Abstract
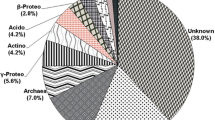
Imidacloprid, used against mango hopper, is a persistent insecticide in soil. Microbes have the ability to remove toxic pesticides from soil surface. Metagenomic is an approach for understanding the diversity and related metabolic activities in any environmental sample without culturing the microbes. Metagenomic analysis of mango orchard soil was carried out using 16S rRNA gene sequencing to understand the impact of imidacloprid on soil microbial population. In control and imidacloprid applied soil samples, representative sequences clustered were 0.142930 and 0.082320 million, respectively. At the kingdom level, 85 and 88 percent represented to bacteria, 2 and 1 percent to archaea, and 13 and 11 percent to unassigned for control and treated metagenomes, respectively. At phylum level, 16 and 17 percent of OTUs (operational taxonomic units) were assigned with Proteobacteria, while 13 and 11 percent of OTUs were unassigned in control and imidacloprid-treated samples, respectively. The other abundant phyla in both the samples were Planctomycetes, Bacteroidetes, and Actinobacteria. At class level, 9 and 11 percent of OTUs were assigned with Planctomycetia in control as well as imidacloprid-treated samples, respectively. A number of OTUs present in control and imidacloprid applied samples are 31,173 and 21,909, respectively, with 18,018 number of OTUs shared between the two samples. The genus Gemmata totally disappeared in imidacloprid applied soil, while those belonging to class Phycisphaerae, genus Prevotella and species copri were identified in imidacloprid treatment. Bacterial community transformation was evident from this study indicating possible microbial bioremediation of imidacloprid in mango orchard soil.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Mango (Mangifera indica L.), a commercially important fruit, suffers from insects infestation throughout its developmental stages which requires spraying of many insecticides to obtain better yield. Hopper (Amritodus atkinsoni, Idioscopus clypealis, and Idioscopus nitidulus) was detected as a serious pest of mango causing up to 50 percent crop loss in cases of severe infestation (Anonymous 2010). This insect is expected to emerge from the last week of February to first week of March. In order to control mango hoppers (if hopper population is more than 5 per panicle) spray of imidacloprid [1-(6-chloro-3-pyridinyl)methyl)-N-nitro-2-imidazolidinimine], a neonicotinoid insecticide, at 0.005% (0.3 mL/L of water) is recommended at early stages of panicle formation (Anonymous 2010). It is a polar compound with good water solubility, relatively non-volatile and persistent in soil with a long residual life varying from 28 to 1250 days depending upon the soil type (Baskaran et al. 1997; Sarkar et al. 2001; Goulson 2013). Since after spraying on trees, a portion of imidacloprid come down in contact with soil flora and fauna, which can contaminate both soil and ground water (through leaching) and thereby have a chance to be accumulated in the food chain. When not exposed to light, imidacloprid breaks down slowly in water and thus has the potential to persist in groundwater for extended periods. A water monitoring study by California Department of Pesticide Regulation have reported imidacloprid in 89 percent of samples with levels ranging from 0.11 to 3.29 μg/L. Nineteen percent of the samples exceeded the USEPA (United States Environmental Protection Agency) threshold level of imidacloprid (1.05 μg/L) for chronic toxicity for aquatic invertebrates (Starner and Goh 2012).
