Abstract
The aim of this study is to propose a method for constructing Artificial Neural Network (ANN) models and evaluating their performance based on the application of two methods for the selection of the ANN topology: the dynamic division method (cross-validation or dynamics-split) (DDM) and the static-split method (SSM). The two methods are compared and applied to predict the amount of organic matter in an up-flow anaerobic sludge blanket (UASB) reactor operated at full scale. The performance of the ANN models was assessed through the coefficient of multiple determination (R 2), the adjusted coefficient of multiple determination (\(R^{2}_{adj}\)), and the root mean square error (RMSE). The comparison reveals that the DDM accurately selects the best model and reliably assesses its quality.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Anaerobic digestion is a complex biochemical process carried out by microorganisms belonging to the Archaea and Bacteria domains that work interactively to promote stable and self-regulating fermentation. The application of anaerobic digestion to treat waste and wastewater conventionally converts organic matter into a mixture of methane, carbon dioxide, hydrogen, and biomass by specific metabolic processes occurring in sequential stages.
The up-flow anaerobic sludge blanket (UASB) reactor, developed in the 1970s by Lettinga and co-workers in the Netherlands [63], is one of the most popular anaerobic systems used in the treatment of various types of wastewater [28, 30–33]. The application of a UASB reactor for domestic wastewater treatment has several benefits, especially when applied in warm climates, as is the case for most Brazilian municipalities. In these conditions, there are multiple benefits, such as high application rates of organic loads, short hydraulic retention times, and low energy demands. These positive aspects therefore characterize a compact system with low operation costs, and low sludge production. Despite the advantages, UASB reactors have a limited capacity to convert organic matter and thus require a post-treatment step to satisfy the requirements of Brazilian environmental legislation for treated effluent [10].
Anaerobic digestion in UASB reactors is very susceptible to fluctuations in process inputs (organic load, flow, temperature, and presence of inhibitors) in the environment of the system (decreasing in pH value, alkalinity consumption, and accumulation of volatile fatty acids) and output (composition, biogas production, removal of organic matter, which is usually measured as biological oxygen demand (BOD), total organic carbon (TOC), and chemical oxygen demand (COD). Therefore, to better understand the process and achieve higher pollutant removal, a UASB reactor must be continuously monitored and controlled by means of costly laboratory analyses.
Mathematical models associated with an analysis of the response and the stability or instability of the system are powerful tools for the elucidation and determination of the behavior of transient phenomena. They are used to represent the main aspects of any system, improving its understanding, the formulation and validation of some hypotheses, the prediction of the system’s behavior under different conditions, and finally, reducing the experimental information, costs, risk, and time [14]. In order to achieve better performances in anaerobic digestion in UASB reactors, several investigations have been made in the fields of phenomenological modeling [11, 27, 35–37, 49, 52]. Among all the existing models, the Anaerobic Digestion Model No.1 (ADM1) developed by the IWA Task Group for Mathematical Modelling of Anaerobic Digestion Processes was designed to reach a common basis for anaerobic digestion model development and validation studies [2]. However, its large number of parameters (86 parameters) and unmeasurable states (especially a large number of types of biomasses) hinder its use in the development of control applications [22]. These quantities vary widely depending on the type of process and the operational and environmental conditions. The kinetics involved in the anaerobic process are extremely complex due to the intricate interaction network formed from a mixture of complex substrates and the various microbial populations involved. Statistical models are typically used to model a system when the system is not so complicated, but when the complexity of a system increases, other methods are required. In such cases, artificial neural networks have been successfully used. Furthermore, when the behavior has to consider linguistic rules, a fuzzy method can be a reliable alternative. Recent research has considered uncertainties in Artificial Neural Network (ANN) models such as those reported by Srivastav et al. [57] and Shrestha et al. [56].
ANN has been investigated and successfully applied to identification and control problems over the last twenty years. In predicting organic matter systems in particular, ANN have been used to help the control engineer to determine the required loading capacity in the subsequent treatment units [3, 12, 13, 20, 23, 25, 26, 29, 62, 64, 66, 68]. The main advantages of ANN are that they require no knowledge of the reaction mechanisms and experimental measurements for a multitude of parameters to monitor the operating conditions and performance of an anaerobic treatment process (UASB) at a large scale facility. However, artificial neural network models have also been criticized for a lack of dependence upon physical relationships and a poor capacity for extrapolation. Thus, ANNs can be applied, for example, in the prediction of organic matter in cold climate regions because the model is built from observations of input-output in the desired process.
