Abstract
The hybrid tulip tree (Liriodendron chinense (Hemsl.) Sarg. × Liriodendron tulipifera L.) is one of the most valuable ornamental plants in China. Recently, two leaf anthracnose disease types have emerged on tulip trees in a park in Beijing, China. One type is yellow halo (chlorosis ring) anthracnose characterized by many small round necrotic lesions each of which is circled by a thick chlorosis ring. Lesion spots remain separate from each other even in fallen decaying leaves. Infected leaves turn entirely yellow on trees and then fall immaturely. The other type is non-yellow halo anthracnose characterized by large and irregular necrotic lesions without thick yellow belt margins. Lesions often merge into larger ones during disease development. Infected leaves do not turn yellowish or drop early. The disease pathogens were identified as Colletotrichum gloeosporioides sensu stricto strains with multi-loci phylogeny inferences and morphological differences in cultural colonies, conidia, and appressoria. The two types of Colletotrichum anthracnose diseases were recorded as novel on Liriodendron hosts based on differential characteristics in pathogenic strains, hosts, and disease symptoms. Finally, comprehensive comparisons among all reported leaf diseases on Liriodendron trees were performed according to other reported literature.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colletotrichum species, with various necrotrophic, hemibiotrophic, and endophytic lifestyles (Jefferies et al., 1990; Kamle & Kumar, 2016; Li et al., 2022; Moreira et al., 2021; Scindiya et al., 2021; Weir et al., 2012), are some of the most important plant pathogens worldwide (Bhunjun et al., 2021; Dean et al., 2012; Jayawardena et al., 2021). The Colletotrichum gloeosporioides species complex is especially important due to its ubiquitous occurrence (Choub et al., 2021; Jefferies et al., 1990; Kamle & Kumar, 2016; Khan et al., 2021; Weir et al., 2012). Considering heterogeneity in the species complex, Weir et al. (2012) applied a polyphasic approach that combined morphological characteristics with eight-loci phylogenetic analysis, and segregated the complex into 22 species plus one subspecies, including C. gloeosporioides sensu stricto (s.s.) (Jayawardena et al., 2016; Liu et al., 2016; Weir et al., 2012).
Liriodendron tulipifera L. and Liriodendron chinense (Hemsl.) Sarg. are ancient relict trees, and the only remaining species of Liriodendron. Furthermore, they are important building and ornamental materials because of their excellent texture, unique leaves, and large, beautiful flowers. Hybrid Liriodendron is known as an inter-species hybrid between L. chinense and L. tulipifera, showing greater vigor and better fitness to various environments than its parent lines, and has been planted widely in China (Wang, 1997; Wang et al., 2021). Liriodendron spp., including the hybrid species, usually exhibit resistance to biotic or abiotic stresses (Wang, 1997; Wang et al., 2021). However, in recent years, some fungal diseases have been observed. For example, several anthracnoses or brown-spot diseases caused by Colletotrichum and other fungi, have been frequently reported on Liriodendron foliage in the field (Choi et al., 2012; Kliuchevych et al., 2019; Wang et al., 2013; Zhu et al., 2019). The Colletotrichum gloeosporiodies species complex was recorded as pathogens on L. chinense in Korea (Choi et al., 2012) and in Ukraine (Kliuchevych et al., 2019), on L. tulipifera in Argentina (Lori et al., 2004), and on hybrid L. chinense × tulipifera in southern China (Zhu et al., 2019). Other Colletotrichum pathogens such as Cerastium acutatum were reported on L. tulipifera in Argentina (Lori et al., 2004), and Chaetoceros siamense on L. chinense × tulipifera in China (Zhu et al., 2019). Apart from Colletotrichum spp., Venturia liriodendra caused a leafspot disease on L. tulipifera in the USA (Hanlin, 1987), and Physoderma sp. caused a brown-spot disease on L. chinense in China (Wang et al., 2013).
More recently, two anthracnose diseases have been observed on hybrid Liriodendron tree foliage growing at Liangshan Park in Beijing, northern China. One disease is symptomatic with round lesion spots circled with yellow halos. Each lesion spot possessed obviously unequal necrotic areas between the upper and lower cuticles, presenting a plain surface on the upper cuticle and a rough surface on the lower cuticle. The rest of the spotted leaves eventually turned yellow and immaturely dropped off. The other disease is characterized by irregular parched necrotic lesions without clear yellow halos, but seldom causes infected leaves to yellow or prematurely drop off. All hybrid Liriodendron trees at Liangshan Park showed infection, and the two anthracnose diseases are being found continually throughout Beijing. Besides describing the symptoms, comprehensive comparisons were also performed to distinguish these two anthracnose diseases based on phylogeny, pathogenicity, and biological features of all recorded pathogens and Liriodendron spp. hosts.
