Abstract
Phomopsis vexans is a phytopathogenic fungus that causes Phomopsis blight of eggplant. Because spore germination is required for infection, preventing it may be a viable approach to disease management. The proteome during the conversion of spores to mycelium was analyzed in the current study to gain more insight into this process. A total of 3337 proteins were discovered and functionally classified. There were 929 proteins with significant fold changes (P value ≤ 0.05, Fold change > 2) among them. which were grouped into 6 clusters based on their relative expression similarities. Sphingolipid metabolism, Glycosylphosphatidylinositol (GPI)-anchor biosynthesis, Riboflavin metabolism, and Glutathione metabolism were found to be significantly upregulated during the germ tube emerging stage, according to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment. Proteins in these pathways may be important in spore germination and may be antifungal targets. In addition, eight potential effectors were discovered. This is the first report on proteomic analysis of Phomopsis vexans spore germination. It can aid in understanding the mechanisms of host invasion and shed light on Phomopsis vexans control.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Eggplant (Solanum melongena) is a vegetable crop known for its taste and nutritional benefits. Phomopsis blight of eggplant is one of the most serious diseases caused by Phomopsis vexans. It was first reported in the United States and has since spread globally (Akhtar & Chaube, 2006; Halsted, 1891; Harter, 1914). Leaf spots, branch-brown spots, withered leaves, and fruit rot are all symptoms of Phomopsis blight (Mahadevakumar & Janardhana, 2016; Rohini et al., 2016). It is one of the most devastating diseases of eggplant diseases, causing significant yield and quality losses (Bhanushree et al., 2022). Outbreaks pose a serious threat to growers’ and breeders’ because it can reduce outputs by 20% to 30%. (Thesiya et al., 2019). Growers are currently controlling it primarily through chemical and disease resistance resources (Bhat et al., 2019; Jannat et al., 2018). Finding the right control target is the cornerstone of effective management, regardless of the control technique used.
Spore germination is a critical step in pathogenic infection (Baltussen et al., 2020). Therefore, understanding the molecular basis of spore germination is important for developing control strategies. Under the right conditions, resting spores will initiate isotropic growth. Following spore swelling, a germ tube emerges, forming fungal hyphae and resulting in filamentous growth (Sephton-Clark & Voelz, 2018). Several structural and metabolic changes occur in fungal spores during this process. As an example, translation of mRNA stored in spores is initiated. The spore's lipids and carbohydrates begin to degrade. The spore's cell wall and membrane structure begin to change to accommodate swelling and pola growth. Changes begin to occur in the lipids and gylan metabolite pathways, which are two important components of cell walls and membranes (Baltussen et al., 2020). Proteins involved in these processes are potential targets for pathogen control.
Proteomics is a reliable method for examining the protein abundance of fungi. This technique has been widely used to study the spore germination process and identify key proteins and pathways. Proteomic analysis of Aspergillus nidulans reveals that energy metabolism, protein synthesis, and protein folding are major physiological functions of proteins that vary quantitatively during the early stages of conidia germination (Oh et al., 2010). Proteins in biological processes such as cellular energy production, oxidative metabolism, stress, fatty acid synthesis, protein synthesis, and protein folding changed expression during Colletotrichum acutatum germination (El-Akhal et al., 2013). Proteomic analysis identified and validated four enzymes in Fusarium oxysporum’s ergosterol biosynthesis pathway. They hold promise as new targets for combating Fusarium wilt in bananas (Deng et al., 2015). Only four hours after spore germination of Moniliophthora perniciosa proteins associated with fungal filamentation, such as septin and kinesin, can be detected (hag). A transcription factor capable of forming polyketides hybrids was induced 2 hag, and polyketide synthase was detected 4 hag (Mares et al., 2017). Alicyclobacillus acidoterrestris spore germination was linked to proteins involved in cell wall hydrolysis, intracellular substance synthesis, and signal transduction (Xu et al., 2019). The transcription and translation factors, elongation factor Tu, histones (H2A and H15), proteasome, and DJ-1_PfpI were found to be upregulated during the germination of Spongospora subterranean resting spores (Balotf et al., 2021). These finding form the foundation for the search for disease-resistant target proteins. However, no study of the Phomopsis vexans proteome exists.
The Tandem mass tag (TMT) approach was used in this study to investigate the proteomic profile of Phomopsis vexans at three germination stages. This study represents the first report of a comprehensive proteomic analysis of Phomopsis vexans spore germination. The findings presented here can be used as a baseline for understanding global protein expressions in this fungus’s early life cycle and for developing new effective fungicides.