Hybridization of ANN and fuzzy has been implemented to increase the capability of Fuzzy and ANN. Fuzzy-logic [8, 61], adaptive neuro-fuzzy inference systems [16, 45] are some methods proposed in the literature. No application in bioprocesses has been found in the literature.
The use of ANN modeling involves two main steps: fitting the parameters with the measured data and selecting the best model. The parameter fitting step uses an adjustment strategy where the capabilities of the network are improved by modifying all the weights. The selection of a network structure with optimal choices of the free design parameters, such as the number of hidden layers, the number of hidden nodes, the activation function, and the input representation, are crucial for the performance of the network in application [53]. For example, a network with too few hidden nodes cannot represent the system satisfactorily and a network with too many hidden nodes overlearns the particular runs presented in the training set instead of learning the underlying dynamics of the process.
A variety of methods can be used to decide how to choose the best model or best set of models [34, 48]. A common way is the static-split method (SSM) which evaluates the network performance on the training data itself by means of statistical methods. In SSM, the available data are divided into three sets; training, validation and testing. The training set is used to adjust the connection weights. The validation set is used to check the performance of the network at various stages of learning, and training is stopped once the error in the validation set increases. The testing set is used to evaluate the performance of the model once training has been successfully accomplished. However, the main limitation in the application of the SSM method is the need for large amounts for data for training, validation, and testing, especially in systems where measured data are limited, expensive, or hard to collect. Very small training and validation sets lead to unsatisfactory training of the network, and a very small test set cannot accurately evaluate the performance of the model [53].
A promising alternative to conventional SSM is cross-validation, or the dynamic division method (DDM), also called cross-validation or dynamics-split [53, 59, 65]. DDM consists of a cyclic allocation of the available data to the training and the test set, which allows the use of an arbitrarily large part of the data in each cycle as the training set and uses the complete data once in the test set. This method can be applied to many algorithms in (almost) any framework, such as regression [59, 60], leave-one-out [19], V-fold cross validation [58], and density estimation [51]. Since these algorithms can be computationally demanding or even intractable, derived closed-form formulas for the Loo estimator of the risk of histograms or kernel estimators are used [5, 51]. These results have been recently extended to the leave-p-out cross-validation by Celisse and Robin [9]. However, the main limitation in the application of the DDM method is the need for repeated training networks, which require computational effort.
The aim of this article is to propose a method for the construction and testing of models based on ANNs. The proposed method is based on the application of the SSM and DDM for the selection of the network topology to assess which is the most appropriate strategy to obtain the models that best represent the process. The main difference and innovation of the proposed method lies in the fact that the test data set is split by using statistical criteria to ensure that it is representative of the population. This same consideration applies to the training set and validation (all are representative of the population). As a case study, both methods were applied to predict the organic matter concentration (COD) of effluent from a UASB reactor operating at full scale.
2 Method
2.1 Model Structure: Characteristics of the Used Neural Network
Since Rosenblattf [50] first employed them, ANNs have been used to model and simulate nonlinear systems of a diverse nature in digital computers or in hardware boards. According to Masson and Wang [41], the topology of an artificial neural network can be expressed through a directed graph characterized by a set of vertices, a set of directed arcs, and a set of weights assigned to these arcs (W) (Fig. 1a). Each vertex in the graph represents a processing unit. A processing unit has u 1, u 2,..., u R inputs. Based on these inputs and the set of synaptic weights, W 1,1,..., W S, R , the neurons are evaluated, generally through an activation function applied to a weighted sum of inputs using the synaptic weights as weighting factors [41]. A network with a single layer having S neurons with an arbitrary activation function and with R inputs is shown in detail in Fig. 1, which also illustrates its processing unit (neuron). Neural networks frequently have one or more hidden layers of sigmoid neurons (for example, tansig, or logsig) [43, 44, 69]:
-
one hidden layer with S I sigmoid neurons with biases b 1, which are associated with each neuron;
-
one layer with S L output neurons activated by a linear function with biases b 2, which are associated with each neuron.