Materials and methods
Fungal pathogens isolation
Hybrid Liriodendron fungal isolates used in this study were randomly collected in August 2020 from diseased leaves of trees growing in Liangshan Park in Beijing, according to the two symptom types observed during the peak disease period. The two symptom types were not observed to co-occur on the same tree. Sampled leaves were rinsed with tap water and surface-sterilized for 1 min in 75% ethanol. Then, symptomatic tissues were cut into segments (3 × 3 mm) along lesion margins. The segments were further surface-disinfested in 3% NaClO for 1 min, 75% ethanol for 30s, rinsed three times in sterile distilled water, air-dried under aseptic conditions, and finally placed on potato dextrose agar (PDA). The cultures were incubated at 25 °C in the dark for 2 to 5 days. Hyphal tips growing from the segments were further transferred to new PDA Petri dishes to form pure colonies. Single-spore cultures were obtained from the pure colonies and examined morphologically (Li et al., 2007). Spores that sporulated from the monosporic colony were used to test pathogenesis.
Pathogenicity assay
Thirty-six Colletotrichum isolates were obtained from samples of the two anthracnose diseases. The isolated colonies from the same symptom samples shared almost identical morphological characteristics. Therefore, FPYF3060 and FPYF3062 strains were randomly assigned for pathogenicity assay and morphological description to represent all isolates from the yellowish ring and non-yellow ring symptoms, respectively. The pathogenicity assay was carried out on detached, healthy mature leaves of hybrid Liriodendron (Than et al., 2008). Wounds on the leaf surfaces were made with a sterile needle for spore inoculation. Twenty μL of conidial suspension (106 spores ml−1) was inoculated onto the wounds. Sterile distilled water was used for control treatments. All inoculated and control leaves were incubated in Petri dishes (150 mm diameter) containing sterilized wet cotton at 25 °C humidity in the dark to observe disease development. Lesions in treated leaves were recorded for seven days after inoculation. The pathogenicity trial was repeated. For matching Koch’s postulates, randomly selected lesions tissues were used to re-isolate fungal pathogens for each of the two FPYF3060 and FPYF3062 isolates that were inoculated, according to the procedure described above. The re-isolated FPYF3061 and FPYF3063 fungal isolates were cultured on PDA for microscopically comparing morphological and molecular phylogeny with the original FPYF3060 and FPYF3062 isolates, respectively. The four isolates were further checked for their corresponding identities using morphological characterizations and phylogeny. All treatments and tests were designed with replicate leaves for each isolate and repeated three times.
Morphological analysis
Mycelial discs (5 mm diameter) were taken from the edges of 6-day-old pure culture, placed into petri dishes containing PDA amended with 0.1% yeast extract (PDAY), and incubated at 25 °C in the dark (Han et al., 2016). Growth rate and colony morphology were assessed daily for six, and eighteen days, respectively. Appressoria were generated by conidia suspension on the surface of a hydrophobic slide (Fisher Scientific, Pittsburgh, USA) in vitro. Conidia and appressoria morphological characteristics were observed and measured with a U-TV0.63XC microscope (Olympus, Tokyo, Japan) and Carl Zeiss Microimaging Gmbh 37,081 (Gottingen, Germany).
Molecular identification and phylogenetic analysis
Four isolates (FPYF3060- FPYF3063) were further characterized by DNA sequencing for species verification and identity. Genomic DNA was extracted from each isolate using the CTAB method (Wang et al., 2017). The four-strain genomes were used as templates for PCR amplifying rDNA-ITS (ITS) sequence with ITS-1 (Gardes & Bruns, 1993) and ITS-4 (White et al., 1990) primers, partial glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene with GDF and GDR primers (Templeton et al., 1992), partial calmodulin (CAL) gene with CL1C and CL2C primers (Weir et al., 2012), partial actin (ACT) gene with ACT-512F and ACT-783R primers (Carbone & Kohn, 1999), partial β-tubulin (TUB2) gene with T1 and T2 primers (O’Donnell & Cigelnik, 1997), and partial chitin synthase (CHS-1) gene with CHS-79F and CHS-345R primers (Carbone & Kohn, 1999) (Table 1). PCR amplifications were performed with a Bioer LifeEco thermal cycler (BIOER Co., Ltd. Hangzhou, China) in a 25 μl reaction volume. PCR mixtures contained 1 μl DNA template, 0.5 μl of each primer, and 12.5 μl 2× PCR Taq Master Mix (TIANGEN, Beijing, China). PCR reactions for ITS amplification were performed under the following conditions: initial denaturation at 94 °C for 3 min, followed by 35 cycles of 30 s at 94 °C, 45 s at 55 °C plus extension for 1 min at 72 °C, with a final extension step at 72 °C for 5 min. PCR conditions for other loci were the same as for ITS amplification except for the annealing temperatures: ACT at 58 °C, TUB2 at 55 °C, GAPDH at 60 °C, CHS-1 at 58 °C, and CAL at 59 °C. PCR products were sequenced with an ABI 3730XL automatic sequencer (Applied Biosystems, USA) by BGI Genomics Co., Ltd. Raw DNA sequences obtained (forward and reverse) were edited and spliced with BioEdit 7.2.5 (Hall, 1999) and then aligned in MAFFT 7 (https://mafft.cbrc.jp/alignment/software/) to form clean sequences for species identification through BLASTn in the NCBI database. All the sequences generated in this study with their accession numbers were deposited in GenBank (Table 2). For phylogenetic inference, all reference sequences from 53 Colletotrichum isolates, representing 52 taxa in the C. gloeosporioides species complex, from the pathogens recorded on Liriodendron, and from the C. boninense (ICMP 17904) outgroup were retrieved from GenBank (Choi et al., 2012; Liu et al., 2013; Weir et al., 2012; Zhu et al., 2019) and listed with their NCBI accession numbers in Table 2. A four loci phylogenetic tree generated by neighbor-joining using ITS, GAPDH, ACT, and TUB2 sequences in MEGA 7 (Stecher et al., 2016), was constructed based on the Tamura three-parameter model. Relative branch stability was assessed by bootstrapping with 1000 replications. To further discriminate native pathogens from those reported, the phylogeny relationship based on the four genes phylogenetic tree was also tested by ML and MP inferences. The MP tree was constructed using the subtree-pruning-regrafting (SPR) search option with 1000 random sequence additions (level = 1). For the ML tree, the following settings were used: the Tamura 3-parameter model, gamma-distributed (5 categories), and heuristic method using SPR-extensive (Hu et al., 2015). To discriminate our strains from C. gloeosporioides s.s. species (Table S1), we also constructed a six-loci phylogenetic tree based on ITS, GAPDH, ACT, TUB2, CAL, and CHS-1, and a single-locus phylogenetic tree with ITS, or GAPDH sequences with the N-J method.
