Abstract
The current study investigated how well an aqueous extract of Rubia cordifolia and a Bacillus amyloliquefaciens bacterium strain (DW6 OR083409) protected Vigna unguiculata L. plants from Fusarium oxysporum infection. In vitro study revealed that Rubia cordifolia aqueous extracts at 10% and 30% did not exhibit antifungal activity against F. oxysporum isolate, likewise no inhibition towards F. oxysporum as a presence of B. amyloliquefaciens DW6. Molecular identification characteristics confirmed the fungal pathogen being F. oxysporum AWEKA, based on the 18s rRNA sequence. B. amyloliquefaciens was found to produce indole-3-acetic acid, gibberellic acid, and hydrogen cyanide at concentrations being 203.67 ± 5.6, 335.6 ± 7.5, and 218 ± 6.4 µg/ml, respectively. In vivo, the growth of plants enhanced the induced resistance of cowpea plants against F. oxysporum during treatments with the biotic agents. The activity of defense-related enzymes was also enhanced, where Bacillus culture showed the highest increase, followed by the R. cordifolia at 30% extract and bacterial supernatant, respectively. SEM investigation of infected cowpea roots revealed notable differences in xylem vessels, including tylose formation and obstruction of vessels. Plasmolysis of parenchymal cells and hydrolysis of some cells were observed following the fungicide treatment. Both Bacillus amyloliquefaciens DW6 and the 30% aqueous extract of R. cordifolia proved effective in enhancing the induced resistance of cowpea against F. oxysporum, leading to a reduction in the damage caused by Fusarium root infection. Interestingly, this is the first report attaining the boosting of Vigna unguculata’s immune system towards F. oxysporum using aqueous extract of R. cordifolia and endophyte bacterium; B. amyloliquefaciens.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cowpea (Vigna unguiculata L.) is an annual legume crop valued for its edible seeds and animal feed. It serves as a significant source of proteins, essential vitamins, minerals, and amino acids in tropical regions (Enyiukwu et al., 2018; Falade et al., 2018). While primarily grown for its grain, a small percentage is cultivated for fresh pods in eastern Asia and as green leafy vegetables, fodder, or fresh pods in Africa (Boukar et al., 2015). With a well-established root system, cowpeas can reach heights of 80 cm for erect varieties and up to 2m for climbing cultivars. However, commercial production is greatly affected by pathogenic organisms and pests (Ogbuji & Isalar, 2021). Cowpeas are susceptible to various pests and diseases that hinder crop establishment, reduce forage production, and diminish seed yield (Nigam & Singh, 2023). Virulent diseases can also suppress nodulation in the root system, leading to negligible nitrogen fixation levels (Chihaoui et al., 2012). Fusarium oxysporum and Rhizoctonia solani are the main culprits behind cowpea wilt disease (Nene et al., 1989), posing a significant threat due to the chlamydospores' long-term presence in the soil which potentially limits crop rotation as a disease management strategy.
The imminent global population surge, projected to reach 9.1 billion by 2050 (Gonzalo et al., 2016), has created an urgent need to bolster food production. However, arable land is dwindling due to both abiotic and biotic factors. Consequently, industrial agriculture made extensive use of chemical fertilizers and fungicides which has caused ecological harm. As an alternative, biological agents are being increasingly favored over chemical treatments (Abo-Elyousr et al., 2021b; Pilkington et al., 2010).
The soil-borne fungus Fusarium oxysporum f.sp. lycopersici (Sacc.), is liable for producing Fusarium wilt disease in various vegetable crops, causing a loss in the harvest in greenhouses (Kannan et al., 2019). This pathogen poses a challenge due to its soil-borne nature and its ability to attack vascular tissues, making disease control difficult. Moreover, the emergence of new pathogen strains that can overcome host resistance further complicates the situation (Reis et al., 2005). Extensive research has been dedicated to the biological management of this pathogen (Aydi Ben Abdallah et al., 2016).
Plant growth-promoting rhizobacteria (PGPR) are important soil bacteria that inhabit the area around plant roots, enhancing plant growth and health (Abdelaziz et al., 2023), such as A. chevalieri and A. egyptiacus which induced plant immunity of Vicia faba (Daigham et al., 2023). These bacteria exhibit various beneficial traits, such as nitrogen fixation, mineral solubilization, and the assembly of growth-promoting chemicals (Dahal et al., 2017; Gouda et al., 2018; Mowafy et al., 2021, 2023).
Biotechnology-based biocontrol agents are gradually replacing chemical fungicides for the control of soil-borne fungal pathogens like Fusarium oxysporum, Rhizoctonia solani, and A. solani due to their perceived safety and lower environmental impact (Arfaoui et al., 2007; Attia et al., 2020; George et al., 2015). Recent studies have demonstrated the efficacy of Bacillus, Pseudomonas, and Rhizobium strains as bioagents against plant diseases (Kanthaiah & Velu, 2019).
Some resistance mechanisms are always active, while others are inducible, only activating when the plant faces a pathogenic attack or interacts with beneficial microbes (Abo-Elyousr et al., 2021a; Pieterse et al., 2012). Understanding the molecular basis of plant-pathogen interactions is crucial for developing effective biocontrol measures to manage Fusarium wilt disease. To defend themselves, plants rely on a complex network of genes. These genes code for hormone production to regulate growth under attack, pathogenesis-related proteins to directly combat invaders, and terpenoid/polyketide synthases to create antimicrobial compounds. Additionally, peroxidase and lignin synthase genes strengthen cell walls, while transcription factors act as master switches controlling defense gene activation. Calcium signaling genes facilitate communication within the plant, and UDP-glucosyltransferase and ubiquitin ligase genes help modify and target proteins for various defense functions (Wang et al., 2015).
Rubia cordifolia L. is a perennial herbaceous climbing plant belonging to the Rubiaceae family, characterized by its long, cylindrical, red roots. Widely distributed across tropical Asia, Africa, China, India, Japan, Tropical Australia, and Malaysia (Wen et al., 2022), Rubia cordifolia has a long history of traditional use in addressing various ailments, including abnormal uterine bleeding, purpura lupus erythematosus, hemorrhage syndrome, arthritis, kidney stones, hemostasis, and psoriasis (Wang et al., 2020). Studies have identified more than 100 different compounds in R. cordifolia, including anthraquinones, naphthoquinones, triterpenoids, and polysaccharides (Natarajan et al., 2018). Furthermore, it exhibits diverse biological activities such as anti-inflammatory, anticancer, immunosuppressive, and antioxidant effects (Balachandran et al., 2021; Lee et al., 2008). Notably, the phytoconstituents of R. cordifolia, particularly anthraquinones, have demonstrated in vivo potential as antifungal and antibacterial agents (Patil et al., 2009; Sittie et al., 1999), and antioxidants (Tripathi et al., 1995), as well as hypertensive and analgesic agents (Hussain et al., 2015).
The experimental work in this study encompasses a multifaceted approach to assess the potential of R. cordifolia extract and B. amyloliquefaciens DW6 bacterial culture as agents that induce fungal resistance to F. oxysporum in cowpea plants. Firstly, the preparation of R. cordifolia extract and bacterial culture sets the stage for subsequent investigations. The composition of R. cordifolia extract is characterized using gas chromatography, providing crucial insights into its chemical profile. Subsequently, both the R. cordifolia extract and bacterial culture underwent in vitro testing for antifungal activity against the fungal pathogen F. oxysporum using the double plate culture method. Following this, the efficacy of the extracts as treatments for cowpea seeds is evaluated to understand their effectiveness in inducing resistance in cowpea plants. Finally, the influence of the treatment on plant growth and yield is meticulously studied and compared to healthy and infested controls, shedding light on the potential benefits of these agents in bolstering plant health and productivity.
Materials and methods
Fungal pathogen
The fungus pathogen, Fusarium oxysporum, isolated under study was acquired from the Plant Pathology Research Institute, Agricultural Research Centre, Giza, Egypt. The pathogen was previously isolated from infected Vigna unguiculate plants and morphologically identified.
Molecular identification of the pathogen
DNA extraction was carried out following the protocol described by Innis et al. (2012). Genomic DNA from the fungal isolate was obtained by collecting hyphal fragments from a 14-day-old culture grown on potato dextrose agar (PDA). These hyphal fragments were meticulously removed from the culture plate using a sterile scalpel and pulverized into a fine powder using liquid nitrogen. The powdered hyphae were then suspended in 500 µl of extraction buffer (comprising 50 mM Tris–HCl pH 8, 150 mM NaCl, 100 mM EDTA, and 2% SDS) and incubated at 65°C for 30 min. DNA extraction involved two rounds of phenol/chloroform/isoamyl alcohol (25:24:1) treatment, followed by precipitation upon the addition of one volume of isopropanol. The resulting DNA pellet was washed with ethanol, dissolved in distilled water, and ultimately stored at -20°C for subsequent sequencing analysis.
The primers ITS 1 (5′ TCC GTA GGT GAA CCT GCGG 3′) and ITS 4 (5′TCC TCC GCT TAT TGA TATGC 3′) were used to amplify the ITS region of rDNA 60. The PCR program consisted of an initial denaturation step at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 68°C for 45 s, and extension at 72°C for 90 s. Agarose gel electrophoresis was performed to confirm the purity of the amplified PCR product's presence and size. The final extension step was performed at 72°C for 8 min.
The ABI Prism 3130xl Genetic Analyzer was used for sequencing the amplified PCR products. The primers (ITS 1 and ITS 4) utilized were the same ones outlined by White et al. (1990). The sequencing procedure was carried out at Macrogen Corp., Korea. The resulting DNA sequences have been deposited in the NCBI GenBank database.
To align and orient multiple sequences, the CLUSTALW algorithm was used. The phylogenetic tree reconstruction was performed using the "build" function of ETE3 v3.1.1 (Huerta-Cepas et al., 2016) available on the GenomeNet platform (https://www.genome.jp/tools/ete/).