By using a sufficient number of neurons (S I ) in the hidden layer of a two-layer network, it is possible to approximate any function with a finite number of discontinuities with an accuracy that is arbitrarily specified [42, 54]. This structure is shown in Fig. 2, and it can be used as a universal approximator of functions.
The applied DDM and SSM consider one hidden layer with a sigmoid transfer function and one output layer with a linear transfer function, suitable for continuous variables such as COD. The proposed architecture is shown schematically in Fig. 3.
2.2 Data Preparation
The data set used for the modeling was obtained from a full-scale UASB reactor treating domestic wastewater [6, 7, 47]. The reactor was operated at a temperature which ranged from 13 to 33 °C for winter and summer, respectively. The input variables (predictors) were the total suspended solids (TSS), volatile suspended solids (VSS), chemical oxygen demand (CODi), alkalinity and volatile fatty acids (VFA) concentration, temperature (T), and pH in the influent, whereas the output variable (predicted) is the chemical oxygen demand (CODe) concentration of the effluent. For each set of predictors and predicted variables, 131 measurements were performed. Table 1 presents the descriptive statistics for these variables.
Due to the large difference in the order of quantities for the values shown in Table 1, the data set for the predictor variables was standardized between 0.1 and 0.9 [15, 70]. The aim of this normalization step is to adjust the input dynamic range of the activation functions of the neural network, in this case, the sigmoid function. Therefore, this procedure equals the numerical magnitude of the input variables because very high values can saturate the activation function, impairing the convergence of the neural network. Values with a greater order of magnitude for an input variable dominate over the weights of the network, making it difficult for the algorithm to converge.
2.3 Separation of the Test Set
There is no clear rule for dividing up the experimental data set, so one can find a neural network with an enhanced generalization ability in any of the cases (DDM and SSM). In contrast, it is necessary that the sets obtained by dividing the experimental data represent the entire population [4, 39, 55]. The experimental data set was initially divided into two groups: a model development group (G1) and a test group (G2). Random samples were taken from the total data set to form each group in order to obtain more robust results in predicting organic matter with temperatures data ranging from 13 to 33 °C. The set G1 was used to define the topology and network training, whereas the set G2 was designed to evaluate the generalization ability of ANNs obtained from both methods. The rule applied for this division was to obtain a set G2 with the minimum number of samples but statistically representative of the population (G1 + G2). The nonparametric Mann-Whitney test (an alternative to Student’s t test) was used to statistically assess the average difference of sets G2 and (G1 + G2) [18, 40, 67]. The confidence level applied to the Mann-Whitney test was 90 %.
2.4 Dynamic division method
The methodology adopted in the DDM establishes which neurons in the hidden layer ANN (S I ) should be determined by symmetrically dividing the data set G1 at random into two subsets A and B (of equal size, i.e., 50 % of the samples in each). Once obtained the subsets A and B, the ideal neural network topology must be evaluated by varying the number of neurons in the hidden layer (i = 1, ... , n). For each i amount of hidden neurons (or topology) i, the network is trained with data subset A and then simulated with data subset B (\(\hat {y}_{i,A,B}\)), generating the loss function (or performance criteria) J i, A, B (the sum of squares of prediction errors from the simulation of the data subset B with a network trained with data set A). The loss function J i, A, B is given by Eq. 1.
Likewise, the loss function J i, B, A is given by Eq. 2. The total loss function of topology i is the sum of the results of each simulation (Eq. 3).
where y A and y B represent the experimental value of the subset A and B, respectively; J i, B, A , is the sum of squares of prediction errors from the simulation of the data subset A with a network trained with data set B; and \(\hat {y}_{i,B,A}\) is the estimate of the simulated with data subset A.
Topologies with different numbers of hidden neurons were evaluated and compared using the total loss of their functions, and the topology with the minimum total loss function was adopted [38]. This is for the network topology with a smaller number of neurons, which best represents the process with the data set G1.