Results
Symptom description
The two anthracnose diseases on hybrid Liriodendron had their own unique characteristic symptoms (Fig. 1). During the disease epidemic period, the yellow halo anthracnose type was symptomized by many small necrotic lesion spots with thick yellow halos on leaves (Fig. 1A). Lesion spots were mostly round or near-round and separated from each other. These features remained, even in fallen, initially decaying leaves (Fig. 1A-E). Lesions had unequal necrotic areas on the upper and lower leaf sides, although less obvious on the upper side. The necrotic surface texture on the upper leaf side was touched flat while it was leathery on the lower surface (Fig. 1F-G). Entire diseased leaves eventually turned yellowish and dropped early (Fig. 1B-C). The distinct yellow halos became more obvious on fallen diseased leaves (Fig. 1D-E). The non-yellow halo anthracnose disease type, was marked by fewer, but larger irregular necrotic lesions on infected leaves (Fig. 1H-I). The yellow halo on lesion margins was not observed, nor was fine ring-like chlorosis. This anthracnose disease type did not turn the entire leaf yellowish and they didn’t drop early (Fig. 1J). Necrotic cuticles in lesions on fallen leaves became fragile and could easily be cracked in the spot center (Fig. 1K). A single necrotic lesion had an identical area on the upper and lower leaf surfaces. Additionally, there were several concentric rings formed in the lesions (Fig. 1L-M). The two anthracnose diseases were not observed on the same tree.
The symptoms of two types of anthracnose diseases on leaves of hybrid Liriodendron. A and B, disease symptom of etiolation (yellow halo) anthracnose on trees in the field. C, heavy defoliation caused by etiolation anthracnose. D and E, the upper and lower sides of fallen diseased leaves of etiolation anthracnose. F and G, the obverse and reverse sides of an etiolation anthracnose leaf. H and I, disease symptoms of non-etiolation (non-yellow halo) anthracnose on trees in the field. J, much less defoliation caused by non-etiolation anthracnose. K, the hole on fallen diseased leaf of non-etiolation anthracnose. L and M, the obverse side and reverse side of non-etiolation anthracnose leaf
Fungal isolate pathogenicity
The FPYF3060 strain isolated from the yellow halo type and the FPYF3062 strain isolated from the non-yellow halo type successfully infected host leaf tissues (Fig. 2). Three days after inoculation, tip black spots occurred in leaf wound sites. These spots then gradually developed into overt circular and black necrotic lesions. On day 7, leaf lesions inoculated with strain FPYF3060 were 12.4 mm (n = 21, SD = 0.28) in average diameter, and produced grainy conidial structures (Fig. 2A). Conversely, leaf lesions inoculated with strain FPYF3062 were 11.7 mm (n = 21, SD = 0.28) in average diameter, and did not sporulate (Fig. 2B).
FPYF3060 and FPYF3062 morphological characteristics
On PDAY, the FPYF3060 strain formed a circular colony with dense white, fluffy aerial mycelia for 18 days under culture conditions. The reverse side of the colony was white (Fig. 3A). The colony average growth rate was 11.0 mm per day and its conidia were hyaline, straight, cylindrical, obtuse at the apex, truncate at the base, and 14.2 ~ 16.9 × 4.5 ~ 5.8 μm (mean 15.6 × 5.1 μm, n = 50) (Fig. 3C). Appressoria were brown, ovoid to irregularly shaped, and 7.4 ~ 8.6 × 6.7 ~ 7.8 μm (mean 8.0 × 7.3 μm, n = 50) (Fig. 3D). On PDAY, the FPYF3062 strain colony was initially white but turned pale gray for 18 days, and the reverse side of the colony was gray in the periphery, deep dark in the middle, and brown/Gy-white in the center. The average growth rate was 12.6 mm per day. The strain initially produced conidial masses around mycelial plugs after incubation for two or three days, and sporulated continuously for 18 days (Fig. 3B). Conidia were cylindrical, straight or slightly curved with round ends, single-celled, and 12.1 ~ 13.6 × 4.3 ~ 4.9 μm (on average 12.9 × 4.6 μm, n = 50) (Fig. 3E). Appressoria were oval-shaped, 7.2 ~ 8.9 × 5.9 ~ 6.7 μm (on average 8.0 × 6.3 μm, n = 50), and brown (Fig. 3F). The morphological characteristics of the re-isolated FPYF3061 strain were identical to the original FPYF3060 strain used for pathogenicity assay. The FPYF3063 strain was also identical to the original FPYF3062 strain challenge agent for the pathogenicity test (data not shown).