Methods and materials
Stains, media, and cultural conditions
A strain of Phomopsis vexans was isolated from an infected branch of an eggplant (Solanum melongena) (Figure S5) grown in Guangzhou, Guangdong Province, China (23°23 ‘24.5"N 113°26 ‘19.4"E). The strain was brought to sporulate after inoculating it into PDB medium and cultivating it for four days at 24 °C and 150 rpm. Spores were collected by filtering the sporulation medium through four layers of gauze. Based on our previous findings, spores were cultured in PDB medium for 4 and 8 h at 28 °C and 150 rpm, representing the hyphal development and mycelium stages, respectively. (Unpublished data).
Spore germination observation
At each stage, a total of 200ul of sporulation medium filtrate was transferred to a clean glass slide and covered with a coverslip. Then samples were then examined under an optical microscope (Leica DM2500).
Total protein extraction
Individual samples were crushed in liquid nitrogen before being lysed in a lysis buffer containing 100 mM NH4HCO3 (pH 8), 6 M Urea, and 0.2% SDS for 5 min on ice. After centrifuging the lysate at 12,000 g for 15 min at 4 °C, the supernatant was transferred to a clean tube. The extracts from each sample were first reduced with 10 mM DTT for 1 h at 56 °C, then alkylated for 1 h in the dark at room temperature with enough iodoacetamide.
The samples were then thoroughly mixed with four times the amount of precooled acetone using a vortexing technique before being incubated at − 20 °C for at least two hours. The precipitate from the samples was collected after centrifugation. After being washed twice with cold acetone, the pellet was dissolved in a dissolving buffer containing 0.1 M triethylammonium bicarbonate (TEAB, pH 8.5) and 6 M urea.
Protein quality test
The instructions for the bradford protein quantification kit were followed to create the BSA standard protein solution, which had gradient concentrations ranging from 0 to 0.5 g/L. A 96-well plate with a capacity of 20μL was filled with BSA standard protein solutions and sample solutions with various dilution multiples. Each gradient was repeated three times. The plate's absorbance at 595 nm was measured after immediately adding 180 μL of G250 dye solution to it and allowing it to stand at room temperature for 5 min. The absorbance of a standard protein solution was used to construct the standard curve, and the protein content of the sample was estimated. 20 μg of protein sample were electrophoresed on a 12% SDS-PAGE gel with the concentration gel at 80 V for 20 min and the separation gel at 120 V for 90 min. The gel was stained with Coomassie Brilliant Blue R-250 and then lightened so that the bands could be seen clearly.
TMT labeling of peptides
Each protein sample weighed 120 μg, and was filled to a volume of 100 μL with lysis buffer. The sample was then mixed and digested overnight at 37 °C with 1 μg/μL trypsin and 500 μL of 50 mM TEAB buffer. The digested sample was mixed with an equal amount of 1% formic acid before centrifugation at 12,000 g for 5 min at room temperature. The supernatant was slowly loaded onto the C18 desalting column, washed three times with 1 mL of washing solution (0.1% formic acid, 4% acetonitrile), and eluted twice with 0.4 mL of elution buffer (0.1% formic acid, 75% acetonitrile). The eluents from each sample were mixed and then lyophilized. The sample was reconstituted with 100 μL of 0.1 M TEAB buffer, 41 μL of acetonitrile-dissolved TMT labeling reagent, and shaken for 2 h at room temperature. The process was then halted by the addition of 8% ammonia. Desalted, lyophilized, and combined in the same volume were all labeled sample mixtures.
Separation of fractions
Mobile phases A (2% acetonitrile, pH adjusted to 10.0 using ammonium hydroxide) and B (98% acetonitrile, pH adjusted to 10.0 using ammonium hydroxide) were used to create a gradient. The lyophilized powder was centrifuged at 12,000 g for 10 min at room temperature after being dissolved in solution. The material was fractionated using a C18 column (Waters BEH C184.6 × 250 mm, 5 μm) on a Rigol L3000 HPLC system with the column oven set to 50 °C. The elution gradient was depicted in detail in Table S6. The eluates were collected at a rate of one tube per minute while being observed at UV 214 nm, and then merged into ten parts. All fractions were vacuum-dried before being reconstituted in water containing 0.1% (v/v) formic acid.