2.5 Static-Split Method
The methodology adopted in the static-split method (SSM) establishes that the subdivision of G1 data should be made at random, with two distinct sets statistically representative of the population (G1 + G2). These sets are called the training set and the validation set. The training set should be used to estimate the network parameters (synaptic weights and levels of “bias”), and the validation set must be used iteratively during training to check the efficiency of the network and its generalization capacity. After processing each epoch t (presentation to the neural network for all elements of the training set and the adjustment of synaptic weights and levels of “bias”), the mean squared error of the validation set E Q M v a (t) is estimated (Eq. 4).
where \(\hat {y}_{va_{t}}\) and y t represent, respectively, the predicted and measured values in the validation set evaluated after each epoch presented (training set), k is the number of training examples. The maximum number of epochs, learning rate, and error tolerance value were 1000, 0.05, and 1 × 10−50, respectively. The value E Q M v a (t) for the validation set is expected to decrease at the beginning of the training until a local or global minimum is reached. When E Q M v a (t) values start to show an increasing trend, this is indicative of early overfitting of the ANN. At this point, the training should be stopped by adopting the values (synaptic weights and levels of “bias”) as adjusted parameters of the model [46, 59]. The E Q M o p t value is defined as the smallest error obtained in the validation set for all epochs (t = 1, ... , t max) (Eq. 5).
There is little consistency in the literature about the methods to be applied to the data division into the validation and training data sets. Therefore, to find an appropriate proportion for this division, we varied the proportions of the training and validation sets, performing a sensitivity analysis based on statistical representativeness and evaluating the impact of this ratio on the performance of the model. [55] performed a similar sensitivity analysis by varying the size of the training and validation sets and found that there is a significant impact on the results, even taking into account the mean hypothesis test of the sample statistical property samples at the 95 % confidence interval.
The best network topology and consequently the model that represents the process through the SSM method was estimated by varying the number of neurons in the hidden layer (i = 1, ... , n) for each i and obtaining a single E Q M o p t (i), given in Eq. 6.
2.6 Evaluation of model predictability
Three statistical criteria were used to evaluate the performance of the ANN models by using the DDM and SSM: the coefficient of multiple determination (R 2), the adjusted coefficient of multiple determination (\(R_{adj}^{2}\)), and the root mean square error (RMSE) (Eqs. 7–9, respectively). These were derived by comparing the measured and predicted values of COD.
where S S E is the sum of the quadratic residues \({\sum }_{i=1}^{n}(y_{i}-\hat {y}_{i})^{2}\). The determination coefficient represents the proportion of the overall variance explained by the model. This coefficient makes it possible to evaluate the performance of different models, and is widely used for this purpose. Nevertheless, R 2 can be manipulated by increasing the number of independent variables to explain the dependent variable that would result in an R 2 of one for the training data set. In contrast to R 2, the \(R_{adj}^{2}\) only increases if the additional model parameters (connection weights) improve the results significantly to compensate for the increase in the regression degrees of freedom [17].
The RMSE is a measure of the remaining measurement variance not explained by the model. Some studies have reported the use of these statistical parameters for the evaluation of ANN models [1, 17, 24].
3 Results
3.1 Dynamic division method results
Figure 4 presents the value of the total loss function (J i ), summed over all parameters, defined by Eq. 3, and estimated for each group of hidden neurons tested. Based on the minimum value, the topology comprising seven hidden neurons was then selected and trained again with the whole experimental data set. This procedure resulted in a model with a total of 64 parameters (synaptic weights and biases of the ANN).
The predictive performance was evaluated for data set G2 (10 % of the data). Values of R 2, (\(R_{adj}^{2}\)), and RMSE equal to 0.94, 0.93, and 28.43 mg L−1, respectively, were obtained. Good agreement between the experimental data and model predictions is observed.
3.2 Static-split method results
Figure 5 presents the mean square error of validation (E Q M o p t (i)) for each ANN model by using the data set G1 and varying the number of hidden neurons. By considering the minimum E Q M o p t (i), the topology (Top m i n ) comprising six hidden neurons was then selected and trained again with the entire experimental data set. The topology estimated by the DDM and SSM are very close: six and seven neurons in the hidden layer, respectively. With the purpose using equal requirements when comparing the results, the topology adopted in the implementation and comparison between the methods was seven neurons in the hidden layer.
