Abstract
A population of Xiphinema americanum-group was recovered in association with stone fruit trees in Isfahan province, center of Iran. A reverse taxonomic approach based upon the large subunit ribosomal DNA (LSU rDNA D2-D3) and the mitochondrial cytochrome c oxidase subunit I (COI mtDNA) gene sequences in integration with morphological studies, revealed that the recovered population belongs to Xiphinema santos. The Iranian population was mainly characterized by 1240–1868 μm long females with 60–84 μm long odontostyle, a = 37.2–51.9 and c = 42.8–54.6. It is further characterized by a lip region having a depression in junction with the body, presence of visible endosymbiont bacteria in ovaries under light microscope, dorsally convex and ventrally slightly concave conical tail with a blunt tip and three juvenile developmental stages. This population was similar to the type population in its morphology; however overlapped and extended morphometric data ranges, as well as differences in some indexes were observed. Compared to a Spanish population of this species, the Iranian population had a close morphology, similar morphometric data ranges and identical LSU and COI sequences. In LSU phylogeny, the relationship between the present and some previously sequenced isolates of the species and some isolates of three species X. georgianum, X. laevistriatum and X. citricolum was not resolved. In COI phylogeny, the clade of the Iranian and Spanish populations appeared as an independent lineage inside an unsupported clade including several species. The comparison with other populations of the species was reported and discussed. A second species, X. primum, that is native to Iran, was recovered from a new locality and characterized molecularly.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Xiphinema americanum-group currently includes 60 species (Mobasseri et al. 2019). The group is known with 10 representatives in Iran (Pedram 2018; Mobasseri et al. 2019). Xiphinema santos Lamberti et al. 1993 was originally described from Portugal in association with Vitis sp. (Lamberti et al. 1993) and was later reported from Portugal, Egypt and Spain in association with hop, pine, peach, grapevine and stone pine (Lamberti et al. 1993, 1994, 1996; Gutiérrez-Gutiérrez et al. 2012). In the present study, a population of the species was recovered from Isfahan province in association with stone fruit trees following our samplings to determine the infestation of pomegranate and fruit tree gardens with plant parasitic nematodes, and studied based upon morphological and molecular approaches. The species X. primum Mobasseri et al. 2019 that was originally described from Iran (Mobasseri et al. 2019) was also recovered from a new location in north of Iran and characterized molecularly.
Materials and methods
Sampling and morphological studies
Several soil samples were collected from fruit tree gardens in Isfahan, Tehran and Qom provinces. The samples were especially collected from the rhizosphere of trees showing general weakness symptoms similar to those of nutrient deficiency. Furthermore, 15 soil samples were collected from forests of Golestan province, north of Iran. Nematodes were extracted from soil samples using the tray method (Whitehead and Hemming 1965). A series of 20 and 60 mesh sieves (US standard mesh nos. equal to 850 and 250 μm sized openings, respectively) were used to extract tentative longidorid species. After extraction, the specimens of interest were hand-picked under a Nikon SMZ1000 stereomicroscope, heat-killed by adding boiling 4% formaldehyde solution, and transferred to anhydrous glycerin according to De Grisse (1969). Permanent slides were prepared and morphological characters were studied using an Olympus BX51 light microscope equipped with differential interference contrast (DIC). The light microphotographs were taken using an Olympus DP72 digital camera attached to the microscope. Morphometric values were obtained using a drawing tube attached to a Nikon E600 light microscope. The juvenile developmental stages were identified according to Robbins et al. (1996).