The morphological characteristics of isolates FPYF3060 and FPYF3062 on PDAY in dark for 18 days. A, View of the upper and reverse sides of FPYF3060 colony on PDAY. B, View of the upper and reverse sides of FPYF3062 colony on PDAY. C and D, The conidia (scale bar = 10 μm) and appressoria (scale bar = 20 μm) of FPYF3060. E and F, The conidia (scale bar = 10 μm) and appressoria (scale bar = 20 μm) of FPYF3062
Molecular identification and phylogenetic analysis
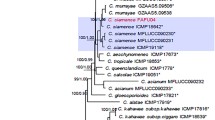
Four-loci phylogenetic inference, based on GAPDH, ACT, TUB2, and ITS region genes, showed that native FPYF3060–3063 isolates were clustered with C. gloeosporioides s.s. in a clade by 98% support, however, they could also be ascribed into different subclades within the clade (Fig. 4). The FPYF3062 and FPYF3063 isolates were grouped into a subclade with 96% bootstrap value. The FPYF3060 and FPYF3061 isolates were grouped into another subclade with 85% bootstrap value, and at a certain distance to the terminal cluster grouped with C. gloeosporioides s.s. type strains. Therefore, the phylogenetic relationships of the native pathogens with yellow and non-yellow halo symptoms suggest that they have phylogenetic linkages within the C. gloeosporioides s.s. clade, but differential phylogenetic linages.
Phylogenetic tree based on neighbor-joining using MEGA7. The tree was built using concatenated data from four sequences of ITS, GAPDH, ACT, and TUB2 of the strains FPYF3060- FPYF3063. C. boninense is used as outgroup. Bootstrap values >70% (1000 replications) marked at the nodes. Bar = 0.02 substitutions per nucleotide position. Our four strains are in bold. Those pathogenic strains reported by Zhu et al. (2019) were marked in green
Discussion
Molecular identifications using ITS sequencing analysis, protein-encoding genes, and phylogenetic inferences determined that native fungal isolates FPYF3060–3063 on anthracnose diseased leaves of hybrid Liriodendron were strains from C. gloeosporioides s.s. (Fig. 4, Fig. S1-S4). Their colony, conidia, and appressoria morphological features were also consistent with descriptions of the C. gloeosporioides s.s. epitype (Cannon et al., 2008). Their pathogenicity on hybrid Liriodendron was verified via Koch’s postulates, with the same strains being successfully re-isolated (Figs. 2 and 3). In the pathogenicity test, symptoms associated with yellow halo disease did not fit well with strain FPYF3060, partly because detached leaf physiology could be inadequate to explain the yellowish occurrence, such as from insufficient light (Liu et al., 2007; Zhu et al., 2019). Furthermore, although the two native strains were determined as C. gloeosporioides s.s. species, their colony characteristics showed many differences in texture, color, and productivities (Fig. 3A-B, Table S2), which were distinct in phylogeny subclades (Fig. 4, Fig. S1-S4). The strain FPYF3062 (FPYF3063) phylogeny even suggested that C. gloeosporioides s.s. could be divided into distinctive genetic groups (Bhunjun et al., 2021; Weir et al., 2012) (Fig. 4). The six-loci phylogenetic tree (Fig. S1) and other phylogenetic relationships (Fig. S2-S4) together showed the same result. Therefore, the Colletotrichum pathogens from the yellow and non-yellow halo anthracnose diseases were different (Fig. 4, Fig. S1, Table S2).