The model performances obtained when different proportions of the experimental data set (G1 + G2) were used for the training set, validation set, and test set (G2) are summarized in Table 2. A code is used to differentiate the proportions of training, validation, and testing. The first number represents the percentage of the data set extracted from the experimental data set (G1 + G2) and used for the test, whereas the second and the third numbers (in brackets) represent the ratio of the training set and validation set extracted from “G1,” respectively. The best results are obtained when 80 and 20 % of G1 were used for training and validation, respectively, with R 2, (\(R_{adj}^{2}\)), and RMSE equal to 0.72, 0.71, and 62.14 mg L−1, respectively. The results also indicate that there is significant variation in the statistical parameters depending on the sample size of the training and validation even when their size is statistically significant.
3.3 DDM versus SSM
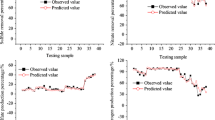
The DDM model was compared with the best SSM model, and the results are shown in Table 3. As can be observed, the results obtained with the DDM are better than those determined with the SSM for predicting COD in the UASB reactor outflow. This result is confirmed by Fig. 6, which shows good agreement between the predicted and observed experimental data values for the DDM model. This result is attributed the ability of the DDM to extract the maximum of the process characteristic during the training of the network. This happens because the adjustment process of the synaptic weights and levels of “bias” is not interrupted until the maximum information about the phenomenology is extracted from the entire G1 data set.
Overtraining of the DDM model is avoided through the use of a proper definition of the network topology and not through the interruption of training, as considered in the SSM. This ensures that the network (during training) extracts the maximum amount of information from the experimental data.
Figure 7 shows a graphical representation of the time series comparing the measured and predicted COD for the G1 and G2 sets using the DDM and SSM for both the G1 and G2 data sets. Although the SSM results follow the variability of the experimental data, the DDM results are closest to the same data ever for the high pics.
4 Conclusions
Anaerobic digestion processes in UASB reactors involve a complex and nonlinear system that is known to be unstable. In such a case, a neural network is used to model this system because this method has proved to be able to capture the nonlinear features and generalized structure of the entire data set.
This study presented a methodology for the construction and testing of ANN models using two methods for selecting the optimal structure of ANNs: the DDM, based on the division of the dynamic G1 data set, and the SSM, based on the split static G1 data set. The two methods were applied to predict the organic matter concentration (COD) effluent from a UASB reactor operated at full scale.
The identification procedure, including the selection of the optimal number of hidden neurons, was efficient and provided consistent results for the DDM and SSM, giving a good description of the system dominant dynamics without degradation of the output prediction.
The DDM showed better results with the highest determination coefficient (R 2 = 0.94) and minimum RMSE, which suggests that ANNs are a useful tool to represent the complexity of the UASB reactor. This method can be successfully applied, particularly when there is a limited amount of data for the construction of ANN models because every data set can be used for training [21]. The disadvantage of this method is the need to train several networks, which requires computational effort.
References
Akratos, C.S., Papaspyros, J.N., & Tsihrintzis, V.A. (2008). An artificial neural network model and design equations for bod and cod removal prediction in horizontal subsurface flow constructed wetlands. Chemical Engineering Journal, 143(1–3), 96–110.
Batstone, D.J., Keller, J., Angelidaki, I., Kalyuzhny, S., Pavlostathis, S., Rozzi, A., Sanders, W., Siegrist, H., & Vavilin, V. (2002). Anaerobic digestion model No. 1 (ADM1). IWA Publishing.
Baughman, R., & Liu, Y.A. (1995). Neural networks in bioprocessing and chemical engineering. Book.
Bowden, G.J., Maier, H.R., & Dandy, G.C. (2002). Optimal division of data for neural network models in water resources applications. Water Resources Research, 38(2), 1–11.
Bowman, A.W. (1984). An alternative method of cross-validation for the smoothing of density estimates. Biometrika, 71, 353–360.
Busato, R. (2004). Desempenho de um filtro anaeróbio de fluxo ascendente como tratamento de efluente de reator uasb: estudo de caso da ete de imbituva. Master’s thesis, Universidade Federal do Paraná (In Portuguese).
Campello, R.P. (2009). Desempenho de reatores anaeróbios de manto de lodo (uasb) operando sob condições de temperaturas t?picas de regiões de clima temperado. Master’s thesis, Universidade Federal do Rio Grande Do Sul. Instituto de Pesquisas Hidráulicas - IPH (In Portuguese).
