Abstract
The causal agent of red root rot disease of common vetch (Vicia sativa) on the far North Coast of New South Wales, Australia, has been identified as Atractiella rhizophila. This basidiomycete infected root tissues directly without specialized appressoria, and developed as a necrotroph in the cortex external to the stele. It has been recorded previously as a non-pathogenic endophyte of Populus spp. and Pinus spp. roots, and a symbiont on Quercus rubra, but this is the first report of A. rhizophila as a plant pathogen.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Common vetch (Vicia sativa cv. Golden Tares) was grown for fodder on red ferrosols of the far North Coast of New South Wales (NSW), Australia, from 1950 to 1970 (Holder et al. 1963; Barnard 1970). It was sown annually into pastures of perennial grasses and naturalized white clover (Trifolium repens). The yields of Golden Tares vetch became increasingly affected by root and stem rot diseases during the 1960s, leading to the use of alternative cultivars such as Morava vetch and other fodder crops.
Red root rot was described by Allen (1972) as a new disease with an unnamed fungal pathogen. In 1965, a voucher specimen of diseased tissue and a living culture of the causal agent were deposited in the NSW Plant Pathology and Mycology Herbarium as DAR 14088. Pathogenicity of DAR 14088 was demonstrated (Allen 1967) in accordance with Koch (1882), firstly, by inoculation of Golden Tares vetch seedlings established on an agar medium in vitro (Date and Vincent 1962) using mycelium in PDA as inoculum and, secondly, by germination of Golden Tares vetch seed in a mixture of mycelial homogenate and steamed loam (Baker 1957). Vetch roots were examined for red root rot disease symptoms periodically and the pathogen was re-isolated from disease lesions. In both tests, an equivalent quantity of agar without mycelium was used as a control. Both methods of inoculation led to identical expression of red root rot symptoms.
A description of DAR 14088, its growth on various nutrient media, responses to incubation temperatures, and pathogenicity to common vetch and other legumes, is given in Allen (1967). No spores, sporophores, clamp connections or septa were observed in the basal hyphae, but simple septa were apparent in hyphal branches referred to as gemmae (Allen 1967). The pathogen was classified provisionally as an oomycete because basal hyphae were coenocytic and young hyphae exhibited cytoplasmic streaming. The culture, now over 50 years old, is no longer viable or suitable for molecular examination, but re-examination of DAR 14088 in 2017 revealed previously unreported septae in the basal hyphae, monilioid hyphal cells, and a rectangular branching habit. These observations indicated that the fungus was a basidiomycete rather than an oomycete. Our aims subsequently were to recover the fungus that caused red root rot disease of V. sativa and, further, to identify the fungus using morphological and molecular methods.
Materials and methods
Isolation of fungi
In March 2019, a red ferrosol associated with white clover roots was sampled from a pasture at Wollongbar NSW (28.817419°S; 153.391257°E) where red root rot had affected Golden Tares vetch during the 1960s. The sampled soil was sieved, sown with Morava vetch, and incubated in a greenhouse at ambient temperatures (20–30 °C) with overhead sprinkler irrigation equivalent to rainfall of 8 mm d-1. The root systems were examined 14 d after sowing and excised diseased root segments, 10–20 mm long, were shaken vigorously in sterilized water for 5–10 s, recovered and blotted dry. This process was repeated four more times on these segments before diseased tissue pieces, 2–3 mm long, were placed on WA containing 50 ppm streptomycin sulphate, 10 ppm oxytetracycline, and 1 ppm prochloraz fungicide. Hyphal tips of fungi that emerged from diseased cortical cells were transferred to PDA. A living culture was deposited in the Queensland Plant Pathology Herbarium as BRIP 70197.
Pathogenicity testing
Pathogenicity of the isolate BRIP 70197 was tested in a Petri dish assay adapted from Muyolo et al. (1993) and Date and Vincent (1962). Morava vetch seed were surface sterilized by immersion in ethanol momentarily to break surface tension, then in 60 mM silver nitrate solution for 3 min followed by 3 M sodium chloride for 1–2 s and finally, by rinsing in sterilized water. The seeds were incubated on sterilized moist sponge cloth at ambient temperatures for 3 d before germinated sprouts were sown into 2 mm diameter wells cut into WA that had been poured into 90 mm diameter Petri dishes to a depth of 5 mm. Mycelium from a PDA culture was placed beside the taproot of a sown vetch seedling, and the inoculated seedlings were incubated for 10 d in darkness. The root systems were examined for disease symptoms and segments of diseased root were surface sterilized by washing in sterilized water as before, immersing in 60 mM silver nitrate for 10 s and 3 M sodium chloride for 1–2 s, and rinsing in sterilized water. Surface sterilized segments of diseased root were incubated for 7 d on WA with antibiotics as before, and hyphal tips were sub-cultured to PDA. Segments of asymptomatic root tissue on inoculated vetch plants were surface sterilized and plated as a control.