Microbial bioremediation can be considered a cost-effective tool for the detoxification of pesticides (Li et al. 2012). Microbial research has generally focused on the study of culturable bacteria; however, only a small proportion of bacterial population is culturable. A vast portion of bacterial community remains unstudied. Molecular identification of dominant culturable microbes isolated from imidacloprid applied soil revealed the presence of Pseudomonas mosselii strain NG1, NCBI (National Centre for Biotechnology Information, USA) accession no. MN227542 (Pseudomonas mosselii n.d) (Bhattacherjee et al. 2020) and Sphingobacterium mucilaginosum strain NG201, NCBI accession no. MN818683 (Sphingobacterium mucilaginosum n.d) (unpublisheddata). For predicting the best strategy for biodegradation, understanding of microbial processes in individual sites is essential because microbes are the primary pollutant degraders in contaminated soil (Jeffries et al. 2018). The published reports on the effect of imidacloprid on soil microorganisms have indicated that imidacloprid can influence the community structure of soil bacteria, ammonia-oxidizing archaea, and ammonia-oxidizing bacteria along with the decrease in biomass and soil enzymatic activities (Cycoń et al. 2013; Cycoń and Piotrowska-Seget 2015a, b; Wang et al. 2014). The influence of imidacloprid on soil microbial communities has proved that it can adversely affect different groups of soil microorganisms (Ahmed and Ahmad 2006; Singh and Singh 2005). These reports suggested that imidacloprid can be potentially risky to the soil biochemical characteristics and microbial activity. Several workers have studied the effects of imidacloprid on the biodiversity of soil microbes in different countries using different techniques, viz., molecular markers like ERIC-PCR (enterobacterial repetitive intergenic consensus-polymerase chain reaction) and RAPD-PCR (random amplified polymorphic DNA-polymerase chain reaction) in contaminated soils of Cameron Highlands, Malaysia (Moghaddam et al. 2011), denaturing gradient gel electrophoresis (DGGE) in saline soils from Yellow River Delta, China (Zhang et al. 2015); next-generation sequencing (NGS) in agrosoddy-podzolic soil of Moscow, Russia (Astaykina et al. 2020); etc. With the help of next-generation DNA sequencing approach, metagenomic analysis can be utilized to obtain more information on the interaction between microbial taxonomy and regarding those bacteria which are contributing to the functioning of soil and are viable but not culturable (Amann et al. 1995; Hugenholtz and Tyson 2008). Until date, the metagenomic analysis for investigating the detailed structure of bacterial communities/populations in soil ecosystem under mango orchards and their response to contamination with imidacloprid treatment have not been reported so far. Thus, the present study was undertaken to assess the influence of imidacloprid application on soil bacterial diversity using 16S ribosomal RNA gene (rDNA)-based metagenomic analysis.
Materials and methods
Soil samples were obtained from imidacloprid-treated (0.005%) and untreated (control) soils as per standard procedure. Soil of mango orchard is loam to sandy loam type with pH 7.6, bulk density between 1.40 and 1.60 g/cm3, particle density between 2.26 and 2.32 g/cm3, and water holding capacity 21.5 percent. The physicochemical properties of soil were organic carbon 0.48 per cent, phosphorus between 15 and 18 mg/kg, potassium 90 and 100 mg/kg, zinc 0.3 and 0.5 mg/kg, copper 0.25 and 0.40 mg/kg, iron 12 and 15 mg/kg, and manganese 7 and 10 mg/kg. DNA for metagenomic analysis was extracted from treated and control soils using genomic DNA isolation research kit according to the manufacturer’s protocol (Chromous Biotech Pvt. Ltd., Bengaluru, India). Twenty-five nanogram DNAs were used to amplify 16S rRNA hyper variable regions V3–V4. The reaction includes KAPA HiFi HotStart Ready Mix and 10-μM final concentration of modified 341F and 785R primers (Klindworth et al. 2013). The PCR program involved an initial denaturation of 95 °C for 5 min followed by 25 cycles of 95 °C for 30 s, 55 °C for 45 s, and 72 °C for 30 s and a final extension at 72 °C for 7 min using primers, viz., forward primer (V3V4F:5′-CCTACGGGNGGCWGCAG-3′) and reverse primer (V3V4R:5′-ACTACHVGGGTATCTAATCC-3′). The amplicons were purified using Ampure beads to remove unused primers, and this was followed by 8 cycles of PCR using Illumina barcoded adapters to prepare the sequencing libraries.