Molecular studies
For DNA extraction, two live Xiphinema americanum-group nematode specimens from the both populations recovered from the both provinces were picked out, washed in distilled water and photographed in temporary slides. Each selected nematode specimen was then transferred to a small drop of Tris-EDTA (TE) buffer (10 mM Tris-Cl, 0.5 mM EDTA; pH 9.0, QIAGEN Inc., Valencia CA, USA) on a clean slide and squashed using a clean slide cover glass with the aid of a plastic material. Each DNA sample was taken from one individual. The suspension was collected by adding 20 μl of TE buffer. DNA samples were stored at −20 °C until used as polymerase chain reaction (PCR) templates. To amplify the near-full-length fragment of the LSU rDNA D2-D3 and partial COI mtDNA, several primer pairs were used in the PCR reactions. Primers used for the amplification of the D2–D3 expansion domains of the LSU rDNA were forward D2A (5′-ACAAGTACCGTGAGGGAAAGT-3′) and reverse D3B (5′-TGCGAAGGAACCAGCTACTA-3′) (Nunn 1992) primers. In some cases, the reverse primer was replaced by the reverse primer KK28S-4 (5′-GCGGTATTTGCTACTACCAYYAMG ATCTGC-3′) (Kiontke et al. 2004). The used primers for amplification of the partial COI mtDNA were: forward primer JB3 (5′-TTTTTTGGGCATCCTGAGGTTTAT-3′) and reverse primer JB4.5 (5′-TAAAGAAAGAACATAATGAAAATG-3′) (Bowles et al. 1992), and forward primer COIF (5′-GATTTTTTGGKCATCCWGARG-3′) and reverse primer COIR (5′-CWACATAATAAGTATCATG-3′) (Lazarova et al. 2006). The PCRs for amplification of the genomic and mitochondrial fragments were done according to Jahanshahi Afshar et al. (2019). The newly obtained sequences were submitted to the GenBank database under the accession numbers given in LSU and COI trees.
Phylogenetic analyses
The recently obtained LSU rDNA D2-D3 and COI mtDNA sequences were manually checked, edited and assembled. These sequences were compared with those of other relevant sequences available in the GenBank database using the BLAST homology search program. The BLAST search using one of the LSU sequences of the Isfahan province population (MN602712) revealed it has maximal identity with several sequences of Xiphinema santos (JQ990029, JQ990030, AY601587 and some others) and the BLAST search using its COI sequence (MN627267) revealed it has maximal identity with a sequence of X. santos (JQ990055). The BLAST search using the LSU sequence of the Golestan province population (MN602714) revealed it has 99.55–99.89% identity with several sequences of X. primum (MF372941–43, MF372947) and the BLAST search using its COI sequence (MN627266) revealed it has 93.07% identity with two sequences of X. primum (MK202795, MK202796). The DNA sequences for reconstructing the phylogenetic trees were selected according to recent studies (Archidona-Yuste et al. 2016; Mobasseri et al. 2019).
Multiple alignments of the LSU and COI datasets were performed using the Q-INS-i algorithm of the online version of MAFFT version 7 (http://mafft.cbrc.jp/alignment/server/) (Katoh and Standley 2013). The Gblocks program (version 0.91b) (Castresana 2000) with all three less stringent parameters, a server tool at the Castresana Lab (http://molevol.cmima.csic.es/castresana/Gblocks_server.html), was used for post-editing of the LSU alignment, i.e., to eliminate poorly aligned regions or divergent positions. The best-fitting substitution model for each of the two datasets was selected according to the Akaike information criterion (AIC) by using PAUP∗/MrModeltest v2.2 (Nylander et al. 2004). A general time reversible model, including among-site rate heterogeneity and estimates of invariant sites (GTR + G + I) was selected and used for both LSU rDNA D2-D3 and COI mtDNA datasets analyses. Bayesian analysis was performed using MrBayes v3.1.2 (Ronquist and Huelsenbeck 2003) with a starting random tree and running the chains for 5 × 106 generations for the both datasets. The Markov chains were sampled every 100 generations for estimating the posterior probabilities of the phylogenetic trees (Larget and Simon 1999) using the 50% majority rule. Of the converged runs, 25% were regarded as ‘burn-in’. Convergence of model parameters and topology were assessed based on the average standard deviation of split frequencies and potential scale reduction factor values. Adequacy of the posterior sample size was evaluated using autocorrelation statistics as implemented in Tracer v.1.6 (Rambaut and Drummond 2009). The output files of the phylogenetic programs used herein were visualized using Dendroscope V.3.2.8 (Huson and Scornavacca 2012) and re-drawn by CorelDRAW software version 17. The Bayesian posterior probability (BPP) values exceeding 0.50 are given on appropriate clades.