Carrasco, E., Rodríguez, J., Al, A.P., Roca, E., & Lema, J. (2004). Diagnosis of acidification states in an anaerobic wastewater treatment plant using a fuzzy-based expert system. Control Engineering Practice, 12, 59–64.
Celisse, A., & Robin, S. (2008). Nonparametric density estimation by exact leave-p-out cross-validation. Computational Statistics and Data Analysis, 52, 2350–2368.
Chernicharo, C.A.L. (2007). Reatores anaeróbios. Belo Horizonte, MG: Departamento de Engenharia Sanitária e Ambiental - DESA (In Portuguese).
Coelho, N., Capela, I., & Droste, R. (2006). Application of adm1 to a uasb treating complex wastewater in different feeding regimes. Water Environment Foundation, 13, 7123–7135.
Costa, A., Henriques, A., Alves, T., Maciel Filho, R., & Lima, E. (1999). A hybrid neural model for the optimization of fed-bath fermentations. Brazilian Journal of Chemical Engineering, 16, 53–63.
Côté, M., Grandjean, B.P., Lessard, P., & Thibault, J. (1995). Dynamic modelling of the activated sludge process: Improving prediction using neural networks. Water Research, 29(4), 995–1004.
Donoso-Bravo, A., Mailiera, J., Martinb, C., Rodríguez, J., Aceves-Larae, C.A., & Wouwera, A.V. (2011). Model selection, identification and validation in anaerobic digestion: A review. Water Research, 45, 5347–5364.
Elsayed, K., & Lacor, C. (2011). Modeling, analysis and optimization of aircyclones using artificial neural network, response surface methodology and cfd simulation approaches. Powder Technology, 212(1), 115–133.
Erdirencelebi, D., & Yalpir, S. (2011). Adaptive network fuzzy inference system modeling for the input selection and prediction of anaerobic digestion effluent quality. Applied Mathematical Modelling, 35, 3821–3832.
Esquerre, K.P.O., Seborg, D.E., Bruns, R.E., & Zori, M. (2004). Application of steady-state and dynamic modeling for the prediction of the bod of an aerated lagoon at a pulp and paper mill: Part i. linear approaches. Chemical Engineering Journal, 104(13), 73–81.
Fay, M., & Proschan, M. (2010). Wilcoxon-mann whitney or t-test? On assumptions for hypothesis tests and multiple interpretations of decision rules. Statistics Surveys, 4, 31–39.
Geisser, S. (1975). The predictive sample reuse method with applications. Journal of the American Statistical Association, 70, 320–328.
Gontarski, C., Rodrigues, P., Mori, M., & Prenem, L. (2000). Simulation of an industrial wastewater treatment plant using artificial neural networks. Computers & Chemical Engineering, 24(2–7), 1719–1723.
Goutte, C. (1997). Note on free lunches and cross-validation. Neural Computation, 9(6), 1245–1249.
Güçlü, D., & Dursun, S. (2008). Prediction of wastewater treatment plant performance using artificial neural networks. CLEAN - Soil, Air, Water, 36(9), 781–787.
Häck, M., & Manfred, K. (1996). Estimation of was tewater process parameters using neural networks. Water Science and Technology, 33, 101–115.
Hamed, M.M., Khalafallah, M.G., & Hassanien, E.A. (2004). Prediction of wastewater treatment plant performance using artificial neural networks. Environmental Modelling & Software, 19(10), 919–928.
Hamoda, M.F., Al-Ghusain, I.A., & Hassan, A.H. (1999). Integrated wastewater treatment plant performance evaluation using artificial neural networks. Water Science and Technology, 40, 55–65.
Harada, L.H.P., da Costa, A.C., & Filho, R.M. (2002). Hybrid neural modeling of bioprocesses using functional link networks. Applied Biochemistry and Biotechnology, 98, 1009–1023.
Kalyuzhnyi, S., Fedorovich, V., Lens, P., Hulshoff Pol, L., & Lettinga, G. (1998). Mathematical modelling as a tool to study population dynamics between sulfate reducing and methanogenic bacteria. Biodegradation, 9, 187–199.
Kato, M.T., Field, J.A., Kleerebezem, R., & Lettinga, G. (1994). Treatment of low strength soluble wastewaters in uasb reactors. Journal of Fermentation and Bioengineering, 77(6), 679–686.