LC–MS/MS analysis
Shotgun proteomics studies for the creation of transition libraries were carried out in the data-dependent acquisition mode using a Q Exactive HF-X mass spectrometer and an EASY-nLCTM 1200 UHPLC system from Thermo Fisher. For the injection of 1 μg of sample, PepMap RSLC C18 analytical column (50 cm × 75 μm) was used, with a linear gradient elution as detailed in Table S7. The isolated peptides were examined using a Thermo Fisher's Q Exactive HF-X mass spectrometer and an ion source called Nanospray FlexTM (ESI). The Automatic Gain Control (AGC) target value was 3 × 106, the maximum ion injection duration was 20 ms, and the overall scan range was m/z 350 to 1800 with a resolution of 120,000 (at m/z 200). In MS/MS, the resolution was 15,000 (at m/z 200) for 6 plex (45,000 for 10 plex, m/z 200), and the automated gain control (AGC) target value was 1 × 105. The top 20 most abundant precursors in the entire scan were chosen, fragmented by higher energy collisional dissociation, and evaluated. The maximum ion injection time was 50 ms, the intensity threshold was 4 × 104, the normalized collision energy was 32%, and the dynamic exclusion time was 30 s. The raw data from MS detection was given the extension “.raw.”
The identification and quantitation of protein
Proteome Discoverer 2.2. (PD 2.2, Thermo) was used to compare the resulting spectra from each fraction with the Phomopsis vexans genome database (unpublished data). The following search criteria were established: the tolerance for precursor ion mass was 10 ppm, and the tolerance for manufacturing mass was 0.02 Da. Carbamidomethyl was listed as a fixed alteration in PD 2.2. The terms “variable modifications” in PD 2.2 were defined as “oxidation of methionine (M),” “acetylation of the N-terminus,” and “TMT 10-plex of tyrosine and lysine.” The discovered protein has an FDR of no more than 1.0% and at least one distinct peptide. Proteins with comparable peptide compositions that could not be differentiated by MS/MS analysis were assigned to the same protein group. The TMT was quantified using Reporter Quantification (TMT 10-plex). Proteins with significantly different quantitation between the experimental and control groups (p < 0.05 and |log2FC|> 1) were classified as differentially expressed proteins (DEP). The Mann–Whitney Test was used to analyze the protein quantitation results.
Annotation of protein and DEP
The non-redundant protein database (including Pfam, PRINTS, ProDom, SMART, ProSiteProfiles, and PANTHER) was analyzed using the InterPro (IPR) analysis using the interproscan-5 program, and the Clusters of Orthologous Groups and KEGG (Kyoto Encyclopedia of Genes and Genomes) databases were used to analyze the protein family and pathway. The enrichment pipeline was used to perform the GO, IPR, and KEGG enrichment analyses.
Total RNA extraction and quantitative RT-PCR
Total RNAs were extracted and purified from three biological replicates at each of the three germination stages using the QIAGEN RNeasy Plant Mini Kit and RNase-free DNase kit (QIAGEN, Germany). To generate cDNA, the PrimeScriptTM RT reagent Kit with gDNA Eraser was used (Takara, Dalian, China). The PCR reactions were carried out in a total volume of 20 μL on a Roche LightCycler 480 Q-PCR equipment, containing 1μL of primer (10μL M), 1μL of cDNA, 8μL PCR-grade water, and 10μL of AceQqPCR SYBR Green Master Mix (Vazyme, Nanjing, China) (Roche, Basel, Switzerland). The PCR protocol began with a 5-min prep step at 95 °C, followed by 45 cycles of 10 s at 95 °C and 30 s at 60 °C. To ensure the purity of the amplified products, a melting curve was created for each sample at the end of each cycle. Table S5 contained data on the qPCR primers and reference genes.
Results
Overview of experimental design and spore visualization by microscopy
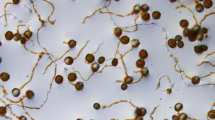
Figure 1 depicts an overview of the experimental design in biological replicate. Three stages were chosen: spore (A, 0 h), germination tube emerging and elongation (B, 4 h), and mature mycelia (C, 8 h). The lipid droplet in the spore vanished during spore germination, according to microscopy observations. At stage B, germination tubes appear. Samples were combined and analyzed with LC–MS/MS after protein isolation, digestion and labeling. Finally, the data was analyzed.
A: Micrograph for Phomopsis vexans at spore stage, B: Micrograph for Phomopsis vexans at germination tube emerging and elongation stage, C: Micrograph for Phomopsis vexans at mature mycelia stage.