Bacillus amyloliquefaciens DW6
Bacillus amyloliquefaciens DW6 (GenBank accession: OR083409) was kindly gifted by the Department of Botany, Faculty of Science, Mansoura University, Egypt, which was previously isolated from Arthrocnemum macrostachyum and grown on Luria–Bertani agar media as detailed by Sun et al. (2006).
B. amyloliquefaciens versus F. oxysporum
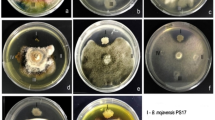
Evaluation of Bacillus amyloliquefaciens DW6 (GenBank accession number; OR083409) against the F. oxysporum pathogen was conducted utilizing the dual culture plate method as described by Mohamadpoor et al. (2022). A one-millimeter-diameter fungal disk, obtained from a five-day-old F. oxysporum culture, was placed one centimeter from the edge of each PDA plate. Conversely, a loopful of B. amyloliquefaciens was streaked one centimeter from the opposite side of the plate. Control plates contained only the fungal pathogen. The plates were then incubated at 25°C until the fungal growth completely covered the control plates, enabling comparison with the dual culture plates.
Rubia cordifolia aqueous extract preparation
Rubia cordifolia seeds (Giza 18 cultivar) were acquired from the Horticulture Research Institute, Agricultural Research Center, Giza, Egypt. For the extraction of active constituents from collected wild plant Rubia cordifolia, a traditional hot infusion method was employed. Dried Rubia cordifolia plant stems (20 g) were mixed with 200 mL of distilled water and shaken in a water bath at 70°C for 30 min. The resulting mixture was filtered, and the filtrate was stored at 4°C for further use, following the method described by El-Zayat et al. (2021).
All plant research and field studies, encompassing both cultivated and wild species, adhered to established guidelines and regulations. This includes protocols set forth by our institution, national and international governing bodies, the IUCN Policy Statement on Research with Threatened Species, and the Convention on International Trade in Endangered Species.
Rubia cordifolia aqueous extract versus F. oxysporum
Concentrations ranging from 10 to 70% of Rubia cordifolia aqueous extract were assessed for their impact on the growth of F. oxysporum AWEKAOR574875. Sterilized flasks containing PDA medium were prepared, with each concentration added separately before solidification. A one mm diameter disk of the previously grown pathogen was inoculated into the center of each PDA plate and then incubated at 25°C for five days. The daily linear growth of the pathogen was monitored and compared to its control, which was grown on the same medium without plant extract (Xu & Kim, 2014).
Identification of the active components of R. cordifolia extract
The volatile components of the extracted R. cordifolia were characterized using GC–MS analysis based on molecular mass and calculated fragments at specific retention times. The identification of the volatile components was based on the accompanying mass spectrum GC–MS using the R. cordifolia database. The structure, type, retention time, molecular weight, chemical formula, and percentage of each component in the extracted R. cordifolia were confirmed. The relative percentage of each volatile component was defined by matching its average peak area to the total areas to identify any distinctive molecules or groups with potential commercial and traditional medicinal uses.
Greenhouse experiment
Giza 18 cultivar of cowpea seeds was acquired from the Horticulture Research Institute, Agricultural Research Center, Giza, Egypt.
The commercial fungicide Topsin-70® Wp obtained from Sigma Company, which contains Thiophanate Methyl, was used at a concentration of 3 g/L to address root rot as stipulated by the Egyptian Ministry of Agriculture. F. oxysporum was cultured on PDA plates at 25 °C for five days. Mycelial plugs were then transferred to a sterilizing medium of sorghum: water: coarse sand mixture (2:2:1 v/v) and incubated at 25 °C for 10 days. The bacterial inoculum was prepared by scraping bacteria from slants using 5 mL of sterilized tap water. one mL of this inoculum was added to a 500-mL flask containing 100 mL of sterilized nutrient broth medium with an initial pH of 7. After sterilization at 121 °C for 15 min, the bacterial inoculum was incubated at 30 °C with shaking (150 rpm) for two days. Following centrifugation at 4 °C for 20 min at 5000 rpm using refrigerated centrifugation, the bacterial cells were rinsed with sterilized distilled water, and the number of viable cells was assessed using serial dilutions and the standard plate method. The concentration of the bacterial suspension was then adjusted to 108 CFU/mL (Youssef et al., 2010). The supernatant (medium with extracellular metabolites) and bacterial cells were stored separately under refrigeration until further use as treatment agents.
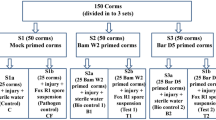
The objective of this experiment was to evaluate the efficacy of R. cordifolia aqueous extract and B. amyloliquefaciens bacterium in enhancing the resistance of cowpea plants to root rot caused by the fungus F. oxysporum, as well as promoting the overall growth. The pot experiment was performed at Tagelzz research station, Agricultural Research Center, Egypt (30° 57′ 25″ N and 31° 35′ 54″ E) in the summer season of 2022. Pots were filled with natural clayey soil, consisting of a 1:2 ratio of sand and clay. Infection with the pathogen was carried out through infestation of the pot soil. Half of the pots, representing 6 treatments, were intentionally infected with F. oxysporum at 0.4% of the soil weight and then watered. These pots were then left for 7 days to ensure uniform distribution of the pathogen within the soil. The remaining half of the pots were kept free from infection, representing 6 treatments. Before planting, cowpea seeds were soaked in either 10 or 30% concentrations of R. cordifolia extract for 10 min. Additionally, other seeds were soaked in bacterial supernatant or culture, while others were subjected to a fungicide. Each treatment contains 10 pots, every pot was seeded with 15 cowpea seeds.
Subsequently, the applied treatments included: (1) Healthy Control (without any treatment), (2) Infected soil, (3) Fungicide; Topsin-70® Wp, (4) Bacterial Supernatant, (5) Bacterial Culture (supernatant and cells), (6) Plant Extraction (10%), (7) Plant Extraction (30%), (8) Infected Soil with F. oxysporum + Fungicide, (9) Infected Soil with F. oxysporum + Bacterial Supernatant, (10) Infected Soil with F. oxysporum + Bacterial Culture, (11) Infected Soil with F. oxysporum + Plant Extract (10%), and (12) Infected Soil with F. oxysporum + Plant Extract (30%). The growing conditions and all other recommended agricultural practices were implemented as recommended for cowpea plants.
Disease assessment
Following 15 days from planting, the percentage of rotted cowpea seeds (un-emerged seeds) due to soil infection was recorded. After 45 days from planting, the post-infected seedlings and the surviving plants were assessed to determine plant survival rates.
Total phenol and flavonoid content of investigated leaves
The total phenolic content in fresh leaves and tubers was estimated by the Folin-Ciocalteu reagent method, following the procedure outlined by Blainski et al. (2013) and Abo-Elyousr et al. (2020). Quantification was achieved by establishing a standard curve of gallic acid, and the results were expressed as mg gallic acid/g of dried extract. Similarly, the total flavonoid content in fresh leaves and tubers was measured using a colorimetric method with aluminum chloride, as detailed by Li et al. (2015). The results were expressed as milligrams of quercetin equivalents (QE) per gram of the dried extract.
Antioxidant activity
The leaf extracts' reducing power was determined following the method outlined by Li et al. (2015), with the color of the reaction mixture assessed using a spectrophotometer at 700 nm. A higher absorbance indicates greater reducing power. Additionally, the antioxidant activity of the leaf extracts was assessed using two different methods: ABTS + cation radical scavenging and DPPH⋅ cation radical scavenging, as per the procedure described by Christodouleas et al. (2014). For the ABTS+ method, the absorbance of the resulting greenish-blue solution was measured at 734 nm, while for the DPPH⋅ method, the absorbance of the solution was measured at 517 nm, with the percentage of inhibition calculated using the decrease in absorbance.
Defense enzymes
Polyphenol oxidase and peroxidase activity in the extract were measured using a spectrophotometric method at 4°C. This method monitors the initial rate of absorbance increase at 410 nm, following the protocol established by Seleim et al. (2014).
Chlorophylls and carotene content
The measurement of total chlorophyll, chlorophyll a, chlorophyll b, and carotene was carried out using the method outlined by (Mackinney, 1941).
Indole-3-acetic acid, gibberellic acid, and hydrogen cyanide determination
The amount of IAA produced by B. amyloliquefaciens was measured using a method described by Loper (1986). Bacterial cultures were grown in LB broth supplemented with tryptophan (1.02 g/L) to assess indole-3-acetic acid (IAA) production. The culture's absorbance at 530 nm was measured, and a standard curve (10–100 μg/mL IAA) was used for quantification (Seleim et al., 2014). Gibberellic acid (GA) concentration was determined following Sharma et al. (2018) by measuring absorbance at 254 nm with a 5% HCl blank. A standard curve (100–1000 μg/mL GA) was used for quantification. Hydrogen cyanide (HCN) production was evaluated using King's B medium with glycine (4.4 g/L) after inoculation and incubation (48 h, 28 °C). A color change in the indicator paper from yellow to reddish-brown confirmed HCN production, as described by Bakker and Schippers (1987).
Vegetative growth and yield trait assessment
After 35 days from planting, three plants per treatment were collected and thoroughly rinsed to remove any soil particles. Various parameters related to vegetative growth were measured, including shoot length, root length, overall plant length, as well as the fresh and dry weights of the root and plant. Additionally, the leaf area was calculated in square centimeters. At the end of the plant life cycle, yield-related parameters were evaluated, including counting the number of pods, measuring both the fresh and dry weights of the pods, and calculating the weight of 100 seeds.