Lee, D., & Park, J. (1999). Neural network modelling for on-line estimation of nutrient dynamics in a sequentially operated batch reactor. Biotechnology, 75, 229–239.
Lettinga, G. (1996). Sustainable integrated biological wastewater treatment. Water Science and Technology, 33, 85–98.
Lettinga, G., Field, J., van Lier, J., Zeeman, G., & Pol, L.H. (1997). Advanced anaerobic wastewater treatment in the near future. Water Science and Technology, 35, 5–12.
Lettinga, G., & Hulshoff Pol, L. (1986). Advanced reactor design, operation and economy. Water Science and Technology, 18, 99–108.
Lettinga, G., & Hulshoff Pol, L. (1991). Uasb process design for various types of wastewater. Water Science and Technology, 21, 87–107.
Liao, K.P., & Fildes, R. (2005). The accuracy of a procedural approach to specifying feedforward neural networks for forecasting. Computers & Operations Research, 32(8), 2151–2169.
Liu, Y., Hai-Lou, X., Shu-Fang, Y., & Joo-Hwa, T. (2003). Mechanisms and models for anaerobic granulation in upflow anaerobic sludge blanket reactor. Water Research, 37, 611–673.
Lopez, I., & Borzacconi, L. (2009). Modelling a full scale uasb reactor using a cod global balance approach and state observers. Chemical Engineering Journal, 146(1), 1–5.
Lyberatos, G., & Skiadas, I. (1999). Modelling of anaerobic digestion - a review. Global Nest: the International Journal, 1, 63–76.
Magalhães, R.S., Fontes, C.H.O., Almeida, L.A.L., Embírucu, M., & Santos, J.M.C. (2010). A model for three-dimensional simulation of acoustic emissions from rotating machine vibration. The Journal of the Acoustical Society of America, 127(6), 3569–3576.
Maier, H.R., & Dandy, G.C. (2000). Neural networks for the prediction and forecasting of water resources variables: a review of modelling issues and applications. Environmental Modelling & Software, 15(1), 101–124.
Mann, H., & Whitney, D. (1947). On a test of whether one of two random variables is stochastically larger than the other. Annals of Mathematical Statistics, 18, 50–60.
Másson, E., & Wang, Y.J. (1990). Introduction to computation and learning in artificial neural networks. European Journal of Operational Research, 47(1), 1–28.
Meade Jr, A.J., & Sonneborn, H.C. (1996). Numerical solution of a calculus of variations problem using the feedforward neural network architecture. Advances in Engineering Software, 27(3), 213–225.
Narendra, K., & Arthasarathyk, P. (1990). Identification and control of dynamical systems using neural networks. IEEE Transactions on Neural Networks, 1, 4–27.
Nikravesh, M., Farell, A., & Stanford, T. (1996). Model identification of nonlinear time variant processes via artificial neural network. Computers & Chemical Engineering, 20(11), 1277–1290.
Perendeci, A., Arslan, S., Tanyolac, A., & Celebi, S. (2009). Effects of phase vector and history extension on prediction power of adaptive-network based fuzzy inference system (anfis) model for a real scale anaerobic wastewater treatment plant operating under unsteady state. Bioresource Technology, 100, 4579–4587.
Prechelt, L. (1998). Automatic early stopping using cross validation: quantifying the criteria. Neural Networks, 11(4), 761–767.
Ramos, R.A. (2008). Avaliação da influência da operao de descarte de lodo no desempenho de reatores uasb em estações de tratamento de esgotos no distrito federal. Master’s thesis, Universidade de Brasília (In Portuguese).
Reich, Y., & Barai, S. (2000). A methodology for building neural networks models from empirical engineering data. Engineering Applications of Artificial Intelligence, 13(6), 685–694.
Ren, T.T., Mu, Y., Yu, H.Q., Harada, H., & Li, Y.Y. (2008). Dispersion analysis of an acidogenic uasb reactor. Chemical Engineering Journal, 142(2), 182–189.
Rosenblatt, F. (1958). The perceptron—a probabilistic model for information-storage and organization in the brain. Psychological Review, 65, 386–408.