On Liriodendron hosts, several leaf spot diseases have been documented including those caused by Colletotrichum spp.. Hanlin (1987) recorded that Venturia liriodendra caused leafspot disease on L. tulipifera (Hanlin, 1987). More recently, Zhu et al. (2019) reported C. gloeosporioides strain G2 and C. siamense strain R3 infections on hybrid Liriodendron growing in southern China. Kliuchevych et al. (2019) detected C. gloesporioides s.s. on L. chinense in Ukraine. Fu et al. (2020) tested the pathogenicity of C. gloeosporioides strain Lc1 isolated from L. chinense host tree leaves, and demonstrated that it was able to cause leaf spots. However, some significant differences exist in the diseases presented in this paper compared to those pathogens or/and symptoms already documented (Tables S2-S3, Fig. S5). For symptoms, only the disease on tulip trees in Ukraine seemed to be highly similar to our yellow halo anthracnose diseases (Fig. S5), but its hosts were L. chinense, not hybrid L. chinense × tulipifera trees. Moreover, the Ukraine Colletotrichum pathogen formed sporulations on the leaves (Kliuchevych et al., 2019) while our Colletotrichum pathogens were not observed to sporulate on necrotic lesions throughout the diseased period even on fallen infected leaves in the field (Fig. 1). However, there was no information on phylogenetic molecular marker sequences (even ITS) available for the Ukraine C. gloesporioides strain (Kliuchevych et al., 2019). There are clearly different symptoms (Table S3, Fig. S5) and pathogens (Table S2) for anthracnose diseases on hybrid Liriodendron trees between southern and northern China (Fu et al., 2020; Zhu et al., 2019). Phylogenetic relationships based on the single locus, such as ITS or GPADH, and multi-loci concatenated sequences of four or six marker molecular sequences make it clear that native FPYF strains from northern China are distinct from pathogens of C. gloeosporioides s.s. strains from southern China (Fig. S1-S4, Fig. 4). Notably, except for phylogeny trees based on single ITS sequences showing uncertain relationships for all C. gloeosporioides s. s. strains (Fig. S6), all multi-loci phylogenetic trees robustly supported that the strains FPYF3060–3063, on the hybrid Liriodendron hosts from northern China have evolved a unique distance phylogeny within the C. gloeosporioides s. s. clade (Fig. S2-S4). Strain FPYF3062 (FPYF2063), the pathogenic agent of non-yellow halo anthracnose, diverged early from almost all reported C. gloeosporioides s. s. strains. Moreover, there were differences in conidia and appressoria size between our two strains (FPYF3060 (FPYF3061) and FPYF3062 (FPYF3063)) and the southern strain C. gloeosporioides G2 (Table S2) (Zhu et al., 2019). These complementary comparisons further confirm that the strain FPYF3060 (FPYF3061) or FPYF3062 (FPYF3063) on hybrid Liriodendron hosts in northern China is not identical to those reported on hosts in southern China (Zhu et al., 2019). As for other pathogens documented on Liriodendron hosts, Colletotrichum gloeosporioides CG2 in Korea cannot be a C. gloeosporioides species, but could be C. aotearoa according to its phylogenetic characterization (Fig. S7). The CG2 strain had smaller conidia and appressoria than strains FPYF3060 and FPYF3062 (Table S2) (Choi et al., 2012). The pathogen Colletotrichum acutatum LPS47188 on L. tulipifera from Argentina also had smaller conidia than our strains (Lori et al., 2004, Table S2), but no data was available for appressoria comparison. The pathogenic strain, Colletotrichum gloeosporioides Lc1, which was isolated from L. chinense hosts and recently genome sequenced (Fu et al., 2020), exhibited a distinctive distance from our strains in phylogenetic relationships (Fig. S2). Finally, the two Colletotrichum anthracnose disease types in this paper were easily discriminated by their different symptoms (Fig. 1). According to our observations, in the field the two types were seldom observed co-occurring on the same tree, although they randomly infected hosts within hybrid Liriodendron populations at the same time. We expect to explore the specificity between the pathogens and hosts in the future.
Conclusions
In view of their polyphyletic nature (Bhat et al., 2018; Bhunjun et al., 2021; Jayawardena et al., 2016; Weir et al., 2012), Colletotrichum gloeosporioides strains on hybrid Liriodendron hosts growing in northern China exhibited an obvious phylogenetic distance with those documented on three tulip tree host species based on detailed molecular phylogenetic analyses (Fig. 4, Fig. S1-S4), conidial features (Fig. 3, Table S2) and unique symptoms (Fig. 3, Fig. S5) (Choi et al., 2012; Fu et al., 2020; Hanlin, 1987; Kliuchevych et al., 2019; Lori et al., 2004; Wang et al., 2013; Zhu et al., 2019). The yellow and non-yellow halo anthracnose diseases on hybrid Liriodendron in northern China also had two distinctive types of foliage spots (Fig. 3, Fig. S5). Although anthracnose disease had been reported on hybrid Liriodendron hosts in Nanjing, a city in south-eastern China (Zhu et al., 2019), the anthracnose pathogens in northern hybrid Liriodendron hosts were distanced from the southern strains in pathogenic genetics (Fig. S2-S6). Moreover, the non-yellow halo anthracnose caused by the C. gloeosporioides s.s. strain FYPF3062 (3063) on hybrid Liriodendron in Beijing in northern China, was symptomatically distinct from reported anthracnose diseases on the same host species in southern regions (Fig. S5) (Choi et al., 2012; Fu et al., 2020; Kliuchevych et al., 2019; Wang et al., 2013; Zhu et al., 2019). Therefore, the two Colletotrichum gloeosporioides diseases on hybrid Liriodendron hosts in northern China were confirmed to be novel types of anthracnose leaf spots. Both diseases could significantly degrade hybrid Liriodendron landscapes in northern China. Recent, frequent reports of Colletotrichum spp. pathogenic diseases emerging on Liriodendron tulip trees are alarming in the present world of global climate change. Urgent, preventative measures are required for Colletotrichum spp. disease management.
References
Bhat, N. N., Mahiya, F., Padder, B. A., Shah, M. D., Dar, M. S., Nabi, A., Bano, A., Rasool, R. S., & Sana, S. (2018). Microsatellite mining in the genus Colletotrichum. Gene Reports, 13, 84–93. https://doi.org/10.1016/j.genrep.2018.09.001
Bhunjun, C. S., Phukhamsakda, C., Jayawardena, R. S., Jeewon, R., Promputtha, I., & Hyde, K. D. (2021). Investigating species boundaries in Colletotrichum. Fungal Diversity, 107, 107–127. https://doi.org/10.1007/s13225-021-00471-z
Cannon, P. F., Buddie, A. G., & Bridge, P. D. (2008). The typification of Colletotrichum gloeosporioides. Mycotaxon, 104, 189–204.