Steps adopted in 16S rRNA gene sequencing and analyses through metagenomics are depicted in Fig. 1. The library was further sequenced on Illumina MiSeq platform using 2 × 250 paired-end (PE) chemistry which generated 0.5 million reads per sample. The quality check of raw reads was carried out by FastQC (v 0.11.7) (Andrews 2017), trimmed (TrimGalore v 0.5.0) (Babraham 2017) to remove adapter contamination, further processed to remove gaps and overhangs (UCHIME algorithm) (Edgar et al. 2011), and filtered using GREENGENES v.13.8-99 (DeSantis et al. 2006). The contigs were then clustered into operational taxonomic units (OTUs). After the classification, OTU abundance was estimated. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was used to predict gene family abundance (Langille et al. 2013). Metagenomes were predicted using predict metagenomes.py script and used for further downstream analysis using Quantitative Insights Into Microbial Ecology (QIIME v.1.9.0) (Kuczynski et al. 2011). Paired end data were given as input in QIIME and OTU were assigned to similar sequences; UCLUST algorithm was used at sequence similarity threshold of 97 percent against Greengenes as the reference database for picking up OTUs. The output files from the QIIME are analyzed for the taxonomic classification using microbiome analyst, which is an online comprehensive statistical, visual, and meta-analysis of microbiome data available at https://www.microbiomeanalyst.ca/faces/home.xhtml. The alpha-diversity (Shannon index) of metagenomic profiles was calculated on biome formatted output tables from FOCUS/SUPERFOCUS (SUbsystems Profile by databasE Reduction using FOCUS) using the QIIME software (Caporaso et al. 2010). Sequence information generated was submitted in the NCBI database, and Sequence Read Archive (SRA) submission hyperlink is https://submit.ncbi.nlm.nih.gov/subs/sra/SUB6674392/overview (Metagenome of mango orchard soil n.d).
Results and discussion
Amplicon sequencing of V3–V4 region of 16S rRNA gene revealed that in treated and control sample, a number of representative sequences clustered were 0.142930 and 0.082320 million, respectively. QIIME analysis of the sequenced data resulted in identification of operational taxonomic units (OTUs) which were used to classify the bacterial population present in the samples at phylum, class, family, order, genus, and species levels. Table 1 depicts the results of rarefaction analysis based on Mothur v.1.21.1 to reveal the diversity indices, including the ACE (abundance-based coverage estimator), Chao, Simpson, and Shannon diversity indices. Alpha-diversity indexes are composite indexes reflecting abundance and consistency. The rarefaction analysis indicated that OTU abundance as reflected by ACE index was higher (58087.57) in control soil compared to that (43642.26) in treated one. The diversity of OTU represented in terms of Shannon index was relatively higher in treated samples (8.98) compared to that in control soil (8.76). The lower OTU abundance in treated soil might be due to the fact that pesticides have inhibitory effect on microorganisms (Yousaf et al. 2013). Muturi et al. (2017) have reported a significantly lower number of OTUs in malathion, carbaryl, and permethrin treated container aquatic habitats compared to control. Using observed species measure, sample C1S22 (imidacloprid treated) was found more diverse compared to the control sample C2S23 (Fig. 2). Higher diversity in pesticide added soil is explained on the basis that depending on biotic and abiotic factors, microorganisms adapt to the environment, and accordingly environmental conditions select for microorganisms featuring specific capabilities (Shah et al. 2012). Kim et al. (2002) have reported the presence of the bacteria with cytochrome genes directly involved in the degradation process of organic carbon compounds. Figure 3 depicting heat map illustrates that the abundance of phyla in imidacloprid-treated (C1S22) samples are relatively diverse.
Total number of OTUs picked was 47,930, out of which 85 and 88 percent were classified with kingdom Bacteria in treated and control sample, respectively. At phylum level, 16 and 17 percent of OTUs were assigned with Proteobacteria in treated and control samples, respectively, whereas 13 and 11 percent were unassigned (Table 2). The other phyla abundant in all the samples were Planctomycetes (13 and 16% of OTUs), Bacteroidetes (13 and 13% of OTUs), Chloroflexi (12 and 13% of OTUs), and Actinobacteria (10 and 7% of OTUs). At class level, the abundant classes were assigned to Planctomycetia (9 and 11% of OTUs), Anaerolineae (9 and 10% of OTUs), Saprospirae (8 and 9% of OTUs), and Alphaproteobacteria (7 and 8% of OTUs) in treated and control samples, respectively. At order level, 8 and 9 percent of OTUs were assigned with Saprospirales in treated and control samples, respectively, followed by Actinomycetales (7 and 5% of OTUs). At family level, 7 and 9 percent of OTUs were assigned with Chitinophagaceae in treated and control samples, respectively. Table 2 indicates that application of imidacloprid in mango orchard soil influenced bacterial community diversity as some bacterial populations increased and some decreased at different levels, while some remained same. At the genus level, the OTU number genera corresponding to Rhodoplanes, order Myxococcales, and class Betaproteobacteria remained the same in imidacloprid applied and control soil but that of class Gammaproteobacteria and order Pedosphaeral decreased in treated soil by 33 percent. The genus Gemmata totally disappeared in imidacloprid applied soil, while those belonging to class Phycisphaerae and genus Prevotella species copri appeared additionally (Figs. 4 and 5). At the genus level, 6 and 7 percent of OTUs were assigned with unassigned genus within the Chitinophagaceae family in treated and control samples, respectively.