Molecular examination
DNA was extracted from mycelium of isolate BRIP 70197 using the ISOLATE II Genomic DNA kit (Meridian Bioscience Incorporated, Ohio, USA) according to the manufacturer’s instructions. The ITS region was amplified with the primers ITS1-f (Gardes and Bruns 1993) and ITS4 (White et al. 1990) and the LSU amplified with primers LR0R and LR5 (Vilgalys and Hester 1990). Purified PCR product was sequenced by Macrogen Incorporated, Seoul, South Korea.
Results
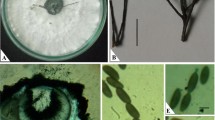
The isolate recovered from Morava vetch (BRIP 70197) had cultural and microscopic characteristics similar to a dried pathogen culture from red root rot of Golden Tares vetch (DAR 14088). On PDA, colonies of BRIP 70197 were composed of both superficial and immersed mycelium, leathery in texture, with an entire margin of translucent hyphae, and a velvety white surface that became slightly wrinkled after 4 wk. The mycelial matt assumed the colour of the media, so departures from the normal white colour were variable. Hyphae were bi-nucleate, hyaline, smooth-walled, and comprised two cell types: basal type, filamentous, 3–5 μm in diameter, with simple septa (Fig. 1a); monilioid type, initially filamentous but 7–18 μm in diameter depending on maturity, irregularly branched, with many simple septa, maturing to spheroidal cells in chains (Fig. 1b), and usually immersed in PDA or host parenchyma. Monilioid cells were uncommon on WA but those on PDA, when separated from the mycelial matt and incubated in water, produced 1–4 basal hyphae per cell. Sporophores, spores, clamp connections, and sclerotia were not observed.
Septated hyphal branch with immature monilioid cells produced on a septated basal hypha of Atractiella rhizophila in PDA [scale bar = 10 μm] (a). Hyphal branch with mature monilioid cells produced on a basal hypha of Atractiella rhizophila in PDA [scale bar = 10 μm] (b). Early red root rot streak lesions on rootlets of Vicia sativa cv. Morava in a Petri dish assay 10 d after inoculation with Atractiella rhizophila [scale bar = 3 mm] (c). Red root rot disease on Vicia sativa cv. Morava in a Petri dish assay 10 d after inoculation with Atractiella rhizophila [scale bar = 10 mm] (d)
In Petri dish assays, BRIP 70197 caused necrotic streaks (Fig. 1c) that expanded and coalesced into lesions typical of red root rot (Fig. 1d). The development of disease symptoms corresponded with the growth of the inoculated pathogen, with symptoms at the margin of fungal growth (Fig. 1c) less severe than symptoms near the point of inoculation (Fig. 1d). Fungal hyphae penetrated the root cortex without evidence of specialized appressoria, killing host cells in advance of penetration, but were restricted by the stele as reported in Allen (1967). Cell death was indicated by granulation and yellowing of the cytoplasm. Monilioid fungal cells developed in the root cortex, similar to those observed in mature cultures on PDA. The same fungus, accessioned as BRIP 70665, was recovered from surface sterilized lesions on inoculated Morava vetch plants but was not isolated from asymptomatic tissue, thereby fulfilling Koch’s postulates. The efficacy of surface sterilization as demonstrated by Bonito et al. (2017) was verified by streaking recently treated root pieces over the surface of PDA and incubating the plates for 10 d.
The ITS and partial LSU region of BRIP 70197 was 99.53% similar to the ex-type sequence of Atractiella rhizophila (GenBank ITS = NR 152538). In fulfilling Koch’s postulates, the sequence amplified from the reisolate from the pathogenicity test (BRIP 70665) was identical with that amplified from the inoculum source (BRIP 70197). A consensus sequence of the ITS and partial LSU of the vetch red root rot pathogen was deposited in GenBank, accession number MN 850289.