Carbone, I., & Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia, 91, 553–556. https://doi.org/10.2307/3761358
Choi, O., Choi, O., Kwak, Y. S., Kim, J., & Kwon, J. H. (2012). Spot anthracnose disease caused by Colletotrichum gloeosporioides on tulip tree in Korea. Mycobiology, 40, 82–84. https://doi.org/10.5941/MYCO.2012.40.1.082
Choub, V., Maung, C. E. H., Won, S. J., Moon, J. H., Kim, K. Y., Han, Y. S., Cho, J. Y., & Ahn, Y. S. (2021). Antifungal activity of cyclic tetrapeptide from bacillus velezensis CE 100 against plant pathogen Colletotrichum gloeosporioides. Pathogens, 10(2), 209. https://doi.org/10.3390/pathogens10020209
Dean, R., Van-Kan, J. A. L., Pretorius, Z. A., Hammond-Kosack, K. E., Di-Pietro, A., Spanu, P. D., Rudd, J. J., Dickman, M., Kahmann, R., Ellis, J., & Foster, G. D. (2012). The top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology, 13, 414–430. https://doi.org/10.1111/j.1364-3703.2012.00822.x
Fu, F. F., Hao, Z., Wang, P., Lu, Y., Xue, L., Wei, G., Tian, Y., Hu, B., Xu, H., Shi, J., Cheng, T., Wang, G., Yi, Y., & Chen, J. (2020). Genome sequence and comparative analysis of Colletotrichum gloeosporioides isolated from Liriodendron leaves. Phytopathology, 110, 1260–1269. https://doi.org/10.1094/PHYTO-12-19-0452-R
Gardes, M., & Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecology, 2, 113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98. https://doi.org/10.1021/bk-1999-0734.ch008
Han, Y. C., Zeng, X. G., Xiang, F. Y., Ren, L., Chen, F. Y., & Gu, Y. C. (2016). Distribution and characteristics of Colletotrichum spp. associated with anthracnose of strawberry in Hubei, China. Plant Disease, 100, 996–1006. https://doi.org/10.1094/PDIS-09-15-1016-RE
Hanlin, R. T. (1987). Venturia liriodendri sp. nov., associated with a leafspot disease of Liriodendron. Mycologia, 79, 464–467. https://doi.org/10.2307/3807473
Hu, M. J., Grabke, A., & Schnabel, G. (2015). Investigation of the Colletotrichum gloeosporioides species complex causing peach anthracnose in South Carolina. Plant Disease, 99, 797–805. https://doi.org/10.1094/PDIS-10-14-1076-RE
Jayawardena, R. S., Hyde, K. D., Damm, U., Cai, L., Liu, M., Li, X. H., Zhang, W., Zhao, W. S., & Yan, J. Y. (2016). Notes on currently accepted species of Colletotrichum. Mycosphere, 7, 1192–1260. https://doi.org/10.5943/mycosphere/si/2c/9
Jayawardena, R. S., Hyde, K. D., de Farias, A. R. G., Bhunjun, C. S., Ferdinandez, H. S., Manamgoda, D. S., Udayanga, D., Herath, I. S., Thambugala, K. M., Manawasinghe, I. S., Gajanayake, A. J., Samarakoon, B. C., Bundhun, D., Gomdola, D., Huanraluek, N., Sun, Y., Tang, X., Promputtha, I., & Thines, M. (2021). What is a species in fungal plant pathogens? Fungal Diversity, 109(1), 239–266. https://doi.org/10.1007/s13225-021-00484-8
Jefferies, P., Dodd, J. C., Jeger, M. J., & Plumbley, R. A. (1990). The biology and control of Colletotrichum species on tropical fruit crops. Plant Pathology, 39, 343–366. https://doi.org/10.1111/j.1365-3059.1990.tb02512.x
Kamle, M., & Kumar, P. (2016). Colletotrichum gloeosporioides: Pathogen of anthracnose disease in mango (Mangifera indica L.). In P. Kumar, V. Gupta, A. Tiwari, & M. Kamle (Eds.), Current trends in plant disease diagnostics and management practices (pp. 207–219). Springer-Cham. https://doi.org/10.1007/978-3-319-27312-9_9
Khan, M. R., Chonhenchob, V., Huang, C., & Suwanamornlert, P. (2021). Antifungal activity of propyl disulfide from neem (Azadirachta indica) in vapor and agar diffusion assays against anthracnose pathogens (Colletotrichum gloeosporioides and Colletotrichum acutatum) in mango fruit. Microorganisms, 9(4), 839. https://doi.org/10.3390/microorganism9040839
Kliuchevych, M. M., Chumak, P. Y., Vigera, S. M., & Stolyar, S. G. (2019). First detection of Colletotrichum gloesporioides (Penz.) pens. & Sacc. On Liriodendron chinense (Hemsl.) Sarg. In Ukraine. Modern Phytomorphology, 13, 9–12.
