Abstract
During nematode surveys in southern Spain and Italy 14 populations of Xiphinema species tentatively identified as Xiphinema americanum-group were detected. Morphological and morphometrical studies identified three new species and six known Xiphinema americanum-group species, viz.: Xiphinema parabrevicolle n. sp., Xiphinema parapachydermum n. sp., Xiphinema paratenuicutis n. sp., Xiphinema duriense, Xiphinema incertum, Xiphinema opisthohysterum, Xiphinema pachtaicum, Xiphinema rivesi, and Xiphinema santos. The Xiphinema americanum-group is the most difficult Xiphinema species group for diagnosis since the morphology is very conservative and morphometric characters often overlap. This group includes vectors of several important plant pathogenic viruses that cause significant damage to a wide range of agricultural crops. Molecular characterisation of these species using D2-D3 expansion regions of 28S rRNA, 18S rRNA, ITS1-rRNA and the protein-coding mitochondrial gene, cytochrome oxidase c subunit 1 was carried out and maximum likelihood and Bayesian inference analysis were used to reconstruct phylogenetic relationships among these species and with other Xiphinema americanum-group species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dagger nematodes of the genus Xiphinema comprise phytopathogenic species that damage a wide range of wild and cultivated plants through direct feeding on root cells and transmission of several plant pathogenic viruses (Taylor and Brown 1997). This transmission is governed by a marked specificity between plant viruses and their Xiphinema spp. vectors. In fact, only nine out of the approximately 260 known species of Xiphinema have been shown to transmit nepoviruses (genus Nepovirus, family Secoviridae, Subfamily Comovirinae) (Decraemer and Robbins 2007). The Xiphinema americanum-group, which comprises a complex of about 50 species, many of them with a cosmopolitan distribution, appears to be the most difficult Xiphinema species group for species diagnosis based on morphology and morphometrics (Coomans et al. 2001) since their morphology is quite conserved and morphometrics overlap. This biases quarantine regulations and protection methods especially since the Xiphinema americanum-group includes the vectors of several important plant viruses that cause significant damage to a wide range of crops, including Tobacco ringspot virus (TRSV), Tomato ringspot virus (TomRSV), Cherry rasp leaf virus (CRLV), and Peach rosette mosaic virus (PRMV) (Taylor and Brown 1997). For this reason, accurate identification of the X. americanum-group species is essential, since several species of this group are listed as A1 (Xiphinema americanum Cobb 1913, Xiphinema californicum Lamberti and Bleve-Zacheo 1979, Xiphinema bricolensis Ebsary et al. 1989) and A2 (Xiphinema rivesi Dalmasso 1969) quarantine organisms by the European and Mediterranean Plant Protection Organisation (EPPO).
Tarjan (1969) introduced the notation ‘Xiphinema americanum-group’ to stress the complexity of the problem and suggested that populations identified as Xiphinema americanum represented a complex of several species. This group was morphologically defined by Loof and Luc (1990) and Lamberti et al. (2000) with the following common characters: spiral or C-shaped small body, lip region more or less rounded and continuous or separated from the rest of the body by an incisure which can ranged from a depression to a constriction, female reproductive system with two equally developed genital branches, usually with short uteri without uterine differentiation, all features grouped per system vulva positioned at 40–60 % of the body length from anterior end and short conical to broadly convex-conoid tail. Even so, controversies exist concerning the nature of the ‘true’ X. americanum-group i.e. only grouping those species which show the presence of symbiotic bacteria with males absent or rare. In fact, Luc et al. (1998) proposed that morphologically the X. pachydermum-group including five species (X. brevisicum Lamberti et al. 1994, X. longistilum Lamberti et al. 1994, X. mesostilum Lamberti et al. 1994, X. microstilum Lamberti et al. 1994, and X. pachydermum Sturhan 1983) comprise a group outside the X. americanum-group, since males are frequent, females do not show symbiontic bacteria associated with the ovaries (1 exception X. mesostilum), oviduct with normal structure clearly separated from the uterus by a well developed sphincter muscle and unipartite uteri relatively long. The action by Luc et al. (1998) was confirmed by the morphological phylogeny carried out by Coomans et al. (2001) in contradiction to Lamberti and Ciancio (1993) and Lamberti et al. (2000) who considered the X. americanum species group in a large sense including the X. pachydermum-group. Thus, species differentiation remains difficult and species diversity and taxonomic validity of species controversial (Lamberti et al. 2000; Gozel et al. 2006). Lamberti and Ciancio (1993) analyzed the species diversity of the X. americanum-group using a hierarchical cluster analysis of morphometrics