Results and description
Xiphinema santos Lamberti, Lemos, Agostinelli & D’Addabo 1993 (Figures 1 & 2, Tables 1 & 2)
Female
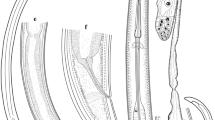
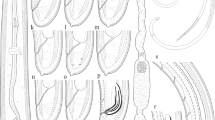
Body cylindrical, tapering gradually towards the extremities, ventrally curved, varying from an open to closed C, the posterior end more ventrally bent when heat-killed. Cuticle 2.0–3.5 μm thick along the body, 1.5–2.0 μm thick in post labial region, 2–4 μm thick at mid-body, 3–4 μm thick just anterior to anus and 5–12 μm thick at tail tip. Lateral chord 10–16 μm wide at mid-body, occupying 27–38% of corresponding body diam. Lip region anteriorly rounded, having a depression in junction with the body, slightly expanded, 11–12 μm wide and 4–5 μm high. Odontostyle 1.4–2.1 times longer than the odontophore, the latter with well-developed flanges. The guiding sheath 1–9 μm long. The pharyngeal bulb 55–76 μm long and 13–23 μm wide, its glandularium 51–67 μm long, occupying about 1/3 to 1/5 of the total pharynx length, with three pharyngeal nuclei, the larger dorsal gland nucleus (DN) located at 18.9–25.0%, the two smaller ventrosublateral nuclei (S1N) located at 58–63% of pharyngeal bulb (location of glands’ nuclei according to Loof and Coomans 1972). Cardia rounded, 4–8 × 6–11 μm in size, intestine simple. Prerectum often indistinct, with great range in length (138–227 μm long) and rectum 13–22 μm long, or 0.6–1.1 times the anal body diameter. Reproductive system didelphic-amphidelphic with two seemingly equally developed branches, each branch ca. 145 μm long, including a ca. 50 μm long tubular uterus and a ca. 100 μm long ovary with visible endosymbiont bacteria. Vulva a transverse slit, vagina 16–19 μm long with short pars distalis vaginae, pars proximalis vaginae and ovejector ca. 35 μm wide and 6 μm high. Tail short, conoid, weakly curved ventrally with a rounded terminus, bearing two pairs of caudal pores.
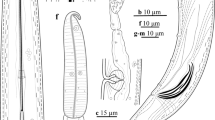
Light microphotographs of Iranian population of Xiphinema santos Lamberti et al. 1993. a: Entire of female, b & c: Female anterior body region, d: Vulval region, e-h: Tail of the first, second and third stage juveniles and female, respectively, i: Anterior ovary and endosymbiont bacteria. Scale bars: a = 100 μm, b-i = 10 μm)
Correlation of functional and replacement odontostyle to body length in juvenile developmental stages and females of Iranian population of Xiphinema santos Lamberti et al. 1993. The replacement odontostyle in each juvenile stage is equal to the functional odontostyle in the next stage, and in J3, is equal to the functional odontostyle length in females
Male
Not found.
Juveniles
Morphologically similar to females except for smaller body and narrower tail. Three juvenile developmental stages were detected. The scatter diagram separating juveniles and females is given in Fig. 2. All three juvenile developmental stages were separated from each other using body length and length of replacement and functional odontostyle. In the first juvenile developmental stage, the tip of the replacement odontostyle is close to the base of the functional odontostyle, and in two rest stages, it is distantly located. All three juvenile stages have conical dorsally convex and ventrally concave tail.
Habitat and locality
The Iranian population of Xiphinema santos was recovered in association with stone fruit trees in Aran va Bidgol, Isfahan province (GPS coordinates are N 34°02′30.173″, E 51°28′49.193″, altitude: 907 m.a.s.l.).
