (2010) A molecular diagnostic for tropical race 4 of the banana Fusarium wilt pathogen. Plant Pathol 59:348–357
Guo Y, Guo HY, Zhang L, Xie HY, Zhao X, Wang FX, Ma SL, Tao JP (2005) Genomic analysis of antihepatitis B virus (HBV) activity by small interfering RNA and lamivudine in stable HBV-producing cells. J Virol 79:14392–14403
Han XY, Yang Y, Wang XM, Zhou J, Zhang WH, Yu CL, Cheng C, Cheng Y, Yan CQ, Chen JP (2014) Quantitative trait loci mapping for bacterial blight resistance in rice using bulked segregant analysis. Int J Mol Sci 15:11847–11861
Heslop-Harrison JS, Schwarzacher T (2007) Domestication, genomics and the future for banana. Ann Bot 100:1073–1084
Hwang SC, Ko WH (2004) Cavendish banana cultivars resistant to Fusarium wilt acquired through somaclonal variation in Taiwan. Plant Dis 88:580–588
Lescot M, Piffanelli P, Ciampi AY, Ruiz M, Blanc G, Leebens-Mack J, da Silva FR, Santos CMR, D’Hont A, Garsmeur O, Vilarinhos AD, Kanamori H, Ronning CM, Matsumoto T, Cheung F, Haas BJ, Althoff R, Arbogast T, Hine E, Pappas GJ Jr, Sasaki T, Souza MT Jr, Miller RNG, Glaszmann JC, Town CD (2008) Insights into the Musa genome: syntenic relationships to rice and between Musa species. BMC Genom 9:58
Li CY, Chen S, Zuo CW, Sun QM, YeQ Huang BZ (2011a) The use of GFP-transformed isolates to study infection of banana with Fusarium oxysporum f. sp. cubense race 4. Eur J Plant Pathol 131:327–340
Li WM, Qian CM, Mo YW, Hu YL, Xie JH (2011b) Tolerance of banana for fusarium wilt is associated with early H2O2 accumulation in the roots. Afr J Biotechnol 10:11378–11387
Li CY, Deng GM, Yang J, Viljoen A, Jin Y, Kuang RB, Zuo CW, Lv ZC, Yang QS, Sheng O, Wei YR, Hu CH, Dong T, Yi GJ (2012) Transcriptome profiling of resistant and susceptble cavendish banana roots following inoculation with fusarium oxysporum f. sp. cubense tropical race 4. BMC Genom 13:374
Li CQ, Shao JF, Wang YJ, Li WB, Guo DJ, Yan B, Xia YJ, Peng M (2013a) Analysis of banana transcriptome and global gene expression profiles in banana roots in response to infection by race 1 and tropical race 4 of Fusarium oxysporum f. sp. cubense. BMC Genom 14:851
Li XS, Bai TT, Li YF, Ruan XL, Li HP (2013b) Proteomic analysis of Fusarium oxysporum f. sp. cubense tropical race 4-inoculated response to Fusarium wilts in the banana root cells. Proteome Sci 11:41
Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 143:1871–1880
Lu Y, Liao DJ, Pu JJ, Qi YX, Xie YX (2013) Proteome analysis of resistant and susceptible Cavendish banana roots following inoculation with Fusarium oxysporum f. sp. cubense. Physiol Mol Plant P 84:163–171
Ma L, Jiang S, Lin G, Cai JH, Ye XX, Chen HB, Li MH, Li HP, Takac T, Samaj J, Xu CX (2013) Wound-induced pectin methylesterases enhance banana (Musa spp. AAA) susceptibility to Fusarium oxysporum f. sp. cubense. J Exp Bot 64:2219–2229
Pelloux J, Rustérucci C, Mellerowicz EJ (2007) New insight into pectin methylesterase structure and function. Trends Plant Sci 12:267–277
Ssali RT, Kiggundu A, Lorenzen J, Karamura E, Tushemereirwe W, Viljoen A (2013) Inheritance of resistance to Fusarium oxysporum f. sp. cubense race 1 in bananas. Euphytica 194:425–430
Stover RH (1990) Fusarium wilt of banana: some history and current status of the disease. In: Ploetz RC (ed) Fusarium wilt of banana. APS Press, St Paul, pp 1–7
Su HJ, Hwang SC, Ko WH (1986) Fusarial wilt of Cavendish bananas in Taiwan. Plant Dis 70:814–818
Subramaniam S, Maziah M, Sariah M, Puad MP, Xavier R (2006) Bioassay method for testing Fusarium wilt disease tolerance in transgenic banana. Sci Hortic 108:378–389
Swarupa V, Ravishankar KV, Rekha A (2013) Characterization of tolerance to Fusarium oxysporum f. sp. cubense infection in banana using suppression subtractive hybridization and gene expression analysis. Physiol Mol Plant P 3:1–7
Swarupa V, Ravishankar KV, Rekha A (2014) Plant defense response against Fusarium oxysporum and strategies to develop tolerant genotypes in banana. Planta 239:735–751
Takken F, Rep M (2010) The arms race between tomato and Fusarium oxysporum. Mol Plant Pathol 11:309–314
van den Berg N, Berger DK, Hein I, Birch PR, Wingfield MJ, Viljoen A (2007) Tolerance in banana to Fusarium wilt is associated with early up-regulation of cell wall-strengthening genes in the roots. Mol Plant Pathol 8:333–341
van Loon LC, Rep M, Pieterse CM (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162
Vikili NG (1965) Fusarium wilt resistance in seedlings and mature plants of Musa species. Phytopathology 55:135–140
Volpi C, Janni M, Lionetti V, Bellincampi D, Favaron F, D’Ovidio R (2011) The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol Plant Microbe Interact 24:1012–1019
Wang Z, Zhang JB, Jia CH, Liu JH, Li YQ (2012) De novo characterization of the banana root transcriptome and analysis of gene expression under Fusarium oxysporum f. sp. cubense tropical race 4 infection. BMC Genom 13:650
Wolf S, Mouille G, Pelloux J (2009) Homogalacturonan methyl esterification and plant development. Mol Plant 2:851–860
Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30:e15
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (No. 31101535, 31261140366), Hainan Provincial Natural Science Foundation of China (313056), the Earmarked Fund for Modern Agro-industry Technology Research System (nycytx-33-20) and the Chinese Special Fund of Basic Scientific Research Project for State Level and Public Welfare-scientific Research Institutes (1251022011002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We’d really like to thank Dr. Joelle Hoggan for helping us polish the manuscript.
Conflict of interest
The authors declare that they have no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, W., Ge, X., Wu, W. et al. Identification of defense-related genes in banana roots infected by Fusarium oxysporum f. sp. cubense tropical race 4. Euphytica 205, 837–849 (2015). https://doi.org/10.1007/s10681-015-1418-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-015-1418-z