The application of molecular methods to studies of nematode population structure and systematics has revealed that some long-assumed single species are in fact cryptic species that are morphologically indistinguishable but may be phylogenetically distant to one another (Gutiérrez-Gutiérrez et al. 2010; Barsi and De Luca 2008; Wu et al. 2007; Oliveira et al. 2005, 2006; Ye et al. 2004). During the last decade sequences of nuclear ribosomal DNA (rDNA) have been used for molecular characterisation and reconstruction of phylogenetic relationships within the Longidoridae and more particular within Xiphinema (Gutiérrez-Gutiérrez et al. 2011a; Ye et al. 2004; He et al. 2005). The D2-D3 expansion segments of the 28S rDNA gene provided rather homogeneous results for Xiphinema species and appeared a useful tool for species differentiation (He et al. 2005). Wu et al. (2007) clearly distinguished Xiphinema hunaniense Wang and Wu 1992 from Xiphinema radicicola Goodey 1936 by D2-D3 rDNA sequences, and Oliveira et al. (2005) could differentiate Xiphinema brevicolle Lordello and Costa 1961 from Xiphinema diffusum Lamberti and Bleve-Zacheo 1979 by the internal transcribed spacer 1 (ITS1) region of rDNA. These species are very similar, showing only minor morphological differences e.g. in lip and tail region shape, and were previously synonymised by Loof et al. (1996) and Luc et al. (1998), respectively. Similarly, other species have been separated from almost identical species by molecular methods (Meza et al. 2011; Oliveira et al. 2006). The protein-coding mitochondrial gene, cytochrome oxidase c subunit 1 (COI), has also been shown to be applicable for genetic bar-coding (ECBOL, http://www.ecbol.org) and phylogeny of plant-parasitic nematodes (Lazarova et al. 2006; Palomares-Rius et al. 2009). Nevertheless, few studies have been carried out using mitochondrial DNA for Xiphinema species diagnostic since it revealed low intraspecific diversity and large interspecific divergences (Gutiérrez-Gutiérrez et al. 2011b; Kumari et al. 2010; Lazarova et al. 2006).
Consequently, current availability of molecular techniques may help to provide tools for differentiating Xiphinema americanum-group species and can significantly improve and facilitate the routine identification of these nematodes. Polyphasic identification, based on an integrative strategy of combining molecular techniques with morphology and measurements for species diagnosis have proven a tool beyond doubt in nematode identification within this group (Gutiérrez-Gutiérrez et al. 2010, 2011a; Palomares-Rius et al. 2009). For this reason, we have conducted an extensive nematode survey in cultivated and natural environments in southern Spain and one incidental sample in Italy, with the following objectives: i) to characterise morphologically and morphometrically species belonging to the Xiphinema americanum-species group and to compare them with previous records; ii) to molecularly characterise the sampled Xiphinema americanum-group populations using the D2-D3 expansion segments of 28S rRNA, ITS1, partial 18S rRNA, and COI gene sequences; and iii) to study the phylogenetic relationships of the identified Xiphinema americanum-group with other previously sequenced species.
Material and methods
Nematode population sampling
Nematode surveys were conducted from 2010 to 2011 during the spring season in cultivated and natural environments in southern Spain and one incidental sample in Italy, including carob tree (Ceratonia siliqua L.), cherry tree (Prunus avium L.), citrus (Citrus aurantium L.), grapevine (Vitis vinifera L.), grasses, lentisc (Pistacia lentiscus L.), marram grass [Ammophila arenaria (L.) Link], olive (Olea europaea L.) and Stone pine (Pinus pinea L.) (Table 1). Samples were collected with a shovel from the upper 50 cm of soil of four to five plants arbitrarily chosen in each locality. Nematodes were extracted from 500 cm3 of soil by centrifugal flotation (Coolen 1979) and a modification of Cobb’s decanting and sieving (Flegg 1967) methods. In some cases, additional soil samples were collected afterwards from the same locality for completing the necessary specimens for morphological and/or molecular identification.