Xiphinema primum Mobasseri, Hutchinson, Jahanshahi Afshar & Pedram 2019
A population of this species was recovered in the present study from Golestan province (GPS coordinates are N 36°46′9″, E 54°34′34.079″) in association with forest trees and sequenced for its LSU D2-D3 and COI mtDNA regions. The recovered population was in accordance with the type population and the newly generated sequences for this population are presented in corresponding trees. This finding shows the species could be more prevalent in the country than already assumed.
Molecular characterization and phylogenetic relationships
To determinate the phylogenetic affinities of the Iranian population of Xiphinema santos and the newly recovered population of X. primum with other species in the Longidoridae Thorne 1935 (and mostly with those species belonging to the X. americanum-group), two separate LSU rDNA D2-D3 and COI mtDNA datasets were prepared for Bayesian inference (BI). For LSU phylogeny (Bayesian tree), a total number of 81 sequences, including the newly generated sequences and sequences of Xiphinema non-americanum group (X. index Thorne and Allen 1950 and X. diversicaudatum (Micoletzky 1927) Thorne 1939), a Tylencholaimidae Filipjev 1934 member (Tylencholaimus teres Thorne 1939) and a Qudsianematidae Jairajpuri 1965 member (Ecumenicus monohystera (de Man 1880) Thorne 1974), as outgroups (accession numbers in Fig. 3), were selected. The LSU dataset was composed of 722 characters of which 308 were variable. The average nucleotide composition was: A: 23.5%, T: 18.7%, C: 25.8%, G: 32.0%. Figure 3 represents the Bayesian phylogenetic tree inferred using the above-mentioned dataset. In this tree, the two sequences of the Iranian isolate of X. santos (MN602712, MN602713) and some sequences from the previously sequenced isolates of the species as well as some isolates of three species X. georgianum Lamberti and Bleve-Zacheo 1979, X. laevistriatum Lamberti and Bleve-Zacheo 1979 and X. citricolum Lamberti and Bleve-Zacheo 1979 formed a highly supported clade (0.94 BPP). The phylogenetic relationships of these species were not resolved due to polytomy. The newly generated LSU sequence of X. primum also fell into the clade including original sequences of the species.
Bayesian 0.50 majority rule consensus tree inferred using LSU rDNA D2-D3 sequences of Iranian population of Xiphinema santos Lamberti et al. 1993. The Bayesian posterior probability values are given for the clades. The newly generated sequences are in bold
A total number of 66 sequences, including the newly generated sequences, plus three sequences of Xiphinema non-americanum group spp. (X. index, X. diversicaudatum and X. setariae Luc 1958) as outgroups, were selected for COI mtDNA Bayesian phylogeny (accession numbers in Fig. 4). The COI dataset was composed of 391 characters of which 222 were variable. The average nucleotide composition was as follows: A: 25.4%, T: 37.1%, C: 15.1%, G: 22.4%. Figure 4 represents the Bayesian phylogenetic tree inferred using this dataset. Based upon this tree, the newly generated sequence of the Iranian population of X. santos (MN627267) formed a highly supported clade (0.99 BPP) with the Spanish isolate (JQ990055). All currently available COI sequences of X. primum formed a clade too.
Bayesian 0.50 majority rule consensus tree inferred using COI mtDNA sequences of Iranian population of Xiphinema santos Lamberti et al. 1993. The Bayesian posterior probability values are given for the clades. The newly generated sequences are in bold
Discussion
A relatively high density population (around 50 individuals/200 g soil) of X. santos was recovered from the rhizosphere of stone fruit trees in a garden in Aran va Bidgol. Compared to the type population, the Iranian population has a close morphology, but some differences in morphometric data ranges i.e. shorter body (1.6 (1.2–1.9) vs. 1.8 (1.7–2.0) mm) and smaller a (44.3 (37.2–51.9) vs. 54 (54–59)) were observed. For most other indexes, the ranges were overlapped; however, present study extended the morphometric data ranges for the species (Table 1). Compared to other populations reported from Portugal (Lamberti et al. 1994) and Egypt (Lamberti et al. 1996), again extension in indexes was observed (Table 1). Compared to the Spanish population, more similarity in some indexes like body and odontophore length and c ratio was observed. In summary, X. santos has a wide intraspecies ranges for body length of females (1.2–2.0 mm). Such a wide range for body length was also observed for the third juvenile developmental stage of the Egyptian population of the species (Lamberti et al. 1997). The wide range for body length has already been reported for some species like Xiphinema sp.1 (isolate CD47) by Orlando et al. (2016) (L = 1193–2268 or 1.2–2.3 mm), X. plesiopachtaicum Archidona-Yuste et al. 2016 (L = 1.5–2.1 mm) and X. parabrevicolle Gutiérrez-Gutiérrez et al. 2012 (L = 1.4–2.0 mm). The present study showed that X. santos also belongs to the group of species having a wide body length range. The species has three juvenile developmental stages (Lamberti et al. 1997) and the reported four juvenile developmental stages by Gutiérrez-Gutiérrez et al. (2012) is amended herein.