Overview of the P.vexans proteome and functional annotation of the identified proteins
The results of principal component analysis (PCA) revealed that the sample scan was properly replicated (Fig. 2). Interestingly, samples at stages A and C were quite similar on PC2, whereas B stage could be distinguished from A and C on both PC1 and PC2, indicating some germination stage differences.
3337 proteins were identified and quantified in total (Table S1). gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and IPR databases have annotated 2228, 3328 and 3013 proteins, respectively (Figure S1). Proteins are classified into different classes based on gene ontology analysis (Fig. 3). More than 500 proteins can be divided into two major molecular function categories: ATP binding and protein binding. The remainder is primarily rich in binding, catalytic, and oxidoreductase activity. Proteins are primarily annotated into the functional categories of biological process of oxidation–reduction, metabolic, and translation. These findings indicate that the proteins identified in the proteome mostly perform binding, and catalysis tasks related to metabolism, redox, and translation.
According to Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation results, proteins are primarily enriched in primary metabolic pathways such as carbohydrate metabolism, lipid metabolism, energy metabolism, and amino acid metabolism. Proteins are primarily enriched in folding sorting and degradation pathways and translation pathways among genetic information processing pathways. Proteins in the cellular processes category are primarily enriched in transport and catalytic pathways. The signal transduction pathway is a major enriched pathway in the environmental information processing category.
According to the InterPro (IPR) annotation results, proteins are mostly annotated with WD40 repeat, protein kinase, and ATPase domains (Fig. 4). The majority of proteins are found in the nucleus, cytoplasm, and mitochondria, according to subcellular localization analysis (Fig. 5).
Differentially abundant proteins in P.vexans during spore germination
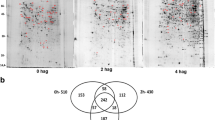
There was a significant difference in relative expression of 929 proteins during spore germination (P value ≤ 0.05, Fold change > 2)(Table S2). 410(up, 211; down, 199), 586 (up, 225; down, 361), and 443 (up,124; down, 319) proteins with a significant change in B vs A, C vs A and C vs B were discovered among them. (See Fig. 6 and Table S3).
Proteins with significant fold changes were classified into six clusters based on their relative expression similarities (Fig. 6). Proteins in clusters 1 and 4 had higher expression at the B stage, whereas proteins in clusters 2 and 5 had the opposite trend. Proteins that showed consistent up- or down-regulation were classified as cluster 3 or cluster 6, respectively. Table S3 displays the KEGG enrichment results for these proteins. Proteins in clusters 1 and 4 may be important in spore germination because they are upregulated at the B stage. Various types of N-glycan biosynthesis pathways are significantly enriched in cluster 1, as are Sphingolipid metabolism, Glycosylphosphatidylinositol (GPI)-anchor biosynthesis, Riboflavin metabolism, and Glutathione metabolism pathways in cluster 4.
The annotated proteins in the Sphingolipid metabolism pathway form a complete pathway for Ceramide synthesis. Among these, 3-dehydrosphinganine reductase (KDSR; EC:1.1.1.102), sphinganine C4-monooxygenase (SUR2; EC:1.14.18.5), and sphingolipid 4-desaturase (DEGS; EC:1.14.19.17) expression was significantly increased during germination tube formation (Figure S2). Furthermore, the expression profile of sphinganine-1-phosphate aldolase (SGPL1; EC:4.1.2.27), which synthesizes phosphoethnolamine in the same pathway as SGPL1, was consistent with the above three findings (Figure S2). GPI ethanolamine phosphate transferase 1 (PIG-N, EC:2.7.-.-) and GPI-anchor transamidase subunit T (PIG-T) are upregulated at the B stage of the GPI-anchor biosynthesis pathways (Figure S3). They could aid in the accumulation of GPI-anchors. Identified proteins form an integral pathway from Ribulose 5-phosphate to flavin adenine dinucleotide (FAD) in the Riboflavin metabolism. At stage B, riboflavin kinase (RFK; EC:2.7.1.26) and 3,4-dihydroxy 2-butanone 4-phosphate synthase (RIB3; EC:4.1.99.12) were significantly upregulated (Figure S4). All of the proteins mentioned above may be important in spore germination and are potential antimicrobial targets.
Eight potential effectors were identified based on Nanopore sequencing (data not shown) and proteomic data from Phomopsis vexans. Among them, Effector 4 demonstrated the most intense signal and the most consistent increasing trend during spore germination (Table S4). None of the eight effectors had previously been reported.