Nematode morphological identification
Specimens for light microscopy were killed by gentle heat, fixed in a solution of 4 % formaldehyde + 1 % propionic acid and processed to pure glycerine using Seinhorst’s method (1966). Specimens were examined using a Zeiss III compound microscope with Nomarski differential interference contrast at powers up to 1,000× magnification. Measurements were done using a drawing tube attached to a light microscope and, unless otherwise indicated in text. All measurements were expressed in micrometres (μm). For line drawing of the new species, light micrographs were imported to CorelDraw software version X5 and redrawn. All other abbreviations used are as defined in Jairajpuri and Ahmad (1992). In addition, a comparative morphological and morphometrical study on type specimens of some species were conducted with specimens kindly provided by Dr. A. Troccoli, from the nematode collection at the Istituto per la Protezione delle Piante, Sede di Bari, Consiglio Nazionale delle Ricerche, (C.N.R.), Bari, Italy (viz. X. rivesi from Portugal) and Dr Z.A. Handoo, from the USDA Nematode Collection, Beltsville, MD, USA (viz. Xiphinema intermedium Lamberti and Bleve-Zacheo 1979 T-3449p, Xiphinema rivesi from France T-3440p, Xiphinema tenuicutis Lamberti and Bleve-Zacheo 1979 slides T-3447p, T-3448p, and T-3449p). Within the framework of the Synthesis project BE-TAF 1769, a collaboration between FERA and RBINS) type material of the Xiphinema pachydermum group was studied amongst them Xiphinema santos Lamberti et al. 1993 and Xiphinema duriense Lamberti et al. 1993 from nematode collection of Rothamsted Experimental Station, Harpenden, England.
Nematode molecular identification
For molecular analyses, two live nematodes from each sample were temporary mounted in a drop of 1 M NaCl containing glass beads and after taking measurements and photomicrographs of diagnostic characters the slides were dismantled and DNA extracted. Nematode DNA was extracted from single individuals and PCR assays were conducted as described by Castillo et al. (2003). The D2-D3 expansion segments of 28S rDNA was amplified using the D2A (5′-ACAAGTACCGTGAGGGAAAGTTG-3′) and D3B (5′-TCGGAAGGAACCAGCTACTA-3′) primers (Castillo et al. 2003; He et al. 2005; Palomares-Rius et al. 2008). The ITS1 region was amplified using forward primer 18S (5′TTGATTACGTCCCTGCCCTTT-3′) and reverse primer rDNA1 (5′-ACGAGCCGAGTGATCCACCG-3′) as described in Wang et al. (2003). Finally, the 18S rDNA gene was amplified using the SSU_F_07 (5′-AAAGATTAAGCCATGCATG-3′) and SSU_R_81 (5′- TGATCCWKCYGCAGGTTCAC-3′) primers (http://www.nematodes.org/barcoding/sourhope/nemoprimers.html). The portion of the COI gene was amplified as described by Lazarova et al. (2006) using primers COIF (5′-GATTTTTTGGKCATCCWGARG-3′) and COIR (5′-CWACATAATAAGTATCATG-3′).
PCR products were purified after amplification using ExoSAP-IT (Affmetrix, USB products), quantified using a Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA) and used for direct sequencing in both directions using the primers referred above. The resulting products were purified and run on a DNA multicapillary sequencer (Model 3130XL genetic analyser; Applied Biosystems, Foster City, CA, USA), using the BigDye Terminator Sequencing Kit v.3.1 (Applied Biosystems, Foster City, CA, USA), at the SCAI, University of Córdoba sequencing facilities (Córdoba, Spain). The newly obtained sequences were submitted to the GenBank database under accession numbers indicated on the phylogenetic trees and Table 1.