Li, M. F., He, J., Ding, L., Kang, J. C., Zhang, Q., & Zheng, Q. W. (2007). Single spore strains without producing fruit body isolated from Cordyceps militeris and their RAPD analysis. Southwest China Journal of Agricultural Sciences, 20, 547–550.
Li, N., Xu, D., Huang, R., Zheng, J., Liu, Y., Hu, B., Gu, Y., & Du, Q. (2022). A new source of Diterpene lactones from Andrographis paniculata (Burm. F.) Nees-two endophytic Fungi of Colletotrichum sp. with antibacterial and antioxidant activities. Frontiers in Microbiology, 13. https://doi.org/10.3389/fmicb.2022.819770
Liu, F., Damm, U., Cai, L., & Crous, P. W. (2013). Species of the Colletotrichum gloeosporioides complex associated with anthracnose diseases of Proteaceae. Fungal Diversity, 61, 89–105. https://doi.org/10.1007/s13225-013-0249-2
Liu, F., Wang, M., Damm, U., Crous, P. W., & Cai, L. (2016). Species boundaries in plant pathogenic fungi: A Colletotrichum case study. BMC Evolutionary Biology, 16, 81. https://doi.org/10.1186/s12862-016-0649-5
Liu, G., Kennedy, R., Greenshields, D. L., Peng, G., Forseille, L., Selvaraj, G., & Wei, Y. (2007). Detached and attached Arabidopsis leaf assays reveal distinctive defense responses against hemibiotrophic Colletotrichum spp. Molecular Plant-Microbe Interactions, 20, 1308–1319. https://doi.org/10.1094/MPMI-20-10-1308
Lori, G. A., Alippi, A. M., & Dimenna, S. (2004). First report of species of Colletotrichum causing leaf blotch of Liriodendron tulipifera in Argentina. Plant Disease, 88, 1381. https://doi.org/10.1094/PDIS.2004.88.12.1381A
Moreira, R. R., Zielinski, E. C., Castellar, C., Bergamin Filho, A., & May De Mio, L. L. (2021). Study of infection process of five species of Colletotrichum comparing symptoms of glomerella leaf spot and bitter rot in two apple cultivars. European Journal of Plant Pathology, 159(1), 37–53. https://doi.org/10.1007/s10658-020-02138-y
O’Donnell, K., & Cigelnik, E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus fusarium are nonorthologous. Molecular Phylogenetics and Evolution, 7, 103–116. https://doi.org/10.1006/mpev.1996.0376
Scindiya, M., Malathi, P., Kaverinathan, K., Sundar, A. R., & Viswanathan, R. (2021). Knock-down of glucose transporter and sucrose non-fermenting gene in the hemibiotrophic fungus Colletotrichum falcatum causing sugarcane red rot. Molecular Biology Reports, 48(3), 2053–2061. https://doi.org/10.1007/s11033-021-06140-3
Stecher, G., Kumar, S., & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. https://doi.org/10.1093/molbev/msw054
Templeton, M. D., Pikkerink, E. H. A., Solon, S. L., & Crowhurst, R. N. (1992). Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene, 122, 225–230. https://doi.org/10.1016/0378-1119(92)90055-T
Than, P. P., Jeewon, R., Hyde, K. D., Pongsupasamit, S., Mongkolporn, O., & Taylor, P. W. J. (2008). Characterization and pathogenicity of Colletotrichum species associated with anthracnose on chilli (Capsicum spp.) in Thailand. Plant Pathology, 57, 562–572. https://doi.org/10.1111/j.1365-3059.2007.01782.x
Wang, J. X., Ma, L. J., Zhang, L. Q., & Mao, S. F. (2013). Pathogen identification of the brown-spot disease of Liriodendron chinensis. Scientia Silvae Sinica, 49, 189–191. https://doi.org/10.11707/j.1001-7488.20130628
Wang, K. Y., Strobel, G. A., & Yan, D. H. (2017). The production of 1,8-cineole, a potential biofuel, from an endophytic strain of Annulohypoxylon sp. FPYF3050 when grown on agricultural residues. Journal of Sustainable Bioenergy Systems, 7, 65–84. https://doi.org/10.4236/jsbs.2017.72006
Wang, P., Dong, Y., Zhu, L., Hao, Z., Hu, L., Hu, X., Wang, G., Cheng, T., Shi, J., & Chen, J. (2021). The role of γ-aminobutyric acid in aluminum stress tolerance in a woody plant, Liriodendron chinense× tulipifera. Horticulture Research, 8, 80. https://doi.org/10.1038/s41438-021-00517-y
Wang, Z. R. (1997). Genetic resources preservation and breeding prospect of Liriodendron chinense. Forest Science and Technology, 9, 8–10.
Weir, B. S., Johnston, P. R., & Damm, U. (2012). The Colletotrichum gloeosporioides species complex. Studies in Mycology, 73, 115–180. https://doi.org/10.3114/sim0011
White, T. J., Bruns, T., Lee, S., & Taylor, J. W. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, & T. J. White (Eds.), PCR protocols: A guide to methods and applications (pp. 315–322). Academic Press.