The abundance transcripts may have a significant impact on the expression of any gene encoding proteins. Changes in a protein's expression may be caused by changes in its transcript. To investigate the relationship between transcripts and the quantitative proteome data, we performed RT-qPCR analysis on ten differentially expressed proteins. Most genes’ relative expression levels match the expression trends of their encoded proteins, such as HP2, HP3, SER, and GLY. Furthermore, other proteins, such as HP1, ISWI, and others, have gene expression patterns that are diametrically opposed in the B and C stages. This could be due to a post-transcriptional regulatory mechanism (Fig. 7).
Relative expression of selected proteins and genes encoding them. (Pro, Proteome; HP1, hypothetical protein GGTG_04657; HP2, hypothetical protein S7711_01809; HP3, hypothetical protein W97_08647; ISWI, putative iswi chromatin-remodeling complex atpase isw2 protein; HP4, hypothetical protein GLRG_01695; SER, potential serine carboxypeptidase; GLY, glycosyl hydrolase family 3 N terminal domain-containing protein; HP7, hypothetical protein SMAC_01828; DRA, DRAP deaminase; ZIN, putative zinc finger protein.)
Discussion
The germination of spores is a critical step in phytopathogenic fungal invasion of crops. An in-depth understanding of the molecular mechanism canhelpto identify biological and chemical control targets. P. vexans was studied using quantitative proteomics at three stages (spore, germination tube emergence and elongation, and mature mycelia). We discovered that highly expressed proteins during the germination tube emergence and elongation stage were primarily enriched in five pathways: Sphingolipid metabolism, Glycosylphosphatidylinositol (GPI)-anchor biosynthesis, Riboflavin metabolism, Glutathione metabolism, and Various types of N-glycan biosynthesis.
Sphingolipids are required for the structure of fungal cell membranes (Rollin-Pinheiro et al., 2016). The two main fungal sphingolipids are Inositol phosphoryl-ceramides (IPCs) and glucosylceramide (GlcCer) and they are both important for a variety of biological activities, including fungal virulence control (McEvoy et al., 2020). As a reult, sphingolipid biosynthesis enzymes have recently been studied as potential new targets for the development of antifungal drugs (Gao et al., 2018; Lazzarini et al., 2018; Zhen et al., 2022). Serine palmitoyltransferase (SPT, EC:2.3.1.50), sphingoid base N-stearoyltransferase (CERS, EC: 2.3.1.299), and inositol phosphorylceramide synthase (IPC, EC: 2.7.1.227) are the main target proteins for inhibiting sphingolipid synthesis (Zhen et al., 2022). In this study, the differential proteins sphinganine-1-phosphate aldolase (SPL, EC:4.1.2.27), 3-dehydrosphinganine reductase (KDHR, EC:1.1.1.102), sphinganine C4-monooxygenase (SUR2, EC:1.14.18.5) and sphingolipid 4-desaturase (DES, EC:1.14.19.17) enriched in this pathway were highly expressed at the germination tube emerging and elongation stage. Unfortunately, these four proteins have orthologous genes in mammals with high sequence similarity. As a result, they may not be suitable antifungal targets.
GPI is critical for the integrity of the fungal cell wall because it is responsible for anchoring proteins to the plasma (Fu et al., 2020). The reported inhibitors’ main antifungal targets are glucosaminylphosphatidylinositol acyltransferase (PIG-W, EC:2.3.-.-) (Wiederhold et al., 2019) and PIG-N (Mann et al., 2015). PIG-N and GPI-anchor transamidase subunit T (PIG-T) were found to be differentially expressed proteins in GPI-anchor biosynthesis in this study. This finding suggests that PIG-N inhibitors may be useful in the control of P. vexans. PIG-T may also be an antifungal target. Furthermore, the N-glycan biosynthesis pathway comes before the GPI-anchor biosynthesis pathway. The former pathway is also enriched in proteins that are highly expressed during germination tube elongation in this study, suggesting that it could be a potential antifungal target.