Phylogenetic analyses
D2-D3 expansion segments of 28S rDNA and portion of the COI sequences of different Xiphinema americanum-group species from GenBank were used for phylogenetic reconstruction. Outgroup taxa for each dataset were chosen according to previous published data (He et al. 2005; Lazarova et al. 2006). The newly obtained and published sequences for each gene were aligned using ClustalW (Thompson et al. 1994) with default parameters. Sequence alignments were manually edited using BioEdit (Hall 1999). Phylogenetic analyses of the sequence data sets were performed with maximum likelihood (ML) using PAUP * 4b10 (Swofford 2003) and Bayesian inference (BI) using MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001). The best fitted model of DNA evolution was obtained using jModelTest v. 0.1.1 (Posada 2008) with the Akaike Information Criterion (AIC). The Akaike-supported model, the base frequency, the proportion of invariable sites, and the gamma distribution shape parameters and substitution rates in the AIC were then used in phylogenetic analyses. BI analysis under GTR + G model for D2-D3 expansion segment of 28S rDNA and TMP3uf + G for ITS1 region and TVM + I + G for COI, were run with four chains for 5.0 × 106, 2 × 106, and 1 × 106 generations, respectively. The Markov chains were sampled at intervals of 100 generations. Two runs were performed for each analysis. After discarding burn-in samples and evaluating convergence, the remaining samples were retained for further analyses. The topologies were used to generate a 50 % majority rule consensus tree. Posterior probabilities (PP) are given on appropriate clades. Trees were visualised using TreeView (Page 1996). In ML analysis the estimation of the support for each node was obtained by bootstrap analysis with 200 fast-step replicates.
Results
Morphology and morphometrics of species of the Xiphinema americanum-group
The morphological and morphometrical traits as well as molecular delineation of a Spanish population of Xiphinema pachtaicum (Tulaganov 1938) Kirjanova 1951 from Jerez de la Frontera (Cádiz province, southern Spain) infesting vineyards agreed completely with previous studies and was not repeated here (Gutiérrez-Gutiérrez et al., 2011a, b). This species is one of the most widespread and frequently occurring Xiphinema species in a wide variety of crops and natural habitats in the Iberian Peninsula and other Mediterranean basin countries (Taylor and Brown 1997; Téliz et al. 2007; Gutiérrez-Gutiérrez et al. 2011a, b).
Xiphinema parabrevicolle Footnote 1 n. sp. (Figs. 1, 2, and 3, Tables 2 and 3)
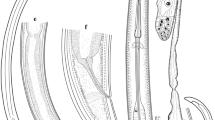
Light micrographs of Xiphinema parabrevicolle n. sp. from southern Italy. a Whole female; b Female neck region; c First-stage juvenile neck region; d–h Anterior regions; i–j Vulval regions; k–n Female tail regions; o–r J1-, J2-, J3-, and J4-juvenile-stages tail, respectively. Abbreviations: a anus; af amphidial fovea; gr guiding ring; rodt replacement odontostyle; V vulva. (Scale bars: a = 100 μm; b–r = 10 μm)
Female
Body medium-sized, robust, forming a close C-shaped to open spiral when killed by heat. Cuticle appearing smooth, but with fine transverse striae on inner cuticle layer, 2.6 ± 0.3 (2.0–3.0) μm thick along body but thicker at tail tip (Table 2). Lip region widely rounded, separated from the rest of the body by depression, 4.8 ± 0.6 (4.0–5.5) μm high. Amphidial fovea large, stirrup-shaped, with wide aperture 12.3 ± 0.3 (12.0–12.5) μm, as a straight transverse slit. Odontophore with basal flanges well developed basal flanges well developed 11.6 ± 0.9 (10.0–12.5) μm wide. Pharyngeal basal bulb 86.2 ± 9.1 (75–102) μm long and 21.7 ± 0.1 (18.5–24.0) μm wide, occupying about 1/4 to 1/5 of the total pharyngeal length (Fig. 3). Glandularium 62.3 ± 5.7 (51–65) μm long. Dorsal pharyngeal gland nucleus in anterior part of bulb, DN 28.3 ± 2.0 (27.0–32.0)% (Fig. 2). Pharyngeal-intestinal valve inconspicuous, hemispherical, 7.9 ± 0.7 (7.0–9.0) μm long. Reproductive system amphidelphic, both branches apparently equally developed, opposed and ovaries reflexed, uteri rather short (43–64 μm) without any differentiation. Ovaries with bacterial symbionts, oviducts relatively long, sphincter little developed. Vulva slightly post-equatorial, a transverse slit-like; vagina 8.0 ± 0.7 (7.0–9.0) μm long with short distal part and well-developed proximal. Prerectum often indistinct. Rectum 23.5 ± 2.1 (20.0–27.0) μm long, or 0.4–0.5 times the anal body diameter. Tail short, dorsally convex-conoid, slightly concave ventrally with conoid-rounded terminus, bearing three pairs of caudal pores (Figs. 1c–d, 3k–n).