Microorganisms are vital for the bioremediation of pesticides. Addition of pesticide affects the growth of certain bacteria while promoting growth of others. Shah et al. (2013) have reported that phylum Proteobacteria is present in higher proportion in heavy metal contamination, waste-water treatments, and other contaminated sites all over the world. The number of OTU representing unassigned phylum supports the observation of relatively higher diversity in treated samples. Shah et al. (2012) have also reported that a sub-community comprising diverse organisms collectively interacts to perform all the metabolic reactions for bioremediation in pesticide-applied soils. Moghaddam et al. (2011) have reported that long-term application of imidacloprid adversely affected the soil bacterial community and the numbers of viable gram-negative bacteria in soil could be reduced due to its application, and presence of its residues in soil could be harmful to selective soil microbial communities. Our findings clearly indicated that application of imidacloprid in mango orchard soil influenced bacterial community diversity as some bacterial populations increased and some decreased at different levels while some remained the same. As shown in Table 2 at the phylum level, in imidacloprid applied soil, the population of OTUs belonging to Proteobacteria, Planctomycetes, Chloroflexi, and Verrucomicrobia decreased, while that of Actinobacteria increased. Cyanobacteria were observed in treated soil, while those were absent in control soil. Kuritz (1998) and Kumar and Singh (2017) have reported Cyanobacteria as agents for the degradation of pesticides and chlorinated organic compounds. Our results are in concurrence with Anhalt et al. (2007) who have reported the biodegradation of imidacloprid in soil by Leifsonia sp. Latter belong to phylum Actinobacteria whose OTU increased by 30 per cent in treated soils. Shetti and Kaliwal (2016) reported that in laboratory studies, application of imidacloprid at 125, 250, 500, and 1000 ppm levels resulted in 9.80, 08.40, 6.73, and 5.60 × 106 colonies (p < 0.05) when compared to 11.05 × 106 colonies in control plates. In the field studies, imidacloprid-treated fields showed a significant (p < 0.05) decline in bacterial counts. Cycoń and Piotrowska-Seget (2015a) have observed a negative effect by imidacloprid applied at the field rate (1 mg/kg soil) dosage for the number of total bacteria in soil as it affected the physiological state of culturable bacteria and caused a reduction in the rate of colony formation. The same authors (2015b) have reported that ammonia-oxidizing archaea and ammonia-oxidizing bacteria community were affected by imidacloprid treatment and concluded that changes in their community structures could be due to an increase in the concentration of N-, which is the most important factor determining the contribution of these microorganisms to soil nitrification process. The nitrification rate was decreased, while the ammonification rate was stimulated by imidacloprid application. Cycoń et al. (2013) have suggested the evolution of bacteria capable of degrading imidacloprid among indigenous microflora, which is similar to our findings where Sphingobacterium mucilaginosum strain NG201 was evolved with imidacloprid degradation potential. Significant increase in bacterial population was observed in imidacloprid-treated groundnut soil (Singh and Singh 2005). Astaykina et al. (2020) have observed the changes in the abundances of the phyla of Actinobacteria and Proteobacteria in agrosoddy-podzolic soil of Moscow, Russia, after the application of three pesticides (herbicide metribuzin, insecticide imidacloprid, and fungicide benomyl) and imidacloprid stimulated nitrogen fixation in soil. Using 16S rRNA amplicon and shotgun metagenomic sequencing methods, certain bacterial genera like Chromohalobacter, Marinimicrobium, Idiomarina, Salinosphaera, Halomonas, Sphingopyxis, Novosphingobium, Sphingomonas, and Pseudomonas were found to be more abundant in the soil sample from the HCH-dumpsite (450 mg HCH/g soil) (Sangwan et al. 2012). Doolotkeldieva et al. (2018) have reported predominance of bacterial genus Micrococcus belonging to phylum Actinobacteria in soils from dumping zones for obsolete pesticides around the Suzak and Balykchy dumping places of Kyrgyzstan. Using metagenomic analysis, Bhardwaj et al. (2020) have reported the abundance of orders Erysipelotrichales, Selemonadales, Clostridiales, and Thermoanaerobacterales exclusively in soil mesocosm treated with herbicide atrazine. The authors have also mentioned that some bacterial genera like Pseudomonas, Achromobacter, Xanthomonas, Stenotrophomonas, and Cupriavidus had emerged as the dominant members in various bioremediation strategies. They have concluded that inherent microbial flora have the ability to adjust their community and metabolic machinery upon exposure to the herbicide.