Cowpea root anatomy using SEM
To analyze the anatomical changes in cowpea roots in response to the tested pathogen of F. oxysporum, root cross-sections were made and gold-coated. The samples were then investigated using the SEM (TEM-2100, JEOL, Tokyo, Japan) connected to an accelerating voltage of 30 kV (Caldwell & Iyer-Pascuzzi, 2019).
Soil biological features
To measure the activity of nitrogenase in the soil around the roots of plants (rhizosphere soil), the acetylene reduction assay was used. The soil was treated with 1% glucose and the method described by Dart et al. (1972) and Okafor and MacRae (1973) was followed. A Hewlett-Packard 5890 gas chromatography Series 2 plus was used to measure the amount of ethylene produced. The gas chromatograph was equipped with two flame ionization detectors, one electron capture detector, an electron pressure control system, and a Plots/Al2O3 capillary column (50 × 0.53 mm × 15.0 μm film thickness). The injector, column, and detector were set at temperatures of 170, 120, and 220 °C, respectively. The analysis utilized nitrogen as the carrier gas at a flow rate of 10 mL/min. For detection, the flame ionization detector employed hydrogen and air at flow rates of 30 mL/min and 300 mL/min, respectively.
Rhizosphere soil samples were collected, ensuring the absence of any plant debris. Bacterial and fungal counts were determined using the following method: 10 g of soil were mixed with 90 ml of sterile water and shaken for 30 min at 120 rpm. Subsequently, a serial dilution was prepared. The bacterial total count was estimated by incubating in nutrient agar plates at 28°C for 3 days, while the total fungal count was determined by incubating in potato dextrose agar plates at 25°C for seven days. The resulting colony numbers were then counted, and the log rate of colony-forming units per gram of dry soil was calculated.
Statistical analysis
All experiments were conducted in triplicates. The pot experiments were arranged in a one-way randomized block design. CoStat (version 6.4, CoHort Software, Monterey, CA, USA) was used to perform ANOVA and identify significant treatments. The means were compared using Tukey's test, and differences were considered significant at alpha ≤ 0.05. Tukey's test is typically performed as a post-hoc test following an ANOVA for multiple comparisons.
Results
Antifungal activity of B. amyloliquefaciens and R. cordifolia aqueous extract against F. oxysporum
The potential of B. amyloliquefaciens against the growth of F. oxysporum was studied using the double plate method, where no indices of inhibition zones were observed. This bacterium, on the other hand, exhibited productivity of auxin-like substances, i.e., IAA and GA at concentrations of 203.67 ± 5.6 and 335.6 ± 7.5 μg/mL, respectively. Additionally, it produced 218 ± 6.4 µg/mL of hydrogen cyanide. The bacterium was harnessed to induce resistance in cowpeas against wilt root disease caused by F. oxysporum. Moreover, aqueous extracts of R. cordifolia at different concentrations (10% and 30%) were tested against F. oxysporum, with no significant suppression observed. The 10% concentration of R. cordifolia is the minimum concentration that supports the growth of cowpea seeds, while the 30% concentration was found to enhance seed germination more optimally.
Extract GC- MS analysis
The aqueous extract of R. cordifolia plants was analyzed using Gas Chromatography-Mass Spectrometry (GC–MS) to identify its volatile components. The analysis revealed the presence of thirty-five volatile components, that contribute to the plant's medicinal properties. The identification of these components was based on their molecular formulas, peak areas, and retention times, as detailed in Table 1 and Fig. 1. Methyl (7E,10E,13E)-icosa-7,10,13-trienoate was the first component with the least retention time (5.39 min), while Stigmast-5-en-3-ol, (3α,24S)- was the last component identified, with the longest retention time (39.94 min). The GC–MS chromatogram in Fig. 1 illustrates the abundance of each component at its respective retention time. The results in Table 1 specified that 99.97% of the total area percentages were recorded during the scanning retention time. The analysis also highlighted the most abundant volatile molecules, including methyl palmitate (16.08%), bis(6-methylheptyl) phthalate (11.26%), and methyl (E)-octadec-11-enoate (10.45%), among others (Fig. 2). Additionally, the volatile components were categorized as hydrocarbons, fatty acids, lipids, terpenes, steroids, and alkaloids.
Figure 2 classifies the volatile components into four main categories, with fatty acids and lipids comprising the most abundant class, representing 64.21% of the total composition percentage. This category included methyl palmitate (16.08%), methyl (7E,10E)-octadeca-7,10-dienoate (7.61%), and other components. Hydrocarbons ranked second in abundance, constituting 23.4% of the total composition percentage, with bis(6-methylheptyl) phthalate (11.26%) and (E)-1-methoxy-4-(prop-1-en-1-yl) benzene (6.78%) as the major components. Terpenes, steroids, and alkaloids accounted for 6.35% and 5.39% of the total composition percentage, respectively.
Fungal identification
The molecular characteristics of the fungal pathogen isolate under study were identified using PCR amplification to sequence the 18SrRNA gene.
The Genomic DNA of the F. oxysporum isolate was extracted from fresh mycelium. Gel electrophoresis illustrated that the amplified ITS fragment (600pb) of the fungus closely matched the target sequence of related fungi. The phylogenetic tree (Fig. 3) provided vital information. Nucleotide sequencing and Blast assessment revealed that the fungal isolate F. oxysporum shared over 95% sequence similarity with Fusarium oxysporum AWEKA (accession number: OR574875) in the Gene Bank database.
Disease assessment
Table 2 presents the impact of biotic and abiotic agents on damping-off and wilt percentages, as well as the survival rate of plants infected with F. oxysporum, compared to the fungicide Topsin-70® Wp. The data demonstrates the antifungal activity of both biotic and abiotic agents, as well as the Topsin-70® Wp fungicide, against F. oxysporum in infested soil. The fungicide showed the most significant reduction in damping-off and wilt percentages, followed by the bacterial culture, with no significant differences between them. Subsequently, soaking seeds in a 30% concentration of Rubia cordifolia aqueous extract for 10 min was the next most effective treatment for protecting cowpea plants from damping-off and root rot diseases caused by F. oxysporum, followed by the application of bacterial supernatant. In terms of the healthy survival of plants, the highest percentage was observed under fungicide treatment in un-infested soil, followed by the healthy control, and then the bacterial culture, without any significant differences among them.
Growth characters
The study assessed the impact of biotic and abiotic agents, in addition to the fungicide Topsin-70® Wp, on the growth parameters of cowpea plants in soil infested with Fusarium oxysporum. The results, presented in Tables 3 and 4, indicate that disease reduction affected the plants' morphological characteristics. All tested morphological characteristics significantly increased in both treated infested and un-infested soil when compared to the infested control.
Bacterial culture was the most effective, followed by Rubia cordifolia aqueous extract at 30%, and bacterial supernatant. The bacterial culture's effectiveness in promoting plant growth might be endorsed by its capability to dissolve phosphate and secrete IAA, while also indirectly inhibiting fungal growth by secreting antimicrobial pathogens, enzymes, and siderophores.
Cowpea nodulation status and nitrogenase activity
The results shown in Table 5 demonstrate a significant decrease in the number of nodules, as well as the fresh and dry weight of nodules in soil infested with the fungal pathogen, consequently leading to a decrease in nitrogenase enzyme activity. However, all treatments, whether biotic or abiotic, mitigated the adverse effects of fungal infection on these parameters, particularly nitrogenase activity. Bacterial culture exhibited the highest values of nodules, fresh and dry nodule weight, and nitrogenase activity, followed by a 30% concentration of Rubia cordifolia aqueous extract and bacterial supernatant. In contrast, the lowest nitrogenase activity values were observed in the infested control, followed by fungal infection + fungicide and fungicide alone. The fungicide Topsin-70® Wp exacerbated the detrimental impact of fungal infection on nitrogenase enzyme activity.
Yield components
Data from Table 6 revealed that all treatments, including exposure to biotic and abiotic stressors as well as the fungicide application, significantly boosted the number of pods per plant and their fresh weight, regardless of the presence of fungal pathogens in the soil. Interestingly, the combined effect of fungal infection and fungicide treatment resulted in a negligible increase in pod number compared to plants solely infected with the pathogen.
Physiological activities
Photosynthetic pigments
The photosynthetic process is crucial for all living organisms, providing the energy needed for growth and maintenance. All treatments except the fungicide improved these levels compared to the infected control. Chlorophyll in chloroplasts captures light energy, which is essential for photosynthesis. The findings in Table 7 showed a noteworthy increase in chlorophyll a, b, total chlorophylls, and carotenoids in cowpea leaflets in response to biotic, abiotic agents, and fungicide treatments compared to the infested control. The bacterial culture was most beneficial, significantly boosting all photosynthetic pigments. The 30% extract of Rubia cordifolia was also effective, significantly raising all pigments except chlorophyll b. Surprisingly, the fungicide further reduced pigment levels compared to the infected control. In particular, bacterial culture was most effective, followed by 30% Rubia cordifolia extract, then bacterial supernatant with or without infection. All infested treatments had lower values than the healthy control. Topsin-70® Wp fungicide significantly reduced photosynthetic pigments compared to the healthy control.
Total phenol, flavonoids, and defense-related enzymes
The levels of phenols, flavonoids, and defense-related enzymes in cowpea leaves in response to biotic and abiotic agents, with and without infestation by F. oxysporum, were investigated (Table 8). The data revealed that the maximum concentrations of total phenols and flavonoids were observed in the infested control group, followed by the fungicide treatment. In general, the application of R. cordifolia extracts led to a significant increase in both phenols and flavonoids compared to bacterial treatments and healthy control. Specifically, the 30% Rubia cordifolia extract was the most effective, followed by the 10% extract and the bacterial culture. Conversely, the lowest values of these factors were documented in the healthy control group.