Male
Not found.
Juveniles
All four juvenile stages (first-, second-, third- and fourth-stage) were found, and were basically similar to adults, except for their smaller size, longer tails and sexual characteristics. Tail becomes progressively shorter and stouter in each moult, being distinguishable by relative lengths of body and functional and replacement odontostyle (Fig. 3o–r, Table 2).
Type host and locality
Xiphinema parabrevicolle n. sp. was found in a sandy soil around the roots of lentisc (Pistacia lentiscus L.) and marram grass [Ammophila arenaria (L.) Link] at Pantanaggiani, Brindisi province, southern Italy (40°45′01.27″ N latitude, 17°43′13.65″ E longitude).
Type material
Holotype female and 14 female paratypes deposited in the Nematode Collection at the Institute for Sustainable Agriculture, CSIC, Córdoba, Spain. Another seven female paratypes deposited at Royal Belgian Institute of Natural Sciences, Brussels, Belgium (slides RIT808-809), and two female paratypes at the USDA Nematode Collection, Beltsville, MD, USA (collection number T-6125p). Specific D2-D3, ITS1, 18S-rRNA and COI sequences are deposited in GenBank with accession numbers JQ990042, JQ990043, JQ990050, and JQ990059, respectively.
Diagnosis and relationships
Xiphinema parabrevicolle n. sp. is characterized by a medium body size (1,716–2,540 μm), lip region widely rounded, separated from the rest of the body by a depression, odontostyle and odontophore 97.0–113.0 and 54.5–61.0 μm long respectively, vulva position at 52–56 %, female tail short (24.5–30.0 μm), dorsally convex-conoid, slightly concave ventrally with conoid-rounded terminus, c ratio (68.6–95.5), c’ ratio (0.7–0.8), and a specific D2-D3, ITS1, 18S-rRNA and COI sequences.
Morphologically, X. parabrevicolle n. sp. can be distinguished from the most similar species by a number of particular characteristics resulting from its specific alpha-numeric codes (A 2, B 32, C 32, D 1, E 21, F 1, G 2, H 3, I 12, J 2) sensu Lamberti et al. (2000).
Using the polytomous key of Lamberti et al. (2000) and sorting on features A (lip region), C (odontostyle length), D (c’ ratio) and J (tail end shape), X. parabrevicolle n. sp. groups with X. brevicolle, X. diffusum, X. incognitum Lamberti and Bleve-Zacheo 1979, Xiphinema luci Lamberti and Bleve-Zacheo 1979, Xiphinema pseudoguirani Lamberti et al. 1991, Xiphinema paramonovi Romanenko 1981, and Xiphinema taylori Lamberti et al. 1991. Xiphinema parabrevicolle n. sp. differs from topotypes of X. brevicolle by small differences in c’ and a slightly wider lip region (Lamberti et al. 1991). Also, the new species showed some differences with other species in this group in measurements and ratios, including L, c’, V ratio, odontostyle and odontophore length, lip region width, distance from guiding ring to anterior end, tail length and shape, and tail hyaline region (Table 3).