Discussion
Our report provides the first evidence that A. rhizophila causes red root rot of V. sativa cv. Morava. The two cultures (DAR 14088 and BRIP 70197) share similar morphology and pathogenicity, which provides strong circumstantial evidence that A. rhizophila was the cause of red root rot on Golden Tares vetch in the 1960s (Allen 1967). Bonito et al. (2017) examined living cultures and environmental DNA sequences to show that A. rhizophila was widespread across North America and also present in other countries. Bonito et al. (2017) reported that A. rhizophila was a non-pathogenic root inhabitant of several plant species, including Populus and Pinus, and had symbiotic associations with Quercus rubra and possibly Zea mays. Genomic sequencing of A. rhizophila may establish the basis of its life history, which currently points to an endophytic and pathogenic capacity.
Abbreviations
- Difco:
-
a product name of Becton, Dickinson and Company, 7 Loveton Circle, Sparks, Maryland 21,152, USA
- DNA:
-
deoxyribonucleic acid
- GenBank:
-
USA National Centre for Biotechnology Information Nucleotide Database
- ITS:
-
internal transcribed spacer
- LSU:
-
large sub-unit
- M:
-
molar; mol dm-3
- PCR:
-
polymerase chain reaction
- PDA:
-
half-strength Difco potato dextrose agar; 19.5 g l-1 Difco PDA plus 7.5 g l-1 Difco agar
- USA:
-
United States of America
- WA:
-
15 g l-1 Difco agar
References
Allen, R. N. (1967). The root and stem rot disease of vetch. M.Sc.Agric. Dissertation, University of Sydney, 135 pp.
Allen, R. N. (1972). Diseases of Golden Tares in the Richmond Valley of New South Wales. Agricultural Gazette of New South Wales, 81, 244–246.
Baker, K. F. (1957). The UC system for producing healthy container grown plants. Berkeley, USA: University of California Press.
Barnard, C. (1970). Register of Australian herbage plant cultivars, 2nd edition. Canberra, Australia: CSIRO Publishing.
Bonito, G., Hameed, K., Toome-Heller, M., Healy, R., Reid, C., Liao, H.-L., Aime, M. C., Schadt, C. & Vilgalys, R. (2017). Atractiella rhizophila sp. nov., an endorrhizal fungus isolated from the Populus root microbiome. Mycologia, 109, 18–26.
Date, R. A. & Vincent, J. M. (1962). Determination of the number of root nodule bacteria in the presence of other organisms. Australian Journal of Experimental Agriculture and Animal Husbandry, 2, 5–7.
Gardes, M. & Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Molecular Ecology, 2, 113–118.
Holder, J. M., Swain, F. G. & Colman, R. L. (1963). The use of sod-sown vetch (Vicia sativa) as a supplement by dairy cows on the far north coast of New South Wales. Australian Journal of Experimental Agriculture and Animal Husbandry, 3, 153–160.
Koch, H. H. R. (1882). Die Aetiologie der Tuberculose. Berliner Klinische Wochenschrift, 19, 221–230.
Muyolo, N. G., Lipps, P. E. & Schmitthenner, A. F. (1993). Anastomosis grouping and variation in virulence among isolates of Rhizoctonia solani associated with dry bean and soybean in Ohio and Zaire. Phytopathology, 83, 438-444.
Vilgalys, R. & Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology, 172, 4238–4246.
White, T. J., Bruns, T., Lee, S. & Taylor, J. W. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. (In M.A. Innis, D.H. Getfand, J.J. Sninsky & T.J. White (Eds.), PCR Protocols: A guide to methods and applications. (pp. 315–322)). (New York, USA: Academic Press).
Acknowledgements
We thank R. L. Dodman and R. J. Vilgalys for commenting on the text, and D. J. Firth and G. P. Kennedy for technical assistance.
Funding
This work was supported by Biosecurity Queensland, a division of the Queensland Government, Department of Agriculture and Fisheries. Some of the work was conducted while the senior author was employed by the New South Wales Government, Department of Agriculture. The research was not externally funded and the authors derived no commercial benefit from any aspect of the work.
Author information
Authors and Affiliations
Contributions
All three authors contributed to the work, and the senior author prepared the first draft.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
All authors read and approved the final manuscript.
Human studies and participants
There was no involvement of human participants and/or animals in the present study.
Rights and permissions
About this article
Cite this article
Allen, R.N., Bransgrove, K. & Shivas, R.G. Red root rot of Vicia sativa caused by Atractiella rhizophila. Eur J Plant Pathol 157, 293–297 (2020). https://doi.org/10.1007/s10658-020-01985-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-020-01985-z