Riboflavin, also known as vitamin B2, is a water-soluble compound that plays an important role in cell metabolism (Dietl et al., 2018). Its active forms, flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN), act as cofactors in a number of flavocoenzyme-catalyzed processes such as electron transport and fatty acid oxidation (Meir & Osherov, 2018). Because humans cannot synthesize vitamin B2, fungal inhibitors that target this pathway are less dangerous(Meir & Osherov, 2018). It has recently been reported in Candida albicans (Becker et al., 2010) and Aspergillus fumigatus (Dietl et al., 2018) that GTP cyclohydrolase II (RIB1, EC:3.5.4.25) and 5-amino-6-(5-phosphoribosylamino) uracil reductase (RIB2, EC:1.1.1.193) are important for virulence development, and there are inhibitors developed against 6,7-dimethyl-8-ribityllumazine synthase (RIB4, EC:2.5.1.78) (Chen et al., 2005). The enzymes, 3,4-dihydroxy 2-butanone 4-phosphate synthase (RIB3, EC:4.1.99.12) and riboflavin kinase (RFK, EC:2.1.7.26) were found to be highly expressed during the germination tube elongation stage and may be potential antifungal targets in this study. Glutathione metabolism is an important amino acid synthesis pathway that is similar to human-related pathways and may not be a good target choice.
Fungal effectors are typically secreted via the endoplasmic reticulum-Golgi apparatus route (He et al., 2020) and can be either harmful secondary metabolites or proteins that destroy the host plant(Rocafort et al., 2020). As a result, they are critical targets for the control of plant fungal diseases. We discovered eight potential effector proteins when combined our previous study of the P.vexans genome. However, none of them have previously been identified as effectors. More function validation is required before they can be used as an antifungal target.
Conclusion
The comparative TMT proteomic analysis revealed a general profile of protein expression during Phomopsis vexans germination at spore (A) stage, Hyphal growth (B) stage, and full mature mycelia (C) stage. Several germination-related pathways and antifungal targets were discovered. This research could help to unravel the mechanisms underlying Phomopsis vexans germination and infection, laying the groundwork for future Phomopsis blight of eggplant research.
Data Availability
The relevant data are provided in the supplementary materials. Corresponding authors may be contacted for raw data.
References
Akhtar, J., & Chaube, H. (2006). Variability in phomopsis blight pathogen[Phomopsis vexans (Sacc. & Syd.) Harter]. Indian phytopathology, 59(4), 439–444.
Balotf, S., Wilson, R., Tegg, R. S., Nichols, D. S., & Wilson, C. R. (2021). Quantitative proteomics provides an insight into germination-related proteins in the obligate biotrophic plant pathogen Spongospora subterranea. Environmental Microbiology Reports, 13(4), 521–532. https://doi.org/10.1111/1758-2229.12955
Baltussen, T. J. H., Zoll, J., Verweij, P. E., & Melchers, W. J. G. (2020). Molecular mechanisms of conidial germination in Aspergillus spp. Microbiology and Molecular Biology Reviews: MMBR, 84(1), 1–34. https://doi.org/10.1128/MMBR.00049-19
Becker, J. M., Kauffman, S. J., Hauser, M., Huang, L., Lin, M., Sillaots, S., Jiang, B., Xu, D., & Roemer, T. (2010). Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proceedings of the National Academy of Sciences of the UNITED States of America, 107(51), 22044–22049. https://doi.org/10.1073/pnas.1009845107
Bhanushree, N., Saha, P., Tomar, B. S., & Munshi, A. D. (2022). Phomopsis blight in eggplant and strategies to manage through resistance breeding. The Journal of Horticultural Science and Biotechnology, 97(1), 34–45. https://doi.org/10.1080/14620316.2021.1966321