Rudemo, M. (1982). Empirical choice of histograms and kernel density estimators. Journal of Statistics, 9, 65–78.
Saravanan, V., & Sreekrishnan, T. (2006). Modelling anaerobic biofilm reactors - a review. Journal of Environmental Management, 81(1), 1–18.
Schenker, B., & Agarwal, M. (1996). Cross-validated structure selection for neural networks. Computers & Chemical Engineering, 20(2), 175–186.
Selmic, R.R., & Lewis, F.L. (2002). Neural network approximation of piecewise continuous functions: Application to friction compensation. IEEE Transactions on Neural Networks, 13, 745–751.
Shahin, M.A., Holger, R.M., & Jaksa, M.B. (2004). Data division for developing neural networks applied to geotechnical engineering. Geotechnical Engineering, 18, 105–114.
Shrestha, D.L., Kayastha, N., & Solomatine, D.P. (2009). A novel approach to parameter uncertainty analysis of hydrological models using neural networks. Hydrology and Earth System Sciences, 13, 1235–1248.
Srivastav, R.K., Sudheer, K.P., & Chaubey, I. (2007). A simplified approach to quantifying predictive and parametric uncertainty in artificial neural network hydrologic models. Water Resources Research, 43(10), 1–12.
Stone, C. (1984). An asymptotically optimal window selection rule for kernel density estimates. The Annals of Statistics, 12, 1285–1297.
Stone, M. (1974). Cross-validatory choice and assessment of statistical predictions. Journal of the Royal Statistical Society. Series B (Methodological), 36(2), 111–147.
Stone, M. (1977). An asymptotic equivalence of choice of model by cross-validation and akaikes criterion. Journal of the Royal Statistical Society. Series B, 44, 44–47.
Turkdogan-Aydinol, F.I., & Yetilmezsoy, K. (2010). A fuzzy-logic-based model to predict biogas and methane production rates in a pilot-scale mesophilic uasb reactor treating molasses wastewater. Journal of Hazardous Materials, 182, 460–471.
Utojo, U., & Bakshi, B. (1995). Neural networks in bioprocessing and chemical engineering, appendix: Connections between neural networks and multivariate statistical methods: An overview. New York: Academic Press.
van Haandel A.C., & e Lettinga, G. (1994). Tratamento anaeróbio de esgotos: Um manual para regiões de clima quente. Campina Grande - Paraíba (In Portuguese).
Verma, A., Wei, X., & Kusiak, A. (2013). Predicting the total suspended solids in wastewater: a data-mining approach. Engineering Applications of Artificial Intelligence, 26(4), 1366–1372.
Warne, K., Prasad, G., Rezvani, S., & Maguire, L. (2004). Statistical and computational intelligence techniques for inferential model development: a comparative evaluation and a novel proposition for fusion. Engineering Applications of Artificial Intelligence, 17(8), 871–885.
Wilcox, S., Hawkes, D., Hawkes, F., & Guwy, A. (1995). A neural network, based on bicarbonate monitoring, to control anaerobic digestion. Water Research, 29(6), 1465–1470.
Wilcoxon, F. (1949). Some rapid approximate statistical procedures. Stamford, CT: Stamford Research Laboratories.
Zeng, G., Qin, X., He, L., Huang, G., Liu, H., & Lin, Y. (2003). A neural network predictive control system for paper mill wastewater treatment. Engineering Applications of Artificial Intelligence, 16, 121–129.
Zhan, J.X., & Ishida, M. (2001). The multi-step predictive control of nonlinear siso processes with a neural model predictive control (nmpc) method. Computers & Chemical Engineering, 21, 201–210.
Zhao, B., & Su, Y. (2010). Artificial neural network-based modeling of pressure drop coefficient for cyclone separators. Chemical Engineering Research and Design, 88(5–6), 606–613.
Acknowledgments
We would like to thank the Coordination for the Improvement of Higher Education Personnel - CAPES, for its financial support for this research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mendes, C., da Silva Magalhes, R., Esquerre, K. et al. Artificial Neural Network Modeling for Predicting Organic Matter in a Full-Scale Up-Flow Anaerobic Sludge Blanket (UASB) Reactor. Environ Model Assess 20, 625–635 (2015). https://doi.org/10.1007/s10666-015-9450-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10666-015-9450-x