Furthermore, the activity of polyphenol oxidase and peroxidase was significantly elevated in both infested and un-infested soil under the various treatments. Defense enzyme activity (polyphenol oxidase and peroxidase) was generally higher in all treatment groups compared to the healthy control. Among the treatments, seeds treated with the bacterial culture exhibited the highest activity of defense-related enzymes. This was followed by the 30% extract treatment and the bacterial supernatant treatment. The lowest enzyme activity was observed in the healthy control, infested control, and fungicide treatment groups (Table 8).
Antioxidant’s capacity
The study investigated the effect of various treatments on the antioxidant capacity of cowpea leaves infected with the F. oxysporum (Table 9). Compared to the healthy control, cowpea leaves infected with the fungus (treatment 2) exhibited significantly lower ABTS and DPPH inhibition, as well as reduced power. This indicates a decline in the leaves' ability to scavenge free radicals. All applied treatments (except 10% Rubia extract) significantly increased the antioxidant capacity compared to the infested control. This suggests that these treatments might help mitigate the negative effects of fungal infection on the leaves' antioxidant defense system. Among the treatments, the fungicide (Topsin-70® Wp) demonstrated the highest activity in all three antioxidant assays (ABTS, DPPH, reducing power). This suggests its effectiveness in protecting the cowpea leaves from oxidative damage caused by the fungus. The bacterial culture and its supernatant (treatments 4 & 5) also significantly increased antioxidant capacity compared to the infested control. However, their effect was less pronounced than the fungicide. The 30% Rubia cordifolia extract (treatment 7) significantly improved antioxidant activity compared to the infested control. However, the 10% extract (treatment 6) surprisingly resulted in a decrease in all antioxidant parameters. This suggests a concentration-dependent effect of the extract and the need for further investigation into its active components. Even when applied to already infected leaves (treatments 8, 9 & 10), the fungicide and bacterial treatments still offered some improvement in antioxidant capacity compared to the infected control.
Soil microflora
The soil microflora in the rhizosphere, including the number of bacteria and fungi, was analyzed at 60 and 90 days after planting to assess the effects of the agents (Table 10). At 60 days after planting, the soil infested with F. oxysporum showed a higher number of bacteria, with the highest count observed in the untreated soil (log 7.327). Conversely, the lowest bacterial count was found in the soil treated with fungicide (log 6.130). The highest bacterial count (log 6.737) was recorded after 90 days in the treatment with 30% plant extract concentration, while the lowest count was in the infested control. Correspondingly, the fungal counts were higher at 60 days compared to 90 days, with the highest count observed in the treatment with bacterial culture (log 5.973) and the lowest count in the F. oxysporum + extract 10% (log 4.540).
SEM anatomical features
In the infested soil, plants exhibited disease symptoms such as yellowing, necrotic lesions on the vegetative growth, root rot, wilt, and eventual plant death before reaching the flowering stage. Figure 4 depicts obvious variances in the cowpea root structure between cross sections under a scanning electron microscope, comparing a normal healthy plant (Fig. 4a) and an infected plant with F. oxysporum.
The infected root cross-section (Fig. 4b) displayed noticeable differences, including the generation of tyloses in the xylem vessels, with multiple tyloses (mTy) simultaneously arising from most of the vessels, leading to vessel blockage. Additionally, fungal hyphae growth was observed at the edges of the xylem vessels. Moreover, the dimensions of the xylem vessels were altered in the infected roots, potentially as an adaptation to compensate for reduced water absorption. The scanning electron microscope cross-section revealed plasmolysis of parenchyma cells associated with xylem vessels (Bcb) followed by the hydrolysis of some cells and closure of many xylem vessels due to the fungicidal effect (Fig. 3c). In contrast, other treatments, particularly the bacterial culture treatment, and 30% extract treatment, mitigated the injurious effects of F. oxysporum on the root’ structure, as shown in (Fig. 4d and e).
Discussion
Concerning the antibacterial activity of B. amyloliquefaciens and R. cordifolia aqueous extract against F. oxysporum, the data demonstrated no indices of inhibition zones were observed. However, B. amyloliquefaciens species was found to boost plant growth during the production of IAA, and GA. Otherwise, R. cordifolia aqueous extract at the two tested concentrations showed no inhibition against F. oxysporum. The concentration of aqueous extracts of R. cordifolia at 30% supported the germination of seeds. These data are complemented by Luo et al. (2022) who evaluated the antibacterial activity of some chemical composition of aerial parts of Rubia cordifolia, they found that these compounds had no potentiality against E. coli, P. aeruginosa, S. aureus, and Salmonella sp. Additionally, Barlow et al. (2016) investigated the anticancer and antioxidant activity of aqueous extract of R. cordifolia towards the MDA-MB231 breast cancer, whereas this aqueous extract exhibited no activity towards six bacterial species of Gram-positive and Gram-negative bacteria.
The GC–MS analysis of the R. cordifolia extract revealed a fascinating diversity of volatile components, with thirty-five distinct molecules identified. This chemical richness suggests the potential for various biological activities associated with the extract. The high abundance of fatty acids and lipids (64.21%) is particularly interesting in light of previous research. Studies by Basu et al. (2005) demonstrated the antimicrobial properties of R. cordifolia extract against various bacteria. Our findings on the prevalence of fatty acids and lipids align with this, suggesting these components might play a key role in the extract's antimicrobial activity. Additionally, Basu et al. (2005) identified specific R. cordifolia constituents like physcion and emodin with antimicrobial properties, further supporting the potential of the extract in this area. The identified volatile components also hint at possible antioxidant properties. Certain components within the extract might be comparable in abundance to vitamins C and E, well-known for their antioxidant capabilities (Joharapurkar et al., 2003). It's important to acknowledge the complexity of R. cordifolia's chemical composition. Luo et al. (2022) identified anthraquinones in the plant, but these compounds lacked antibacterial activity in their study. Conversely, Okhtia et al. (2020) reported significant antimicrobial activity of the extract. This highlights the potential for diverse bioactivities depending on the specific components present and their synergistic interactions. Furthermore, the potential presence of phenolics and flavonoids aligns with the findings of Kiran et al. (2019) who observed antioxidant activity in the ethanolic extract of R. cordifolia. Additionally, Zeng et al. (2023) identified components within the extract exhibiting anti-inflammatory properties. This finding might explain the reported effectiveness of R. cordifolia in reducing foot swelling and joint damage. In brief, the GC–MS analysis of the R. cordifolia extract unveiled a diverse array of volatile components, suggesting its potential for various biological activities, including antimicrobial, antioxidant, and anti-inflammatory properties. Further investigations are necessary to isolate and characterize the specific components responsible for these activities, along with exploring potential synergistic effects within the extract.
While gel electrophoresis confirmed amplification of the target 18S rRNA gene region, further analysis using a phylogenetic tree was necessary to pinpoint the exact fungal species. This tree acts like a map of evolutionary relationships, allowing us to compare the sequence of our isolate to known fungal species and visualize its closest relatives. The strong nucleotide sequence homology (over 95%) strongly supports the initial identification of the fungal isolate as F. oxysporum AWEKA (accession number: OR574875). This finding allows for a more precise understanding of the fungal pathogen under study. Further investigations could explore the specific virulence factors or pathogenic characteristics associated with this particular strain of F. oxysporum.
The effectiveness of the bacterial culture might be accredited to its aptitude to generate chitinase and glucanase, which are hydrolase enzymes capable of lysing chitin and β-glucan, essential components of fungal cell walls (Table 2). The hydrolysis of cell walls can decrease the integrity of the fungal cell wall, preventing them from infecting cowpea plants. Additionally, chitinase and β-glucanase enzymes are crucial for inhibiting fungal growth (Adams, 2004). Overall, both biotic and abiotic agent treatments reduced the harmful effects of Fusarium, preventing damping-off and root rot infections, except for the Rubia cordifolia aqueous extract at a 10% concentration, which showed an insignificant effect on damping-off under infested soil compared to the infested control. These results align with those obtained by Abbasi et al. (2019), who found that Streptomyces isolates induced resistance in tomato plants against Fusarium oxysporum f.sp. lycopersici race 3, mitigating Fusarium wilt of tomato, such as chlorosis, stunting, and wilting disease symptoms, better than carbendazim (fungicide). These results suggest that both biotic and abiotic agents have potential as eco-friendly approaches for managing Fusarium wilt in cowpea plants.
Furthermore, the study found that there were no substantial variances between the fungicide treatment and the healthy control under un-infested soil. Additionally, the data revealed that seed treatment with the bacterial culture is the most effective at enhancing growth characteristics, followed by the aqueous extract of Rubia cordifolia at 30%. Both tested biotic agents and the 30% extract neutralized the detrimental effects of Fusarium fungal infection on cowpea growth parameters. This is corroborated by Abbasi et al. (2019), who found that tomato plants responded positively to inoculation with Streptomyces isolates compared to infested control plants with Fusarium oxysporum f.sp. lycopersici Race 3. The enhanced growth of the plant is likely attributable to the bacterial isolate's capability to solubilize phosphate, synthesize growth hormones, and counteract the pathogenic fungus (Tables 3 and 4).
The results of Cowpea nodulation status and nitrogenase activity (Table 5) suggest that both the bacterial culture and the 30% plant extract likely work synergically by promoting the growth of beneficial nitrogen-fixing root bacteria. This co-action helps mitigate the negative effects of the fungal infection on cowpea plants, ultimately enhancing their overall nitrogen fixation abilities.