In molecular phylogenetic analyses, similar to our previous study (Mobasseri et al. 2019), the relationships of X. santos with three species X. georgianum, X. laevistriatum and X. citricolum were not resolved in LSU tree due to polytomy. Although X. citricolum is shown to be the most closely related species to X. santos in most LSU analyses (He et al. 2005; Gutiérrez-Gutiérrez et al. 2012; Lazorava et al. 2016; Orlando et al. 2016; Mobasseri et al. 2019), the effect of the alignment/postediting methods on the resulted topology should not be ruled out. The topology of the present LSU tree and dividing of the species into two subclades I and II are similar to the resolved topology by Mobasseri et al. (2019).
The COI mtDNA of Xiphinema americanum-group species has an acceptable interspecies variation (Lazarova et al. 2016; Gutiérrez-Gutiérrez et al. 2012; Orlando et al. 2016; Archidona-Yuste et al. 2016) and could yield better cladogenesis in the corresponding tree. In our COI phylogeny, X. santos has appeared as an independent lineage in an unsupported clade including several other species. The general topology and the subclades resolved in the present COI tree are similar to the COI tree presented by Mobasseri et al. (2019).
This is the first record of X. santos from Iran, corroborating its occurrence out of the Mediterranean Basin, after its several reports from Portugal, North Egypt and Spain (Lamberti et al. 1993, 1994, 1996; Gutiérrez-Gutiérrez et al. 2012).
Conclusion
In this study, Xiphinema santos, a member of X. americanum-group was reported for the first time from Iran, extending its morphometric data ranges like body length and its geographical distribution areas.
References
Archidona-Yuste, A., Navas-Cortés, J. A., Cantalapiedra-Navarrete, C., Palomares-Rius, J. E., & Castillo, P. (2016). Cryptic diversity and species delimitation in the Xiphinema americanum-group complex (Nematoda: Longidoridae) as inferred from morphometrics and molecular markers. Zoological Journal of the Linnean Society, 176, 231–265.
Bowles, J., Blair, D., & McManus, D. P. (1992). Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Molecular and Biochemical Parasitology, 54, 165–174.
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution, 17, 540–552.
De Grisse, A. T. (1969). Redescription ou modifications de quelques techniques utilisées dans l’étude des nematodes phytoparasitaires. Mededelingen Rijksfakulteit Landbouwwetenschappen. Gent, 34, 351–369.
de Man J. G. (1880). Die einheimischen, frei in der reinen Erde und im su ¨ssen Wasser lebende Nematoden, monographischen bearbeitet. Tijdschrift Nederlandse Dierkundige vereniging. 104 p.
Filipjev, I. N. (1934). The classification of free-living nematodes and their relations to parasitic nematodes. Smithsonian Miscellaneous Collection, 89, 1–63.
Gutiérrez-Gutiérrez, C., Cantalapiedra-Navarrete, C., Decraemer, W., Vovlas, N., Prior, T., Rius, J. E. P., & Castillo, P. (2012). Phylogeny, diversity, and species delimitation in some species of the Xiphinema americanum-group complex (Nematoda: Longidoridae), as inferred from nuclear and mitochondrial DNA sequences and morphology. European Journal of Plant Pathology, 134, 561–597.