Remarks
Recently, Monteiro (2010) discussed the nomenclatorial disagreement between the original and widely used epithet brevicolle (=short neck) and the newly proposed by Luc et al. (1998) brevicollum. We concur with Monteiro (2010) that this is an “unjustified emendation”, since the termination colle is used as an adjective (neuter gender), and the International Code of Zoological Nomenclature (ICZN) allows this use. Consequently, the correct original species name must be preserved unaltered as Xiphinema brevicolle Lordello and Costa 1961, as has also been recognized by Sakai et al. (2011).
Xiphinema parapachydermum Footnote 2 n. sp. (Figs. 1, 4, Tables 3, 4)
Light micrographs of Xiphinema parapachydermum n. sp. from southern Spain. a Whole female; b, c Female neck regions; d Anterior region; e Vulval region; f, g Female tail regions; h Whole male; i, j Male tail regions, with ventromedian papillae arrowed. Abbreviations: a anus; gr guiding ring; v vulva. (Scale bars: a, h = 100 μm; b, c, e–j = 10 μm; d = 5 μm)
Female
Body medium-sized, habitus coiled in a more or less closed C to open spiral when killed by heat. Body tapering very gradually towards the extremities. Cuticle smooth, inner finely striated, 1.9 ± 0.2 (1.5–2.0) μm thick along body but thicker at tail tip (Table 4). Lip region flatly rounded, separated from the rest of the body by a constriction, 2.5 ± 0.1 (2.0–2.5) μm high. Amphidial fovea funnel-shaped with slit-like aperture at constriction level, 7.9 ± 0.3 (7.5–8.0) μm. Odontostyle 1.0–1.5 μm wide, with slightly furcate base. Odontophore with basal flanges weakly developed 6.5 ± 0.0 (6.5–6.5) μm wide. Pharyngeal basal bulb 66.7 ± 9.2 (52.0–85.5) μm long and 11.8 ± 2.4 (8.5–15.5) μm wide, occupying about 1/3 to 1/4 of the total pharyngeal length (Fig. 4b). Glandularium 47.8 ± 1.0 (47–49) μm long. Dorsal pharyngeal gland nucleus in anterior part of bulb, DN 30.6 ± 2.8 (29.0–35.0) %. Pharyngeal-intestinal valve broadly rounded, rather indistinct. Reproductive system didelphic, ovaries reflexed, uteri rather short (45–56 μm) without differentiation; anterior genital branch slightly more developed than the posterior. Vulva a transverse slit, clearly posterior to mid-body; vagina 13.2 ± 1.0 (12.0–15.0) μm long with short distal part and well-developed proximal. Ovaries without symbiontic bacteria and oocytes clearly separated; sphincter clearly visible; no sperm present. Prerectum often indistinct. Rectum 19.5 ± 2.0 (17.0–21.5) μm long or 1–0.8 times the anal body diameter. Tail short, but twice longer than anal body diameter (1.5–2.3), dorsally convex-conoid, with acute rounded tip, and often with a dorso-ventral depression; two pairs of caudal pores present (Figs. 1g, 4f–g). Tail hyaline region about 1/3 of the tail length.
Male
Almost as common as females. General morphology similar to that of female except for a rather coiled posterior region. Testes well developed, containing numerous large oval sperms. Spicules well sclerotized, ventrally curved (Fig. 4i,j). Lateral guiding pieces about 1/4 to 1/5 spicules length. A pre-anal pair of supplements 5 to 10 μm anterior to cloacal opening and a row of 5 to 6 single ventromedian supplements, located anterior to the spicule region.
Juveniles
Two juvenile stages were detected (third- and fourth-stage), which were morphologically similar to females from which they differ by their size, longer and more tapering tails and development of reproductive system (Table 4).
Type host and locality
Xiphinema parapachydermum n. sp. was found in a sandy soil around the roots of grapevine (Vitis vinifera L.) at Bollullos par del Condado, Huelva province, southern Spain (37°18′02.71″ N latitude, 6°30′51.55″ W longitude).
Other localities
The species was also detected in sandy soils around the roots of olive (Olea europaea subsp. europaea L.) at Hinojos, and grapevine at Chucena, both in Huelva province, southern Spain.