Zhu, L. H., Wan, Y., Zhu, Y. N., Huang, L., & Liu, C. L. (2019). First report of species of Colletotrichum causing leaf spot of Liriodendron chinense × tulipifera in China. Plant Disease, 103, 1431. https://doi.org/10.1094/PDIS-12-18-2265-PDN
Acknowledgments
This research was funded by DEVELOPMENT PROJECT OF ECOLOGY AND NATURE CONSERVATION INSTITUTE, CAF, grant number 99813-2020 and YOUTH FUND PROJECT OF NATIONAL SCIENCE FOUNDATION OF CHINA, grant number 31901316. Mr. Zheng Wang and Mrs. Shimeng Tan are acknowledged for their support and help in constructing phylogenetic trees. We also appreciate Miss. Danran Bian for her help in pathogenic test.
Availability of data and material
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Code availability
Not applicable.
Funding
DEVELOPMENT PROJECT OF RESEARCH INSTITUTE OF FOREST ECOLOGYY, ENVIRNMENT AND PROTECTION, CAF, grant number 99813–2020 and YOUTH FUND PROJECT OF NATIONAL SCIENCE FOUNDATION OF CHINA, grant number 31901316.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest/competing interests
The authors declare no conflict of interest.
Research involving human participants and/or animals
Not applicable.
Informed consent
Not applicable.
Supplementary Information
Fig. S1
Phylogenetic tree based on neighbor-joining using MEGA7. The tree was built using concatenated data from six sequences of ITS, GAPDH, ACT, CAL, CHS-1, and TUB2 of FPYF3060-FPYF3063 described in this study. C. boninense was used as outgroup. Our four isolates were in bold. Bar = 0.02 substitutions per nucleotide position. (JPG 112 kb)
Fig. S2
Phylogenetic tree of all C. gloeosporioides s. s. strains with allied taxa calculated from glyceraldehyde-3-phosphate dehydrogenase using neighbor-joining method. C. boninense was used as outgroup. Bootstrap values >50% (1000replications) were given at the nodes. Bar = 0.005 substitutions per nucleotide position. Our four isolates were in bold. Those pathogens C. gloeosporioides s. s. strains reported by Zhu et al. (2019) and Fu et al. (2020) were marked in colors. (JPG 99 kb)
Fig. S3
Phylogenetic tree based on maximum-likelihood using MEGA7. The tree was built using concatenated data from sequences of ITS, GAPDH, ACT, and TUB2 of FPYF3060- FPYF3063 described in this study. C. boninense was used as outgroup. Bootstrap values >70% (1000 replications) were given at the nodes. Bar = 0.02 substitutions per nucleotide position. Our four isolates were in bold. Those pathogens reported by Zhu et al. (2019) were marked in green. (JPG 106 kb)
Fig. S4
Phylogenetic tree based on maximum parsimony using MEGA7. The tree was built using concatenated data from sequences of ITS, GAPDH, ACT, and TUB2 of FPYF3060- FPYF3063 described in this study. C. boninense was used as outgroup. Bootstrap values >70% (1000 replications) were given at the nodes. Bar = 0.02 substitutions per nucleotide position. Our four isolates were in bold. Those pathogens reported by Zhu et al. (2019) were marked in green. (JPG 112 kb)
Fig. S5
Symptoms of the anthracnose diseases on the Liriodendron chinense or Hybrid Liriodendron. a-f, Symptom of the anthracnose disease on the L. chinense in Ukraine, reported by Kliuchevych et al. (2019). g, Symptom of the anthracnose disease on the Liriodendron Hybrids in northern China, reported by Zhu et al. (2019). h, Upper side and lower side of yellow halo anthracnose leaf. i, Upper side and lower side of non-yellow halo anthracnose leaf. (JPG 399 kb)
Fig. S6
Phylogenetic tree of all C. gloeosporioides s. s. strains with allied taxa calculated from internal transcribed spacer using neighbor-joining method. C. boninense was used as outgroup. Bootstrap values >50% (1000 replications) were given at the nodes. Bar = 0.005 substitutions per nucleotide position. Our four isolates were in bold. Those pathogens C. gloeosporioides s. s. strains reported by Zhu et al. (2019) and Fu et al. (2020) were marked in colors. (JPG 97 kb)
Fig. S7
Phylogenetic tree of FPYF3060-FPYF3063 and C. gloeosporioides CG2 (Choi et al., 2012) with allied taxa calculated from internal transcribed spacer using neighbor-joining method. C. boninense was used as outgroup. Bootstrap values >50% (1000 replications) were given at the nodes. Bar = 0.005 substitutions per nucleotide position. Our four isolates were in bold. C. gloeosporioides CG2 was marked in blue. (PNG 1786 kb)
Table S1
A list of strains from Colletotrichum gloeosporioides sensu stricto. (DOCX 24 kb)
Table S2
Comparison of the sizes of conidia and appressoria. (DOCX 17 kb)
Table S3
Comparison of the symptoms of anthracnose diseases on Liriodendron spp.. (DOCX 18 kb)
Rights and permissions
About this article
Cite this article
Dou, G., Lü, X., Ren, F. et al. Two distinct leaf anthracnose disease infections in hybrid Liriodendron trees in northern China. Eur J Plant Pathol 163, 775–787 (2022). https://doi.org/10.1007/s10658-022-02514-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-022-02514-w