He, Y., Subbotin, S., Rubtsova, T. V., Lamberti, F., Brown, D. J. F., & Moens, M. (2005). A molecular phylogenetic approach to Longidoridae (Nematoda: Dorylaimida). Nematology, 7, 111–124.
Huson, D. H., & Scornavacca, C. (2012). Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Systematic Biology, 61, 1061–1067.
Jahanshahi Afshar, F., Pourjam, E., & Pedram, M. (2019). Lobocriconema iranense (Van den berg, Eskandari, Tiedt & Karegar, 2010) n. comb. and description of L. nokandense n. sp. (Nematoda: Criconematidae) from Iran. Nematology, 21, 1043–1061.
Jairajpuri, M. S. (1965). Qudsianema amabilis n. gen., n. sp. (Nematoda: Dorylaimoidea) from India. Proceedings of the Helminthological Society of Washington, 32, 72–73.
Kiontke, K., Gavin, N. P., Raynes, Y., Roehrig, C., Piano, F., & Fitch, D. H. (2004). Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proceedings of the National Academy of Sciences of the United States of America, 101, 9003–9008.
Katoh, K., & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780.
Lamberti, F., & Bleve-Zacheo, T. (1979). Studies on Xiphinema americanum sensu lato with descriptions of fifteen new species (Nematoda, Longidoridae). Nematologia Mediterranea, 7, 51–106.
Lamberti, F., Lemos, R. M., Agostinelli, A., & D’Addabo, T. (1993). The Xiphinema americanum-group in the vineyards of the Dao and Douro regions (Portugal) with description of two new species (Nematoda, Dorylaimida). Nematologia Mediterranea, 21, 215–225.
Lamberti, F., Bravo, M. A., Agostinelli, A., & Lemos, R. M. (1994). The Xiphinema americanum-group in Portugal with descriptions of four new species (Nematoda, Dorylaimida). Nematologia Mediterranea, 22, 189–218.
Lamberti, F., Agostinelli, A., & Radicci, V. (1996). Longidorid nematodes from northern Egypt. Nematologia Mediterranea, 24, 307–339.
Lamberti, F., Iovev, T., Choleva, B., Brown, D. J. F., Agostinelli, A., & Radicci, V. (1997). Morphometric variation and juvenile stages of some longidorid nematodes from Bulgaria with comments on the number of juvenile stages of Longidorus africanus, L. closelongatus and Xiphinema santos. Nematologia Mediterranea, 25, 213–237.
Larget, B., & Simon, D. L. (1999). Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Molecular Biology and Evolution, 16, 750–759.
Lazarova, S. S., Malloch, G., Oliveira, C. M. G., Hübschen, J., & Neilson, R. (2006). Ribosomal and mitochondrial DNA analyses of Xiphinema americanum-group populations. Journal of Nematology, 38, 404–410.
Lazarova, S., Peneva, V., & Kumari, S. (2016). Morphological and molecular characterisation, and phylogenetic position of X. browni sp. n., X. penevi sp. n. and two known species of Xiphinema americanum-group (Nematoda, Longidoridae). ZooKeys, 574, 1–42.
Loof, P. A. A., & Coomans, A. (1972). The oesophageal gland nuclei of Longidoridae (Dorylaimida). Nematologica, 18, 213–233.
Luc, M. (1958). Xiphinema de l'ouest Africain: Description de cinq nouvelles espèces (Nematoda; Dorylaimidae). Nematologica, 3, 57–72.
Micoletzky, H. (1927). Neue und seltene freilebende Nematoden aus dem Wolgagebiet (Kama). Zoologischer Anzeiger, 73, 113–123.
Mobasseri, M., Hutchinson, M. C., Jahanshahi Afshar, F., & Pedram, M. (2019). New evidence of nematode-endosymbiont bacteria coevolution based on one new and one known dagger nematode species of Xiphinema americanum-group (Nematoda, Longidoridae). PLoS One, 14, e0217506.