Type material
Holotype female and 14 female and six male paratypes deposited in the Nematode Collection at the Institute for Sustainable Agriculture, CSIC, Córdoba, Spain. Another three female and one male paratypes deposited at Royal Belgian Institute of Natural Sciences, Brussels, Belgium (slides RIT 810, 811), and two female paratypes at the USDA Nematode Collection, Beltsville, MD, USA (collection number T-6126p). Specific D2-D3, ITS1, 18S-rRNA and COI sequences are deposited in GenBank with accession numbers JQ990034-JQ990036, JQ990045, JQ990051, and JQ990056, respectively.
Diagnosis and relationships
Xiphinema parapachydermum n. sp. is characterized by a medium body size (1,411–2,000 μm), lip region flatly rounded, separated from the body by a constriction, odontostyle and odontophore 70.0–87.5 and 36.5–54.5 μm, respectively, vulva position at 55–66 %, female tail short, about twice longer than anal body diameter (1.4–2.3), dorsally convex-conoid, with acute rounded tip, and often with a dorso-ventral depression (26.5–35.5 μm long), c ratio (46.3–75.5), c’ ratio (1.5–2.3), and specific D2-D3, ITS1, 18S-rRNA and COI sequences.
Morphologically, X. parapachydermum n. sp. can be distinguished from the most similar species by a number of particular characteristics resulting from its specific alpha-numeric codes (A 2, B 21, C 12, D 34, E 32, F 21, G 12, H 2, I 23, J 1) sensu Lamberti et al. (2000).
Using the polytomous key of Lamberti et al. (2000) and sorting on features A (lip region), C (odontostyle length), D (c’ ratio) and J (tail end shape), X. parapachydermum n. sp. groups with X. pachydermum, X. brevisicum, X. duriense, X. intermedium, X. microstilum, X. opisthohysterum, and X. tarjanense Lamberti and Bleve-Zacheo 1979. Xiphinema parapachydermum n. sp. differs from X. pachydermum only by small differences in body length, female and male tail shape, and spicules shape (Table 3). Also, the new species showed some differences with other species in this group in measurements and ratios, including L, c’, V ratio, odontostyle and odontophore length, lip region width, distance from guiding ring to anterior end, tail length and shape, tail hyaline region, and presence/absence of males (Table 3).
Xiphinema paratenuicutis Footnote 3 n. sp. (Figs. 1, 5, Tables 3, 5)
Female: Body medium to large sized, forming a close C-shape when killed by heat. Cuticle 2.4 ± 0.3 (2.0–2.5) μm thick along body but thicker at tail tip (Table 5). Lip region expanded and flat at the anterior extremity, separated from body by constriction, 2.9 ± 0.4 (2.5–3.5) μm high. Amphidial fovea aperture 8.5 ± 0.5 (8.0–9.0) μm wide. Guiding apparatus with a double guiding ring. Odontophore with basal flanges moderately developed 7.8 ± 0.3 (7.5–8.0) μm wide. Pharyngeal basal bulb 72.2 ± 8.7 (60–89) μm long, occupying about 1/3 of the total pharyngeal length. Glandularium 63.2 ± 8.1 (55–70) μm long. Dorsal pharyngeal gland nucleus in anterior part of bulb, DN 29.4 ± 4.3 (25.0–34.0)%. Reproductive system amphidelphic, both branches apparently equally developed, opposed and ovaries reflexed; uteri rather long (76–100 μm) without any uterine differentiation but sperm present. Ovaries without symbiont bacteria. Vulva post-equatorial, a transverse slit-like; vagina 13.7 ± 2.4 (7.5–16.5) μm long with short distal part and well-developed proximal (Fig. 5e,f). Prerectum often indistinct. Rectum 17.9 ± 2.1 (15.0–19.5) μm long, or 1.6–2.1 times the anal body diameter. Tail short, conoid, ventrally almost straight with pointed terminus; two pairs of body pores (Fig. 5g–i).
Male
Almost as common as females. General morphology similar to female except for a stronger coiled posterior region. Testes well developed, containing numerous large oval sperm. Spicules well sclerotized, ventrally curved and lamina with clear ventral bulge (Fig. 5k). Lateral guiding pieces about 1/4 spicules length. A precloacal pair of supplements and a row of 5 single ventromedian supplements, located anterior to the retracted spicule.