Fusarium infection significantly reduced the number and weight of nodules and nitrogenase activity, indicating disruption in the cowpea-rhizobia symbiotic relationship. This disruption likely results from Fusarium-induced root damage, competition for nutrients, and altered symbiosis signaling pathways due to phytohormone disruption (Srivastava et al., 2024). However, all treatments (bacterial culture, Rubia cordifolia extract, and bacterial supernatant) mitigated Fusarium's negative effects on nodulation and nitrogenase activity, suggesting their potential to promote a healthy symbiotic relationship even in the presence of the pathogen (Naseri, 2019; Srivastava et al., 2024). The bacterial culture showed the greatest improvement, likely due to its ability to suppress Fusarium, produce growth-promoting substances, or induce plant defense mechanisms. The 30% extract may enhance nodulation by promoting root growth and development or inducing plant defense responses that benefit rhizobia (Singh et al., 2010). Conversely, the fungicide Topsin-70® Wp further reduced nitrogenase activity, suggesting potential toxicity toward rhizobia or disruption of the microbial balance in the rhizosphere (Naseri, 2019).
Notably, treatment with bacterial culture showed the highest increase in cowpea yield productivity, with no significant differences compared to the 30% Rubia cordifolia extract, while the 10% extract exhibited a delayed effect (Table 6). Overall, under infested soil conditions, all the tested treatments resulted in lower productivity compared to the healthy control, except for the bacterial culture, which demonstrated the opposite effect. Similarly, the bacterial culture treatment had a positive impact on all vegetative growth parameters, reflecting its influence on yield parameters through a similar mechanism. Interestingly, the fungicide offered little benefit in managing the fungal infection. In uninfected conditions, only the bacterial culture treatment achieved a similar yield to the healthy control group. This suggests the bacterial culture promotes overall plant health and yield, even under fungal pressure.
Analyzing the impact on physiological activities (Table 7), particularly photosynthetic pigments reveals interesting trends. All treatments except the fungicide improved these pigments compared to infected controls. The bacterial culture was most effective, significantly boosting all pigments, likely due to its growth-promoting effects. The 30% Rubia cordifolia extract also improved most pigments, suggesting it might trigger plant defense mechanisms that enhance chlorophyll and carotenoid production. Surprisingly, the fungicide further reduced pigment levels, highlighting a potential negative impact on plant physiology beyond targeting the fungus.
The significant increase in chlorophyll a, b, total chlorophyll, and carotenoids with the bacterial culture treatment indicates a positive influence on the plant's photosynthetic machinery. This may be due to improved nutrient availability, production of growth-promoting hormones, and induced plant defense mechanisms (Bejandi et al., 2012). The 30% extract, while not directly inhibiting the fungus, increased photosynthetic pigments, possibly by triggering plant defense responses and enhancing carotenoid synthesis. Carotenoids, which protect against oxidative stress, may be stimulated by the extract to mitigate Fusarium-induced stress (Stefan et al., 2013). Conversely, the fungicide Topsin-70® Wp reduced pigment levels, indicating potential negative impacts on the plant's physiological processes. This observation suggests a need for further investigation into the fungicide's phytotoxic effects.
In the context of managing Fusarium wilt in cowpeas, our study highlights the complex trade-offs associated with fungicide use. While Topsin-70® Wp demonstrated the strongest enhancement of antioxidant activity in cowpea leaves, likely by mitigating fungal-induced oxidative damage (current study), its detrimental effects on root xylem integrity, nitrogen fixation via potential rhizobia toxicity, and photosynthetic pigment levels raise concerns about long-term plant health (current study). These observations extend beyond the fungicide's intended target (Fusarium) and suggest potential disruption of crucial physiological processes. While the short-term increase in antioxidant activity might be beneficial, the negative impacts on other plant functions could compromise long-term growth and productivity. Furthermore, broad-spectrum fungicides like Topsin-70® Wp can disrupt the natural soil microbiome, potentially increasing susceptibility to other diseases (Verma et al., 2017). This disruption can occur by targeting not only the pathogenic fungus but also beneficial microbes that play a vital role in plant health and nutrient cycling. Another significant concern is the development of fungicide resistance. Overreliance on a single fungicide can lead to the evolution of resistant fungal populations over time, rendering the treatment ineffective in the future (Lukasko & Hausbeck, 2024). This necessitates the continuous development and implementation of alternative control methods to ensure sustainable disease management.
The bacterial culture treatment resulted in the strongest activity of defense-related enzymes, followed by the 30% extract, suggesting they contribute to the plant's defense response. Similar results were observed in tomato plants, where the catalase and peroxidase enzymes were increased in response to the induction of Streptomyces species on Fusarium oxysporum f.sp. lycopersici race 3 (Abbasi et al., 2019).
The data on total phenolics, flavonoids, and defense enzymes offer valuable insights into the impact of the treatments on cowpea's defense mechanisms against Fusarium wilt. Interestingly, the infested control group displayed the highest concentrations of phenolics and flavonoids, suggesting the plant's natural response to infection (Abo-Elyousr et al., 2022). The fungicide treatment also elevated these compounds, possibly due to a general stress response triggered by the fungicide itself. The most significant observation is the marked increase in phenolics and flavonoids following the application of Rubia cordifolia extracts, particularly the 30% concentration. This suggests that the extract is a potent inducer of the plant's defense system, producing compounds that can combat fungal pathogens (Abo-Elyousr et al., 2020). This mechanism might explain the extract's effectiveness in mitigating Fusarium wilt despite not directly inhibiting fungal growth. The fungicide treatment, in contrast, showed the lowest enzyme activity alongside the healthy and infested control groups. This could be due to the fungicide directly targeting the pathogen, potentially reducing the need for the plant to activate its defense systems. However, this also aligns with the observation of reduced photosynthetic pigments with the fungicide, suggesting potential unintended consequences on plant health beyond its intended target (Abo-Elyousr et al., 2020).
The study confirms the detrimental effect of Fusarium wilt on the antioxidant capacity of cowpea leaves. Compared to healthy controls, infected leaves displayed a significant decline in ABTS and DPPH radical scavenging activity, as well as diminished reducing power. This indicates a weakened ability to combat free radicals, potentially contributing to oxidative stress and tissue damage caused by the fungus. Opportunely, all applied treatments (except the 10% Rubia extract) considerably enhanced the antioxidant capacity compared to the infected control. This suggests their potential to mitigate the negative effects of Fusarium wilt by bolstering the plant's antioxidant defense system. Interestingly, even when applied to already infected leaves, the fungicide and bacterial treatments still offered some improvement in antioxidant capacity compared to the control. This suggests their potential for both preventative and remedial strategies against Fusarium wilt. Generally, this study indicated that fungicides and potentially beneficial microbes may promote the antioxidant defense mechanisms in cowpea leaves against fungal infection. This opens doors for treating cowpea health and potentially improving the nutritional value of the leaves through strategic treatments.
The analysis of soil microflora in the rhizosphere reveals fascinating insights into the potential impact of various treatments on the microbial community and its interaction with Fusarium wilt (Table 10). The presence of F. oxysporum led to changes in the bacterial and fungal profiles of the rhizosphere, with both microbial counts decreasing over 90 days compared to those obtained after 60 days. This effect may be assigned for the occurrence of F. oxysporum, the bacterial culture of B. amyloliquefaciens, and/or the R. cordifolia extract treatment comprising various phyto-components such as hydrocarbons, terpenes, alkaloids, steroids, fatty acids, and lipids. Several factors contribute to the different profiles of bacteria and fungi in the rhizosphere, directly or indirectly related to plant physiology and root exudates. For instance, Fusarium may outcompete other microbes for resources, altering the rhizosphere microbiome's composition. Soil enzyme activity also influences the microbial community (Jangir et al., 2019). Root exudates, which are secretions providing nutrients for beneficial soil microbes, improve soil respiration and promote plant growth by positively altering the microbial community's composition (Adamczyk et al., 2021; Nazir et al., 2017). Particularly, as the plant ages, the decrease in the amount of root exudates, potentially leads to a reduction in the rhizosphere microbe count (Moussa et al., 2017).
The SEM images of cowpea root cross-sections (Fig. 4) provide compelling evidence of the detrimental effects of Fusarium wilt on root anatomy and the potential protective role of certain treatments. Fungal pathogens are known to produce ethylene gas, which induces harmful changes in the anatomical structure of various plant organs. Ethylene promotes the activity of exo- and endo-cellular hydrolytic enzymes, such as pectin methyl esterase, polygalacturonase, and cellulose, which ultimately lead to the maceration (softening and disintegration) of tissues (El-Samra et al., 1994). Fungal pathogens such as Macrophomina phaseolina, Rhizoctonia solani, F. oxysporum, and F. solani can cause deformation in the anatomical structures of the soybean stem's basal portion. This deformation primarily affects the epidermis, cortex, and pith, leading to complete disruption of epidermal cells, severe plasmolysis in cortical cells, and destruction of outer cortical cells (Abd El-Hai et al., 2010). The bacterial culture might produce antifungal compounds or compete with Fusarium for resources, while the extract might stimulate the plant's defense mechanisms.
Bacillus amyloliquefaciens, a ubiquitous soil bacterium, offers a promising alternative approach for managing Fusarium wilt in cowpeas. These microorganisms display remarkable antifungal properties, including a variety of lipopeptides, such as fengycin and bacteriocins. Hence, these compounds directly target fungal pathogens, preventing their growth and spore germination (Imran et al., 2022). Furthermore, B. amyloliquefaciens can colonize plant roots, acting as a biological control agent by competitively excluding the fungal pathogen Fusarium from vital space and nutrients, hindering its establishment (Mousa et al., 2021). In contrast, the 30% Rubia cordifolia extract (Fig. 4e) appears to work indirectly by potentially stimulating the plant's inherent defense mechanisms. Studies have shown that extracts from this plant can induce the production of pathogenesis-related (PR) proteins and secondary metabolites within the plant itself. These PR proteins and metabolites can directly inhibit fungal growth or even attract beneficial microbes that actively suppress the pathogen (Abd El-Wahed et al., 2023).