Bhat, M., Anwar, A., Mughal, M. N., Mohiddin, F., Makhdoomi, M., Bhat, A. H., & Fayaz, U. (2019). Morpho-cultural and pathological variability in Phomopsis vexans causing leaf blight and fruit rot of brinjal in Kashmir. Indian Phytopathology, 72(2), 225–233. https://doi.org/10.1007/s42360-019-00128-7
Chen, J., Illarionov, B., Bacher, A., Fischer, M., Haase, I., Georg, G., Ye, Q. Z., Ma, Z., & Cushman, M. (2005). A high-throughput screen utilizing the fluorescence of riboflavin for identification of lumazine synthase inhibitors. Analytical Biochemistry, 338(1), 124–130. https://doi.org/10.1016/j.ab.2004.11.033
Deng, G. M., Yang, Q. S., He, W. D., Li, C. Y., Yang, J., Zuo, C. W., Gao, J., Sheng, O., Lu, S. Y., Zhang, S., & Yi, G. J. (2015). Proteomic analysis of conidia germination in Fusarium oxysporum f. sp. cubense tropical race 4 reveals new targets in ergosterol biosynthesis pathway for controlling Fusarium wilt of banana. Applied microbiology and biotechnology, 99(17), 7189–7207. https://doi.org/10.1007/s00253-015-6768-x
Dietl, A. M., Meir, Z., Shadkchan, Y., Osherov, N., & Haas, H. (2018). Riboflavin and pantothenic acid biosynthesis are crucial for iron homeostasis and virulence in the pathogenic mold Aspergillus fumigatus. Virulence, 9(1), 1036–1049. https://doi.org/10.1080/21505594.2018.1482181
El-Akhal, M. R., Colby, T., Cantoral, J. M., Harzen, A., Schmidt, J., & Fernández-Acero, F. J. (2013). Proteomic analysis of conidia germination in Colletotrichum acutatum. Archives of Microbiology, 195(4), 227–246. https://doi.org/10.1007/s00203-013-0871-0
Fu, Y., Estoppey, D., Roggo, S., Pistorius, D., Fuchs, F., Studer, C., Ibrahim, A. S., Aust, T., Grandjean, F., & Mihalic, M. (2020). Jawsamycin exhibits in vivo antifungal properties by inhibiting Spt14/Gpi3-mediated biosynthesis of glycosylphosphatidylinositol. Nature Communications, 11(1), 3387. https://doi.org/10.1038/s41467-020-17221-5
Gao, J., Wang, H., Li, Z., Wong, A. H., Wang, Y. Z., Guo, Y., Lin, X., Zeng, G., Liu, H., Wang, Y., & Wang, J. (2018). Candida albicans gains azole resistance by altering sphingolipid composition. Nature Communications, 9(1), 4495. https://doi.org/10.1038/s41467-018-06944-1
Halsted, B. D. (1891). A new egg-plant disease. Bulletin of the Torrey Botanical Club, 18(10), 302–303.
Harter, L. (1914). Fruit rot leaf spot and stem blight of the eggplant caused by phomopsis vexans. Journal of Agricultural Research, 2, 331–338.
He, Q., McLellan, H., Boevink, P. C., & Birch, P. R. J. (2020). All Roads Lead to Susceptibility: The Many Modes of Action of Fungal and Oomycete Intracellular Effectors. Plant Communications, 1(4), 100050. https://doi.org/10.1016/j.xplc.2020.100050
Jannat, R., Shaha, M., Rubayet, M. T., & Sultana, S. (2018). Role of chitosan in induction of defense response against Phomopsis vexans and augmentation of growth and yield of eggplant. Global Journal of Science Frontier Research: C Biological Science, 18(C3), 7–16.
Kalda, T.S., S.V., and Choudhury, B. (1976). Studies on resistance to phomopsis blight in eggplant (Solanum melongena L.). Vegetable science 3(1), 65–70.
Lazzarini, C., Haranahalli, K., Rieger, R., Ananthula, H. K., Desai, P. B., Ashbaugh, A., Linke, M. J., Cushion, M. T., Ruzsicska, B., Haley, J., et al. (2018). Acylhydrazones as antifungal agents targeting the synthesis of fungal sphingolipids. Antimicrobial Agents and Chemotherapy, 62(5), e00156-e218. https://doi.org/10.1128/AAC.00156-18
Mahadevakumar, S., & Janardhana, G. R. (2016). Phomopsis vexans current research and future perspectives (1914–2015). Journal of Botanical Sciences, 5, 4–12.