Conclusion
This study is the first of its kind in ascertaining the role of microbial community variation in mango orchard soils created by imidacloprid application. The results indicate that in control soil, total microbial population abundance is higher, but diversity is lesser. In treated soil, imidacloprid susceptible microbial abundance decreased, while the diversity increased. The number of unassigned bacteria was higher in treated samples which might provide explanation for higher diversity. This suggests the selection and adaptation of potential imidacloprid degrading microbes in treated soils. This study gives insights into microbial transformation phenomenon in the mango rhizosphere soil system which could be exploited for developing microbial bioremediation consortium.
Data availability
Not applicable.
References
Ahmed, S., & Ahmad, M. S. (2006). Effect of insecticides on the total number of soil bacteria under laboratory and field conditions. Pakistan Entomology, 28, 63–68.
Amann, R. I., Ludwig, W., & Schleifer, K. H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiological Reviews, 59(1), 143–169.
Andrews, S. (2017). FastQC: A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 3 May 2017.
Anhalt, J. C., Moorman, T. B., & Koskinen, W. C. (2007). Biodegradation of imidacloprid by an isolated soil microorganism. Journal of Environmental Science and Health. Part. B, 42(5), 509–514.
Anonymous (2010). Management of mango hopper. http://www.cishlko.org/Technologies/MangoHopper_advisory.pdf. Accessed 10 August 2011.
Astaykina, A. A., Streletskii, R. A., Maslov, M. N., Belov, A. A., Gorbatov, V. S., & Stepanov, A. L. (2020). The impact of pesticides on the microbial community of agrosoddy-podzolic soil. Eurasian Soil Science, 53, 696–706. https://doi.org/10.1134/S1064229320050038.
Babraham Bioinformatics – TrimGalore! (2017). Available from: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. Accessed 3 May 2017.
Baskaran, S., Kookana, R. S., & Naidu, R. (1997). Determination of the insecticide imidacloprid in water and soil using high performance liquid chromatography. Journal of Chromatography. A, 787, 271–275. https://doi.org/10.1016/S0021-9673(97)00652-3.
Bhardwaj, P., Singh, K. R., Jadeja, N. B., Phale, P. S., & Kapley, A. (2020). Atrazine bioremediation and its influence on soil microbial diversity by metagenomics analysis. Indian Journal of Microbiology, 60, 388–391. https://doi.org/10.1007/s12088-020-00877-4.
Bhattacherjee, A. K., Garg, N., Shukla, P. K., Singh, B., Vaish, S., & Dikshit, A. (2020). Bacterial bioremediation of imidacloprid in mango orchard soil by Pseudomonas mosselii strain NG1. International Journal of Current Microbiology and Applied Sciences, 9(10), 1150–1159. https://doi.org/10.20546/ijcmas.2020.910.138.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., Fierer, N., Peña, A. G., Goodrich, J. K., Gordon, J. I., Huttley, G. A., Kelley, S. T., Knights, D., Koenig, J. E., Ley, R. E., Lozupone, C. A., McDonald, D., Muegge, B. D., Pirrung, M., Reeder, J., Sevinsky, J. R., Turnbaugh, P. J., Walters, W. A., Widmann, J., Yatsunenko, T., Zaneveld, J., & Knight, R. (2010). QIIME allows analysis of high throughput community sequencing data. Nature Methods, 7(5), 335–336. https://doi.org/10.1038/nmeth.f.303.
Cycoń, M., & Piotrowska-Seget, Z. (2015a). Biochemical and microbial soil functioning after application of the insecticide imidacloprid. Journal of Environmental Sciences, 27(1), 147–158. https://doi.org/10.1016/j.jes.2014.05.034.
Cycoń, M., & Piotrowska-Seget, Z. (2015b). Community structure of ammonia-oxidizing archaea and ammonia-oxidizing bacteria in soil treated with the insecticide imidacloprid. Journal of Biomedicine & Biotechnology, 582938, 1–12. https://doi.org/10.1155/2015/582938.