When kidney bean roots were infected with a mixture of pathogenic fungi (Fusarium oxysporum, Fusarium solani, Rhizoctonia solani, and Pythium ultimum), significant structural damage was noticed. The epidermis was destroyed, and the cortex tissue exhibited separation in some areas. Consequently, there was decomposition and hydrolysis of cellular elements (Abd El-Hai & Ali, 2018). In the case of faba bean plants infected with F. solani, the xylem vessels underwent hydrolysis, leading to their separation and eventual disruption. Cross sections of the infected plants displayed black areas may be due to the degradation and dissolution of cell components, with fungal hyphae found in the intracellular spaces and infected cells (El-Sayed, 2022). Moreover, infection of bean roots with Pythium fungi resulted in dramatic changes in the root cross-section shape, which might be due to the degradation and dissolution of pith cell components, which leads to cell death and the appearance of black areas in the central part of the root cross-section (Ghoniem et al., 2021). According to Beck (2010), fungal infections cause anatomical changes in different plant organs, including swelling, hydrolysis (dissolution) of cell components, and eventually tissue maceration due to increased ethylene production. The study also found that tylosis formation in cowpea roots infected by fungi could block the vessels, preventing pathogen entry and reducing water loss. The changes in the dimensions of xylem vessels in infected roots may represent an adaptive response to water absorption limitations. The positive effects of the bacterial culture and 30% extract treatment on cowpea root structure may be attributed to their antifungal properties, which could mitigate the harmful effects of F. oxysporum.
Conclusion
In this investigation, we explored eco-friendly methods to safeguard Vigna unguiculata from the detrimental effects of Fusarium oxysporum, a fungal pathogen responsible for wilt disease. The study revealed two promising biocontrol agents with distinct modes of action. The 30% aqueous extract of Rubia cordifolia emerged as a powerful inducer of induced resistance in cowpeas. While not directly inhibiting fungal growth in lab tests, it significantly bolstered cowpea's natural defense mechanisms. The extract promoted the production of chlorophyll and carotenoids, crucial for photosynthesis, and boosted the levels of phenolics, flavonoids, and defense enzymes within cowpea leaves. This multi-faceted approach strengthens cowpea's inherent ability to resist Fusarium infection, offering a potential and more sustainable alternative to traditional fungicides. The bacterium Bacillus amyloliquefaciens DW6, although not exhibiting direct antifungal activity in vitro, proved beneficial in vivo experiments. These growth-promoting substances stimulate root development, potentially aiding plants in better withstanding Fusarium's attack. Overall, this study paves the way for developing sustainable cowpea production practices. By harnessing the power of Rubia cordifolia extract and Bacillus amyloliquefaciens DW6, these eco-friendly approaches can be used to protect cowpea crops.
Data availability
The datasets generated and/or analyzed during the current study are available in the current paper, supplementary file, and the GenBank repository under accession number OR574875 which can be found at the following link: https://www.ncbi.nlm.nih.gov/nuccore/OR574875.1?report=GenBank
References
Abbasi, S., Safaie, N., Sadeghi, A., & Shamsbakhsh, M. (2019). Streptomyces strains induce resistance to Fusarium oxysporum f. sp. lycopersici race 3 in tomato through different molecular mechanisms. Frontiers in Microbiology, 10, 1505.
Abd El-Hai, K., & Ali, A. (2018). Amelioration of the structural and biochemical features of kidney bean against root rot and rust diseases. Journal of Plant Protection and Pathology, 9(3), 237–245. https://doi.org/10.21608/jppp.2018.41396
Abd El-Hai, K. M., El-Metwall, M. A., & El-Baz, S. M. (2010). Reduction of soybean root and stalk rots by growth substances under salt stress conditions. Plant Pathology Journal, 9(4), 149–161. https://doi.org/10.3923/ppj.2010.149.161
Abd El-Wahed, M. H., Bereika, M. F., Abo-Elyousr, K. A., & Almasoudi, N. M. (2023). Integration of Pseudomonas fluorescens and Rosemarinus officinalis for controlling of potato bacterial wilt. Egyptian Journal of Biological Pest Control, 33(1), 31.
Abdelaziz, A. M., Hashem, A. H., El-Sayyad, G. S., El-Wakil, D. A., Selim, S., Alkhalifah, D. H. M., & Attia, M. S. (2023). Biocontrol of soil borne diseases by plant growth promoting rhizobacteria. Tropical Plant Pathology, 48(2), 105–127. https://doi.org/10.1007/s40858-022-00544-7
Abo-Elyousr, K. A., Almasoudi, N. M., Abdelmagid, A. W., Roberto, S. R., & Youssef, K. (2020). Plant extract treatments induce resistance to bacterial spot by tomato plants for a sustainable system. Horticulturae, 6(2), 36.
Abo-Elyousr, K. A., Abdel-Rahim, I. R., Almasoudi, N. M., & Alghamdi, S. A. (2021a). Native endophytic Pseudomonas putida as a biocontrol agent against common bean rust caused by Uromyces appendiculatus. Journal of Fungi, 7(9), 745.
Abo-Elyousr, K. A., Al-Qurashi, A. D., & Almasoudi, N. M. (2021b). Evaluation of the synergy between Schwanniomyces vanrijiae and propolis in the control of Penicillium digitatum on lemons. Egyptian Journal of Biological Pest Control, 31, 1–10.
Abo-Elyousr, K. A., Imran, M., Almasoudi, N. M., Ali, E. F., Hassan, S., Sallam, N. M. A., Youssef, K., Abdel-Rahim, I. R., & Khalil Bagy, H. M. (2022). Controlling of Xanthomonas axonopodis pv. phaseoli by induction of phenolic compounds in bean plants using salicylic and benzoic acids. Journal of Plant Pathology, 104(3), 947–957.
Adamczyk, M., Rüthi, J., & Frey, B. (2021). Root exudates increase soil respiration and alter microbial community structure in alpine permafrost and active layer soils. Environmental Microbiology, 23(4), 2152–2168. https://doi.org/10.1111/1462-2920.15383
Adams, D. J. (2004). Fungal cell wall chitinases and glucanases. Microbiology, 150(7), 2029–2035. https://doi.org/10.1099/mic.0.26980-0
Arfaoui, A., El Hadrami, A., Mabrouk, Y., Sifi, B., Boudabous, A., El Hadrami, I., Daayf, F., & Chérif, M. (2007). Treatment of chickpea with Rhizobium isolates enhances the expression of phenylpropanoid defense-related genes in response to infection by Fusarium oxysporum f. sp. ciceris. Plant Physiology and Biochemistry, 45(6–7), 470–479. https://doi.org/10.1016/j.plaphy.2007.04.004
Attia, M. S., El-Sayyad, G. S., Abd Elkodous, M., & El-Batal, A. I. (2020). The effective antagonistic potential of plant growth-promoting rhizobacteria against Alternaria solani-causing early blight disease in tomato plant. Scientia Horticulturae, 266, 109289. https://doi.org/10.1016/j.scienta.2020.109289
Aydi Ben Abdallah, R., Mokni-Tlili, S., Nefzi, A., Jabnoun-Khiareddine, H., & Daami-Remadi, M. (2016). Biocontrol of Fusarium wilt and growth promotion of tomato plants using endophytic bacteria isolated from Nicotiana glauca organs. Biological Control, 97, 80–88. https://doi.org/10.1016/j.biocontrol.2016.03.005
Bakker, A. W., & Schippers, B. (1987). Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas SPP-mediated plant growth-stimulation. Soil Biology and Biochemistry, 19(4), 451–457. https://doi.org/10.1016/0038-0717(87)90037-x
Balachandran, P., Ibrahim, M. A., Zhang, J., Wang, M., Pasco, D. S., & Muhammad, I. (2021). Crosstalk of cancer signaling pathways by cyclic hexapeptides and anthraquinones from Rubia cordifolia. Molecules (Basel, Switzerland), 26(3), 735. https://doi.org/10.3390/molecules26030735
Barlow, R., Barnes, D., Campbell, A., Nigam, P., & Owusu-Apenten, R. (2016). Antioxidant, anticancer and antimicrobial, effects of Rubia cordifolia aqueous root extract. Journal of Advances in Biology & Biotechnology, 5(1), 1–8.
Basu, S., Ghosh, A., & Hazra, B. (2005). Evaluation of the antibacterial activity of Ventilago madraspatana Gaertn., Rubia cordifolia Linn. and Lantana camara Linn.: isolation of emodin and physcion as active antibacterial agents. Phytotherapy Research: An International Journal Devoted to Pharmacological and Toxicological Evaluation of Natural Product Derivatives, 19(10), 888–894.
Beck, C. B. (2010). An Introduction to Plant Structure and Development. Cambridge University Press. https://doi.org/10.1017/cbo9780511844683
Bejandi, T. K., Sharifii, R. S., Sedghi, M., & Namvar, A. (2012). Effects of plant density, Rhizobium inoculation and microelements on nodulation, chlorophyll content and yield of chickpea (Cicer arietinum L.). Annals of Biological Research, 3(2), 951–958.
Blainski, A., Lopes, G. C., & de Mello, J. C. P. (2013). Application and analysis of the folin ciocalteu method for the determination of the total phenolic content from Limonium brasiliense L. Molecules (Basel, Switzerland), 18(6), 6852–6865. https://doi.org/10.3390/molecules18066852
Boukar, O., Fatokun, C. A., Roberts, P. A., Abberton, M., Huynh, B. L., Close, T. J., Kyei-Boahen, S., Higgins, T. J. V., & Ehlers, J. D. (2015). Cowpea. In: De Ron, A. (Ed.), Grain Legumes. Handbook of Plant Breeding, vol 10. Springer. https://doi.org/10.1007/978-1-4939-2797-5_7
Caldwell, D., & Iyer-Pascuzzi, A. S. (2019). A scanning electron microscopy technique for viewing plant− microbe interactions at tissue and cell-type resolution. Phytopathology, 109(7), 1302–1311.