Mann, P. A., McLellan, C. A., Koseoglu, S., Si, Q., Kuzmin, E., Flattery, A., Harris, G., Sher, X., Murgolo, N., Wang, H., et al. (2015). Chemical genomics-based antifungal drug discovery: Targeting glycosylphosphatidylinositol (GPI) precursor biosynthesis. ACS Infectious Diseases, 1(1), 59–72. https://doi.org/10.1021/id5000212
Mares, J. H., Gramacho, K. P., Santos, E. C., da Silva Santiago, A., Santana, J. O., de Sousa, A. O., Alvim, F. C., & Pirovani, C. P. (2017). Proteomic analysis during of spore germination of Moniliophthora perniciosa, the causal agent of witches’ broom disease in cacao. BMC Microbiology, 17(1), 176. https://doi.org/10.1186/s12866-017-1085-4
McEvoy, K., Normile, T. G., & Poeta, M. D. (2020). Antifungal drug development: Targeting the fungal sphingolipid pathway. Journal of Fungi, 6(3), 142. https://doi.org/10.3390/jof6030142
Meir, Z., & Osherov, N. (2018). Vitamin biosynthesis as an antifungal target. Journal of Fungi, 4(2), 72. https://doi.org/10.3390/jof4020072
Oh, Y. T., Ahn, C. S., Kim, J. G., Ro, H. S., Lee, C. W., & Kim, J. W. (2010). Proteomic analysis of early phase of conidia germination in Aspergillus nidulans. Fungal Genetics and Biology: FG and B, 47(3), 246–253. https://doi.org/10.1016/j.fgb.2009.11.002
Rocafort, M., Fudal, I., & Mesarich, C. H. (2020). Apoplastic effector proteins of plant-associated fungi and oomycetes. Current Opinion in Plant Biology, 56, 9–19. https://doi.org/10.1016/j.pbi.2020.02.004
Rohini, G., & H.G., Hariprasad, P., Singh, S.B., and Niranjana, S.R. (2016). Biological control of phomopsis leaf blight of brinjal (Solanum melongena L.) with combining phylloplane and rhizosphere colonizing beneficial bacteria. Biological Control, 101, 123–129. https://doi.org/10.1016/j.biocontrol.2016.05.007
Rollin-Pinheiro, R., Singh, A., Barreto-Bergter, E., & Del Poeta, M. (2016). Sphingolipids as targets for treatment of fungal infections. Future Medicinal Chemistry, 8(12), 1469–1484. https://doi.org/10.4155/fmc-2016-0053
Sephton-Clark, P. C. S., & Voelz, K. (2018). Spore Germination of Pathogenic Filamentous Fungi., 102, 117–157. https://doi.org/10.1016/bs.aambs.2017.10.002
Thesiya, M., Rakholiya, K., Panara, K., & Bhagariya, D. (2019). Evaluation of different fungicides in vitro against Phomopsis vexans causing stem blight and fruit rot in Brinjal. International Journal of Chemical Studies, 7(5), 3259–3263.
Wiederhold, N. P., Najvar, L. K., Shaw, K. J., Jaramillo, R., Patterson, H., Olivo, M., Catano, G., & Patterson, T. F. (2019). Efficacy of delayed therapy with fosmanogepix (APX001) in a murine model of Candida auris invasive candidiasis. Antimicrobial Agents and Chemotherapy, 63(11), e01120-e1219. https://doi.org/10.1128/AAC.01120-19
Xu, X., Ran, J., Jiao, L., Liang, X., & Zhao, R. (2019). Label free quantitative analysis of Alicyclobacillus acidoterrestris spore germination subjected to low ambient pH. Food Research International, 115, 580–588. https://doi.org/10.1016/j.foodres.2018.09.033
Zhen, C., Lu, H., & Jiang, Y. (2022). Novel promising antifungal target proteins for conquering invasive fungal infections. Frontiers in microbiology, 13, 911322. https://doi.org/10.3389/fmicb.2022.911322
Acknowledgements
We would like to thank Xiaoqing Sun and Junlin Huang for collecting fungi samples and for helpful discussions.
Funding
Foundation project: GuangDong Basic and Applied Basic Research Foundation (2020A1515011330, 2021A1515012132, 2021A1515012490, 2020A0505020006), Yue & Gui united fund, GuangDong Basic and Applied Basic Research Foundation (2020A1515410005), Dean Fund of Guangdong Academy of Agricultural Sciences (201812), Modern Agricultural Innovation Team Project of Guangdong Province (2021KJ110, 2021KJ106), Guangdong Academy of Agricultural Sciences Foundation, (R2020PY-JX003, R2021YJ-YB3019). 2021 Provincial Rural Revitalization Strategy Special Fund Planting industry scientific research promotion and testing projects.
Author information
Authors and Affiliations
Contributions
The experiments were created by TL. The data was analyzed and the paper was written by ZH and QY. ZhiliangL, ZhengingL, YL, and HW were in charge of the eggplant field and sampled fruits. Phomopsis vexans was isolated by XX, BS, and CG. The final manuscript has been read and approved by all authors.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Heng, Z., You, Q., Sun, B. et al. Quantitative proteomics provides an insight into germination‐related proteins in the plant pathogenic fungi Phomopsis vexans. Eur J Plant Pathol 166, 65–75 (2023). https://doi.org/10.1007/s10658-023-02643-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-023-02643-w