Cycoń, M., Markowicz, A., Borymski, S., Wójcik, M., & Piotrowska-Seget, Z. (2013). Imidacloprid induces changes in the structure, genetic diversity and catabolic activity of soil microbial communities. Journal of Environmental Management, 131, 55–65. https://doi.org/10.1016/j.jenvman.2013.09.041.
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., Huber, T., Dalevi, D., Hu, P., & Andersen, G. L. (2006). Greengenes, a Chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology, 72(7), 5069–5072. https://doi.org/10.1128/AEM.03006-05.
Doolotkeldieva, T., Konurbaeva, M., & Bobusheva, S. (2018). Microbial communities in pesticide-contaminated soils in Kyrgyzstan and bioremediation possibilities. Environmental Science and Pollution Research, 25, 31848–31862. https://doi.org/10.1007/s11356-017-0048-5.
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., & Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27(16), 2194–2200. https://doi.org/10.1093/bioinformatics/btr38.
Goulson, D. (2013). An overview of the environmental risks posed by neonicotinoid insecticides. Journal of Applied Ecology, 50(4), 977–987. https://doi.org/10.1111/1365-2664.12111.
Hugenholtz, P., & Tyson, G. W. (2008). Microbiology: metagenomics. Nature, 455, 481–483. https://doi.org/10.1038/455481a.
Jeffries, T. C., Rayu, S., Nielsen, U. N., Lai, K., Ijaz, A., Nazaries, L., & Singh, B. K. (2018). Metagenomic functional potential predicts degradation rates of a model organophosphorus xenobiotic in pesticide contaminated soils. Frontiers in Microbiology, 9, 147. https://doi.org/10.3389/fmicb.2018.00147.
Kim, D., Kim, Y. S., Kim, S. K., Kim, S. W., Zylstra, G. J., Kim, Y. M., & Kim, E. (2002). Monocyclic aromatic hydrocarbon degradation by Rhodococcus sp. strain DK17. Applied and Environmental Microbiology, 68, 3270–3278. https://doi.org/10.1128/aem.68.7.3270-3278.2002.
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., & Glöckner, F. O. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research, 41(1), e1. https://doi.org/10.1093/nar/gks808.
Kumar, A., & Singh, J. S. (2017). Cyanoremediation: A green-clean tool for decontamination of synthetic pesticides from agro- and aquatic ecosystems. In J. Singh & G. Seneviratne (Eds.), Agro-environmental sustainability (pp. 59–83). Cham: Springer. https://doi.org/10.1007/978-3-319-49727-3_4.
Kuritz, T. (1998). Cyanobacteria as agents for the control of pollution by pesticides and chlorinated organic compounds. Journal of Applied Microbiology Symposium Supplement, 85, 186S–192S.
Kuczynski, J., Stombaugh, J., Walters, W. A., González, A., Caporaso, J. G., & Knight, R. (2011). Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Current Protocols in Bioinformatics, Chapter 10, Unit 10.7–10.7. https://doi.org/10.1002/0471250953.bi1007s36.
Li, C., Zhang, J., Wu, Z. G., Cao, L., Yan, X., & Li, S. P. (2012). Biodegradation of buprofezin by Rhodococcus sp. strain YL-1 isolated from rice field soil. Journal of Agricultural and Food Chemistry, 60(10), 2531–2537. https://doi.org/10.1021/jf205185n.
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology, 8, 1–10. https://doi.org/10.1038/nbt.2676.
Metagenome of mango orchard soil (n.d.) https://submit.ncbi.nlm.nih.gov/subs/sra/SUB6674392/ overview.
Microbiome Analyst (n.d.) Comprehensive statistical, visual and meta-analysis of microbiome data. https://www.microbiomeanalyst.ca/faces/home.xhtml
Moghaddam, N. S., Zakaria, M. P., Omar, D., Sijam, K., & Khakvar, R. (2011). Effects of imidacloprid on the biodiversity of soil microbes in selected soils of Malaysia. 2nd International Conference on Environmental Science and Development, IPCBEE, 4, 7-10.