Chihaoui, S. A., Mhadhbi, H., & Mhamdi, R. (2012). The antibiosis of nodule-endophytic agrobacteria and its potential effect on nodule functioning of Phaseolus vulgaris. Archives of Microbiology, 194, 1013–1021.
Christodouleas, D. C., Fotakis, C., Nikokavoura, A., Papadopoulos, K., & Calokerinos, A. C. (2014). Modified DPPH and ABTS Assays to Assess the Antioxidant Profile of Untreated Oils. Food Analytical Methods, 8(5), 1294–1302. https://doi.org/10.1007/s12161-014-0005-6
Dahal, B., NandaKafle, G., Perkins, L., & Brözel, V. S. (2017). Diversity of free-Living nitrogen fixing Streptomyces in soils of the badlands of South Dakota. Microbiological Research, 195, 31–39. https://doi.org/10.1016/j.micres.2016.11.004
Daigham, G. E., Mahfouz, A. Y., Abdelaziz, A. M., Nofel, M. M., & Attia, M. S. (2023). Protective role of plant growth-promoting fungi Aspergillus chevalieri OP593083 and Aspergillus egyptiacus OP593080 as biocontrol approach against Alternaria leaf spot disease of Vicia faba plant. Biomass Conversion and Biorefinery. https://doi.org/10.1007/s13399-023-04510-4
Dart, P., Day, J., & Harris, D. (1972). Assay of nitrogenase activity by acetylene reduction. Use of Isotopes for Study of Fertilizer Utilization by Legume Crops, 149, 85–100.
El-Samra, I., Noman, K., Fayed, M., & El-Farnawany, M. (1994). Studies on some histopathological and enzymatic activity aspects during the Rhizoctonia damping-off disease of beans. Journal of Agricultural Science, Mansoura Univ.(Egypt), 19(7), 2209–2225.
El-Sayed, S. (2022). Collaborative potentialities of Trichoderma spp. and Saccharomyces cerevisiae against damping-off and root rot diseases of Faba Bean. Egyptian Journal of Phytopathology, 50(1), 65–78. https://doi.org/10.21608/ejp.2022.134177.1060
El-Zayat, M. M., Eraqi, M. M., Alrefai, H., El-Khateeb, A. Y., Ibrahim, M. A., Aljohani, H. M., Aljohani, M. M., & Elshaer, M. M. (2021). The antimicrobial, antioxidant, and anticancer activity of greenly synthesized selenium and zinc composite nanoparticles using Ephedra aphylla extract. Biomolecules, 11(3), 470.
Enyiukwu, D. N., Amadioha, A. C., & Ononuju, C. C. (2018). Nutritional significance of cowpea leaves for human consumption. Greener Trends in Food Science and Nutrition, 1(1), 1–10. https://doi.org/10.15580/gtfsn.2018.1.061818085
Falade, M., Enikuomehin, O., Borisade, O., & Aluko, M. (2018). Control of cowpea (Vigna unguiculata L. Walp) diseases with intercropping of maize (Zea mays L) and spray of plant extracts. Journal of Advances in Microbiology, 7(4), 1–10. https://doi.org/10.9734/jamb/2017/38156
George, E., Kumar, S. N., Jacob, J., Bommasani, B., Lankalapalli, R. S., Morang, P., & Kumar, B. S. D. (2015). Characterization of the Bioactive Metabolites from a Plant Growth-Promoting Rhizobacteria and Their Exploitation as Antimicrobial and Plant Growth-Promoting Agents. Applied Biochemistry and Biotechnology, 176(2), 529–546. https://doi.org/10.1007/s12010-015-1593-3
Ghoniem, A. A., Abd El-Hai, K. M., El-Khateeb, A. Y., Eldadamony, N. M., Mahmoud, S. F., & Elsayed, A. (2021). Enhancing the potentiality of Trichoderma harzianum against Pythium pathogen of beans using Chamomile (Matricaria chamomilla, L.) flower extract. Molecules (Basel, Switzerland), 26(4), 1178. https://doi.org/10.3390/molecules26041178
Gonzalo, J. A., Alfonseca, M., & Perez, F.-F. M. (2016). Quantitative estimates of the future. World Population Past, Present & Future, by World Scientific Publishing Co. Pte. Ltd., 5 Toh Tuck Link, Singapore 596224, p. 143.
Gouda, S., Kerry, R. G., Das, G., Paramithiotis, S., Shin, H.-S., & Patra, J. K. (2018). Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiological Research, 206, 131–140. https://doi.org/10.1016/j.micres.2017.08.016
Huerta-Cepas, J., Serra, F., & Bork, P. (2016). ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Molecular Biology and Evolution, 33(6), 1635–1638.
Hussain, L., Akash, M. S., Ain, N.-U., Rehman, K., & Ibrahim, M. (2015). The analgesic, anti-inflammatory and anti-pyretic activities of Tinospora cordifolia. Advances in Clinical and Experimental Medicine, 24(6), 957–964.
Imran, M., Abo-Elyousr, K. A., Mousa, M. A., & Saad, M. M. (2022). A study on the synergetic effect of Bacillus amyloliquefaciens and dipotassium phosphate on Alternaria solani causing early blight disease of tomato. European Journal of Plant Pathology, 162(1), 63–77.
Innis, M. A., Gelfand, D. H., Sninsky, J. J., & White, T. J. (2012). PCR protocols: a guide to methods and applications. Academic press.
Jangir, M., Sharma, S., & Sharma, S. (2019). Non-target effects of Trichoderma on Plants and Soil Microbial Communities. In A. Varma, S. Tripathi, & R. Prasad (Eds.), Plant Microbe Interface (pp. 239–251). Springer International Publishing.
Joharapurkar, A. A., Zambad, S. P., Wanjari, M. M. & Umathe, S. N. (2003). In vivo evaluation of antioxidant activity of alcoholic extract of Rubia cordifolia Linn. and its influence on ethanol-induced immunosuppression. Indian Journal of Pharmacology, 35(4), 232–236.
Kannan, K., Shibinaya, N., & Umamaheswari, M. (2019). Control of Fusarium wilt disease in cowpea plant (Vigna Unguiculata L.). using secondary metabolites produced in Bradyrhizobium japonicum. Kongunadu Research Journal, 6(2), 28–36. https://doi.org/10.26524/krj298
Kanthaiah, K., & Velu, R. K. (2019). Characterization of the bioactive metabolite from a plant growth promoting rhizobacteria Pseudomonas aeruginosa VRKK1 and exploitation of antibacterial behaviour against Xanthomonas campestris a causative agent of bacterial blight disease in cowpea. Archives of Phytopathology and Plant Protection, 55(7), 797–814. https://doi.org/10.1080/03235408.2018.1557883
Kiran, F., Khan, M. A., Batool, R., Kanwal, S., Shah, S. L., & Mahmood, T. (2019). Biological evaluation of some important medicinal plants from Poonch valley, Azad Kashmir, Pakistan. Journal of Traditional Chinese Medicine, 39(6), 753–763.
Lee, J.-E., Hitotsuyanagi, Y., Kim, I.-H., Hasuda, T., & Takeya, K. (2008). A novel bicyclic hexapeptide, RA-XVIII, from Rubia cordifolia: Structure, semi-synthesis, and cytotoxicity. Bioorganic & Medicinal Chemistry Letters, 18(2), 808–811. https://doi.org/10.1016/j.bmcl.2007.11.030
Li, J.-E., Fan, S.-T., Qiu, Z.-H., Li, C., & Nie, S.-P. (2015). Total flavonoids content, antioxidant and antimicrobial activities of extracts from Mosla chinensis Maxim. cv. Jiangxiangru. LWT Food Science and Technology, 64(2), 1022–1027. https://doi.org/10.1016/j.lwt.2015.07.033
Loper, J. E. (1986). Influence of bacterial sources of indole-3-acetic acid on root elongation of sugar beet. Phytopathology, 76(4), 386. https://doi.org/10.1094/phyto-76-386
Lukasko, N. T., & Hausbeck, M. K. (2024). Resistance to seven site-specific fungicides in Botrytis cinerea from greenhouse-grown ornamentals. Plant Disease, 108(2), 278–285.
Luo, H., Qin, W., Zhang, H., Ren, F.-C., Fang, W.-T., Kong, Q.-H., Yang, L., Zhang, J.-M., Fang, C.-W., & Hu, J.-M. (2022). Anthraquinones from the aerial parts of Rubia cordifolia with their NO inhibitory and antibacterial activities. Molecules (Basel, Switzerland), 27(5), 1730.
Mackinney, G. (1941). Absorption of light by chlorophyll solutions. Journal of Biological Chemistry, 140(2), 315–322.
Mohamadpoor, M., Amini, J., Ashengroph, M., & Azizi, A. (2022). Evaluation of biocontrol potential of Achromobacter xylosoxidans strain CTA8689 against common bean root rot. Physiological and Molecular Plant Pathology, 117, 101769. https://doi.org/10.1016/j.pmpp.2021.101769
Mousa, M. A., Abo-Elyousr, K. A., Abdel Alal, A. M., & Alshareef, N. O. (2021). Management fusarium wilt disease in tomato by combinations of bacillus amyloliquefaciens and peppermint oil. Agronomy, 11(12), 2536.
Moussa, Z., El-Hersh, M. S., & El-Khateeb, A. Y. (2017). Induction of potato resistance against bacterial wilt disease using Saccharomyces cerevisiae. Biotechnology (Faisalabad), 16(2), 57–68. https://doi.org/10.3923/biotech.2017.57.68
Mowafy, A. M., Fawzy, M. M., Gebreil, A., & Elsayed, A. (2021). Endophytic Bacillus, Enterobacter, and Klebsiella enhance the growth and yield of maize. Acta Agriculturae Scandinavica, Section B—Soil & Plant Science, 71(4), 237–246.