Nunn, G. B. (1992). Nematode molecular evolution: an investigation of evolutionary patterns among nematodes based upon DNA sequences. Ph.D. Thesis, University of Nottingham, Nottingham, UK.
Nylander, J. A., Ronquist, F., Huelsenbeck, J. P., & Nieves-Aldrey, J. L. (2004). Bayesian phylogenetic analysis of combined data. Systematic Biology, 53, 47–67.
Orlando, V., Chitambar, J. J., Dong, K., Chizhov, V. N., Mollov, D., Bert, W., & Subbotin, S. A. (2016). Molecular and morphological characterisation of Xiphinema americanum-group species (Nematoda: Dorylaimida) from California, USA, and other regions, and co-evolution of bacteria from the genus Candidatus Xiphinematobacter with nematodes. Nematology, 18, 1015–1043.
Pedram, M. (2018). Nematodes of the family Longidoridae. In: Ghaderi, R., Kashi, L. & Karegar, A. (Eds). Plant-parasitic nematodes in Iran. Science reference in collaboration with the Iranian Society of Nematology, pp. 627–67. (In Persian).
Rambaut, A., & Drummond, A. J. (2009). Tracer version 1.5 [computer program] http://beast.bio.ed.ac.uk/.
Robbins, R. T., Brown, D. J. F., Halbrendt, J. M., & Vrain, T. C. (1996). Compendium of juvenile stages of Xiphinema species (Nematoda: Longidoridae). Russian Journal of Nematology, 4, 163–171.
Ronquist, F., & Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572–1574.
Thorne, G. (1935). Notes on free-living and plant-parasitic nematodes. II-higher classification groups of Dorylaimoidea. Proceedings of the Helminthological Society of Washington, 2, 96–98.
Thorne, G. (1939). A monograph of the nematodes of the superfamily Dorylaimoidea. Capita Zoological, 8, 261 pp.
Thorne, G. (1974). Nematodes of the Northern Great Plains. Part II. Dorylaimoidea in part (Nemata: Adenophorea). South Dakota State University Agriculture Experimental Station Technical Bulletin 41, 120 pp.
Thorne, G., & Allen, M. W. (1950). Paratylenchus hamatus n. sp, and Xiphinema index n. sp., two nematodes associated with fig roots, with a note on Paratylenchus anceps cobb. Proceedings of the Helminthological Society of Washington, 17, 27–35.
Whitehead, A. G., & Hemming, J. R. (1965). A comparison of some quantitative methods for extracting small vermiform nematodes from soil. Annals of Applied Biology, 55, 25–38.
Acknowledgements
The financial support of Tarbiat Modares University is appreciated. The authors acknowledge the critically reading of the MS by Dr. Mohammad Reza Atighi. The presented findings in this paper are part of the results of the project number 0-16-16-94176 by the first author entitled “Determination of infestation of pomegranate orchards and nurseries to plant parasitic nematodes with an emphasis on root knot nematodes”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors certify that they do not have any conflict of interest, the presented data are original and the presented data are not published, and are not under evaluation for publication elsewhere, all prevailing local, national and international regulations and conventions, and normal scientific ethical practices, have been respected. They also certify that the final version of this manuscript has been reviewed by all authors.
Research involving human participants and/or animals
The soil samples analyzed in this study were collected under the direct monitoring of the garden owner or from the natural public regions that are not under protection against soil samplings.
Informed consent
The authors certify that they have accepted the principles of ethical standards and that guidelines of professional conduct have been followed. The funders had no role in designing of this study, collecting and analyzing of the data, preparing the manuscript and are not involved in evaluating or publishing process of this research.
Rights and permissions
About this article
Cite this article
Jahanshahi Afshar, F., Shahryari, F., Mokhtassi-Bidgoli, A. et al. Occurrence of Xiphinema santos Lamberti, Lemos, Agostinelli & D’Addabo 1993 (Nematoda: Longidoridae), a X. americanum-group member in Iran. Eur J Plant Pathol 157, 281–291 (2020). https://doi.org/10.1007/s10658-020-01980-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-020-01980-4