Muturi, E. J., Donthu, R. K., Fields, C. J., Moise, I. K., & Kim, C. H. (2017). Effect of pesticides on microbial communities in container aquatic habitats. Scientific Reports, 7, 44565. https://doi.org/10.1038/srep44565.
Pseudomonas mosselii (n.d.) strain NG1 16S ribosomal RNA gene, partial sequence. https://www.ncbi.nlm.nih.gov/nuccore/1736685383
Sangwan, N., Lata, P., Dwivedi, V., Singh, A., Niharika, N., Kaur, J., Anand, S., Malhotra, J., Jindal, S., Nigam, A., Lal, D., Dua, A., Saxena, A., Garg, N., Verma, M., Kaur, J., Mukherjee, U., Gilbert, J. A., Dowd, S. E., Raman, R., Khurana, P., Khurana, J. P., & Lal, R. (2012). Comparative metagenomic analysis of soil microbial communities across three hexachlorocyclohexane contamination levels. PLoS ONE, 7(9), e46219. https://doi.org/10.1371/journal.pone.0046219.
Sarkar, M. A., Roy, S., Kole, R. K., & Chowdhury, A. (2001). Persistence and metabolism of imidacloprid in different soils of West Bengal. Pest Management Science, 57, 598–602. https://doi.org/10.1002/ps.328.
Shah, V., Jain, K., Desai, C., & Madamwar, D. (2012). Molecular analyses of microbial communities involved in bioremediation. In T. Satyanarayana & B. N. Johri (Eds.), Microbes in environmental management and biotechnology (pp. 221–247). Amsterdam: Springer.
Shah, V., Zakrzewski, M., Wibberg, D., Eikmeyer, F., Schlüter, A., & Madamwar, D. (2013). Taxonomic profiling and metagenome analysis of a microbial community from a habitat contaminated with industrial discharges. Microbial Ecology, 66(3), 533–550. https://doi.org/10.1007/s00248-013-0253-9.
Shetti, A. A., & Kaliwal, B. B. (2016). Effect of imidacloprid on bacterial soil isolate Bacillus weihenstephanensis. In S. Trdan (Ed.), Insecticides resistance (pp. 275–294). Slovenia: IntechOpen. https://doi.org/10.5772/61503.
Singh, J., & Singh, D. K. (2005). Bacterial, azotobacter, actinomycetes, and fungal population in soil after diazinon, imidacloprid, and lindane treatments in groundnut (Arachis hypogaea L.) fields. Journal of Environmental Science and Health. Part. B, 40(5), 785–800. https://doi.org/10.1080/03601230500189725.
Sphingobacterium mucilaginosum (n.d.) strain NG201 16S ribosomal RNA gene, partial sequence. https://www.ncbi.nlm.nih.gov/nuccore/1783385310
Starner, K., & Goh, K. S. (2012). Detections of imidacloprid in surface waters of three agricultural regions of California, USA, 2010-2011. Bulletin of Environmental Contamination and Toxicology, 88(3), 316–321. https://doi.org/10.1007/s00128-011-0515-5.
Wang, F., Yao, J., Chen, H., Yi, Z., & Choi, M. M. F. (2014). Influence of short-time imidacloprid and acetamiprid application on soil microbial metabolic activity and enzymatic activity. Environmental Science and Pollution Research, 21, 10129–10138. https://doi.org/10.1007/s11356-014-2991-8.
Yousaf, S., Khan, S., & Aslam, M. (2013). Effect of pesticides on the soil microbial activity. Pakistan Journal of Zoology, 45, 1063–1067.
Zhang, Q., Xue, C., & Wang, C. (2015). Effects of imidacloprid on soil microbial communities in different saline soils. Environmental Science and Pollution Research, 22, 19667–19675. https://doi.org/10.1007/s11356-015-5154-7.
Code availability
Software codes are provided in the materials and methods section.
Funding
The authors are grateful to the Council of Science & Technology, Uttar Pradesh, Lucknow, India, for providing financial support of the present study in the form of a project grant vide Council’s Letter No. CST/AAS/D-1542 dated 02/08/2017.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Garg, N., Bhattacherjee, A.K., Shukla, P.K. et al. Influence of imidacloprid on bacterial community diversity of mango orchard soil assessed through 16S rRNA sequencing-based metagenomic analysis. Environ Monit Assess 193, 102 (2021). https://doi.org/10.1007/s10661-021-08885-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-021-08885-7