Mowafy, A. M., Khalifa, S., & Elsayed, A. (2023). Brevibacillus DesertYSK and Rhizobium MAP7 stimulate the growth and pigmentation of Lactuca sativa L. Journal of Genetic Engineering and Biotechnology, 21(1), 17.
Naseri, B. (2019). Legume root rot control through soil management for sustainable agriculture. Sustainable Management of Soil and Environment, 217–258. https://doi.org/10.1007/978-981-13-8832-3_7
Natarajan, S., Mishra, P., Vadivel, M., Basha, M. G., Kumar, A., & Velusamy, S. (2018). ISSR characterization and quantification of purpurin and alizarin in Rubia cordifolia L. populations from India. Biochemical Genetics, 57(1), 56–72. https://doi.org/10.1007/s10528-018-9875-4
Nazir, N., Kamili, A. N., Zargar, M. Y., Khan, I., Shah, D., Tyub, S., & Dar, R. (2017). Studies on Bacillus sp, as an efficient plant growth promoting Rhizobacteria from Taxus wallichiana Zucc. an endangered conifer of Kashmir Himalaya. International Journal of Current Microbiology and Applied Sciences, 6(7), 41–50. https://doi.org/10.20546/ijcmas.2017.607.006
Nene, Y., Haware, M., Reddy, M., Philps, J., Castro, E. L., Kotasthane, S., Gupta, O., Singh, G., Shukia, P., & Sah, R. (1989). Identification of broad based and stable resistance to wilt and root-rots in chickpea. Indian Phytopathology, 42, 499–505.
Nigam, R., & Singh, J. (2023). Response of pH on indegenous biocontrol activity of fungal and bacterial biological agents against Rhizoctonia solani. Journal of Eco-Friendly Agriculture, 18(2), 365–369. https://doi.org/10.48165/jefa.2023.18.02.27
Ogbuji, N. G., & Isalar, O. F. (2021). Effect of Some Fungal and Bacterial Organisms on the Growth of Cowpea (Vigna unguiculata (L.) Walp) Seedlings. Journal of Applied Life Sciences International, 11–19. https://doi.org/10.9734/jalsi/2021/v24i1230274
Okafor, N., & MacRae, I. C. (1973). The influence of moisture level, light, aeration and glucose upon acetylene reduction by a black earth soil. Soil Biology and Biochemistry, 5(1), 181–186. https://doi.org/10.1016/0038-0717(73)90108-9
Okhtia, Z. A., Al-Sudani, B. T., & Abdalah, M. E. (2020). Study the effect of Rubi a Cordifolia extract on different type of cancer cell lines and different microbial. Sys Rev Pharm, 11, 994–1000.
Patil, R., Mohan, M., Kasture, V., & Kasture, S. (2009). Rubia cordifolia: A review. Oriental Pharmacy and Experimental Medicine, 9(1), 1–13. https://doi.org/10.3742/opem.2009.9.1.001
Pieterse, C. M. J., Van der Does, D., Zamioudis, C., Leon-Reyes, A., & Van Wees, S. C. M. (2012). Hormonal Modulation of Plant Immunity. Annual Review of Cell and Developmental Biology, 28(1), 489–521. https://doi.org/10.1146/annurev-cellbio-092910-154055
Pilkington, L. J., Messelink, G., van Lenteren, J. C., & Le Mottee, K. (2010). “Protected Biological Control” – Biological pest management in the greenhouse industry. Biological Control, 52(3), 216–220. https://doi.org/10.1016/j.biocontrol.2009.05.022
Reis, A., Costa, H., Boiteux, L. S., & Lopes, C. A. (2005). First report of Fusarium oxysporum f. sp. lycopersici race 3 on tomato in Brazil. Fitopatologia Brasileira, 30(4), 426–428. https://doi.org/10.1590/s0100-41582005000400017
Seleim, M., Abo-Elyousr, K., Mohamed, A., & Al-Marzoky, H. (2014). Peroxidase and polyphenoloxidase activities as biochemical markers for biocontrol efficacy in the control of tomato bacterial wilt. J. Plant Physiol. Pathol, 2(1), 2.
Sharma, S., Sharma, A., & Kaur, M. (2018). Extraction and evaluation of gibberellic acid from Pseudomonas sp.: Plant growth promoting rhizobacteria. Journal of Pharmacognosy and Phytochemistry, 7(1), 2790–2795.
Singh, P. K., Singh, M., & Vyas, D. (2010). Biocontrol of fusarium wilt of chickpea using arbuscular mycorrhizal fungi and Rhizobium leguminosorum biovar. Caryologia, 63(4), 349–353.
Sittie, A., Lemmich, E., Olsen, C., Hviid, L., Kharazmi, A., Nkrumah, F., & Christensen, S. (1999). Structure-activity studies: in vitro antileishmanial and antimalarial activities of anthraquinones from Morinda lucida. Planta Medica, 65(03), 259–261. https://doi.org/10.1055/s-2006-960473
Srivastava, P., Sahgal, M., & Paul, S. (2024). Nitrogen-fixing Rhizobium-legume symbiosis in agroecosystems. In Plant Endophytes and Secondary Metabolites (pp. 35–54). Elsevier. https://doi.org/10.1016/B978-0-443-13365-7.00016-6
Stefan, M., Munteanu, N., Stoleru, V., & Mihasan, M. (2013). Effects of inoculation with plant growth promoting rhizobacteria on photosynthesis, antioxidant status and yield of runner bean. Romanian Biotechnological Letters, 18(2), 8132–8143.
Sun, L., Lu, Z., Bie, X., Lu, F., & Yang, S. (2006). Isolation and characterization of a co-producer of fengycinsand surfactins, endophytic Bacillus amyloliquefaciens ES-2, from Scutellaria baicalensis Georgi. World Journal of Microbiology and Biotechnology, 22(12), 1259–1266. https://doi.org/10.1007/s11274-006-9170-0
Tripathi, Y. B., Shukla, S., Sharma, M., & Shukla, V. K. (1995). Antioxidant property of Rubia cordifolia extract and its comparison with vitamin E and parabenzoquinone. Phytotherapy Research, 9(6), 440–443. https://doi.org/10.1002/ptr.2650090611
Verma, P., Yadav, A. N., Kumar, V., Singh, D. P., & Saxena, A. K. (2017). Beneficial plant-microbes interactions: biodiversity of microbes from diverse extreme environments and its impact for crop improvement. Plant-Microbe Interactions in Agro-Ecological Perspectives: Volume 2: Microbial Interactions and Agro-Ecological Impacts, 543–580. https://doi.org/10.1007/978-981-10-6593-4_22
Wang, P., Liu, X., Guo, J., Liu, C., Fu, N., & Shen, H. (2015). Identification and Expression Analysis of Candidate Genes Associated with Defense Responses to Phytophthora capsici in Pepper Line “PI 201234.” International Journal of Molecular Sciences, 16(5), 11417–11438. https://doi.org/10.3390/ijms160511417
Wang, K., Gao, L., Zhang, Q., Zhang, Y., Yao, W., Zhang, M., Tang, Y., Ding, A., & Zhang, L. (2020). Revealing the mechanisms and the material basis of Rubia cordifolia L. on abnormal uterine bleeding with uniting simultaneous determination of four components and systematic pharmacology approach-experimental validation. Journal of Pharmaceutical and Biomedical Analysis, 189, 113475. https://doi.org/10.1016/j.jpba.2020.113475
Wen, M., Chen, Q., Chen, W., Yang, J., Zhou, X., Zhang, C., Wu, A., Lai, J., Chen, J., Mei, Q., Yang, S., Lan, C., Wu, J., Huang, F., & Wang, L. (2022). A comprehensive review of Rubia cordifolia L.: Traditional uses, phytochemistry, pharmacological activities, and clinical applications. Frontiers in Pharmacology, 13, 965390–965390. https://doi.org/10.3389/fphar.2022.965390
White, T. J., Bruns, T., Lee, S., & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols (pp. 315–322). Elsevier. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
Xu, S. J., & Kim, B. S. (2014). Biocontrol of Fusarium crown and root rot and promotion of growth of tomato by Paenibacillus strains isolated from soil. Mycobiology, 42(2), 158–166.
Youssef, K., Ahmed, Y., Ligorio, A., D'Onghia, A., Nigro, F., & Ippolito, A. (2010). First report of Penicillium ulaiense as a postharvest pathogen of orange fruit in Egypt. https://doi.org/10.1111/j.1365-3059.2010.02337.x
Zeng, W., Fang, Y., Mo, S., Shen, C., Yang, H., Luo, G., Xiao, L., Zhan, R., & Yan, P. (2023). The Underling Mechanisms Exploration of Rubia cordifolia L. Extract Against Rheumatoid Arthritis by Integrating Network Pharmacology and Metabolomics. Drug Design, Development and Therapy, 439–457.
Funding
This research work is derived from a research grant funded by the Research, Development, and Innovation Authority (RDIA)—Kingdom of Saudi Arabia—with grant number (13445-Tabuk-2023-UT-R-3–1-SE).
Author information
Authors and Affiliations
Contributions
Conceptualization, AAG, ASA, MSEH, HG, AYE, WIAS, and AE; methodology, analysis, and data curation, AAG, HME, YAE, and NME; writing-original draft preparation, AAG, MSEH, WIAS, and AE; discussion, review and editing, AAG, ASA, AYE, HG, NME, YAE, KME, and HME; project supervision and administration, AAG, MSEH, WIAS and AE. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ghoniem, A.A., Elattar, K.M., Alotaibi, A.S. et al. Enhanced resistance of Vigna unguiculata to Fusarium oxysporum via Rubia cordifolia extract and growth-promoting endophytic Bacillus amyloliquefaciens DW6. Eur J Plant Pathol (2024). https://doi.org/10.1007/s10658-024-02922-0
Accepted:
Published:
DOI: https://doi.org/10.1007/s10658-024-02922-0