Juveniles
All four juvenile stages were detected (first-, second-, third- and fourth-stage), which were morphologically similar to females from which differ by their size and tails longer and more tapering (Table 5).
Type host and locality
Xiphinema paratenuicutis n. sp. was found in a sandy soil around the roots of undetermined grasses (graminaceae) at Andújar, Jaén province, southern Spain (38°11′06.84″ N latitude, 4°02′18.96″ W longitude).
Type material
Holotype female and 14 female and 11 male paratypes deposited in the Nematode Collection at the Institute for Sustainable Agriculture, CSIC, Córdoba, Spain. Another three females and one male paratype deposited at Royal Belgian Institute of Natural Sciences (slide RIT 843), Brussels, Belgium, and two female paratypes at the USDA Nematode Collection, Beltsville, MD, USA (collection number T-6124p). Specific D2-D3 sequence is deposited in GenBank with accession number JQ990041.
Diagnosis and relationships
Xiphinema paratenuicutis n. sp. is characterized by a medium to large body size (1,888–2,205 μm), lip region expanded and anteriorly flat, separated from body by a constriction, odontostyle and odontophore 72–83 and 42–47 μm resp., vulva position at 55–60 %, female tail short, conoid, ventrally almost straight with pointed terminus (25–35 μm long), c ratio (56.7–79.9), c’ ratio (1.0–1.7), and specific D2-D3 sequence.
Morphologically, X. paratenuicutis n. sp. can be distinguished from the most similar species by a number of particular characteristics resulting from its specific alpha-numeric codes (A 2, B 23, C 2, D 21, E 32, F 21, G 23, H 2, I 21, J 1) sensu Lamberti et al. (2000).
Using the polytomous key of Lamberti et al. (2000) and sorting on features A (lip region), C (odontostyle length), D (c’ ratio) and J (tail end shape), X. paratenuicutis n. sp. groups with X. tenuicutis, X. bricolensis, X. californicum, Xiphinema mesostilum Lamberti et al. 1994, X. oxycaudatum, Xiphinema parvum Lamberti et al. 1991, and X. peruvianum. Xiphinema paratenuicutis n. sp. differs from X. tenuicutis only by small differences in V ratio and presence of males (Table 3). Also, the new species showed some differences with other species in this group in measurements and ratios, including L, c’, V ratio, odontostyle and odontophore length, lip region width, distance from guiding ring to anterior end, tail length and shape, and tail hyaline region (Table 3). Further detailed study of paratypes of X. tenuicutis (Fig. 6n–s) from USDA Nematode Collection (slides T-3447p, T-3448p, and T-3449p) showed the new species also differs by lip region (expanded and flat at the anterior extremity and separated from body by a constriction vs conoid-rounded and separated by a slight depression from the rest of the body), female tail shape (short, conoid, ventrally almost straight with pointed terminus vs elongated conoid with ventrally curved pointed terminus, Figs. 5 and 6), and some differences in measurements and ratios (Table 3).
Light micrographs of paratypes of Xiphinema santos Lamberti et al. 1993 (a–f) from Rothamsted; Xiphinema intermedium Lamberti and Bleve-Zacheo 1979 (g–m) from USDA-Nematode collection; paratypes of Xiphinema tenuicutis Lamberti and Bleve-Zacheo 1979 (n–s) from USDA-Nematode collection; paratypes of Xiphinema rivesi Dalmasso 1969 (t–v) from USDA-Nematode collection; and X. rivesi from Portugal (w–y) deposited in the Nematode Collection of Bari, Italy. a, d, j Vulval regions; b–c, g–i, n–o, t, w Anterior regions; d–f, k–m, q–s, u–v, x–y Female tail regions. (Scale bars: a = 50 μm; b–y = 10 μm)
Xiphinema duriense Lamberti et al. 1993 (Fig. 7, Table 6)
Light micrographs of Xiphinema duriense Lamberti et al. 1993 from southern Spain. a Whole female; b–e Female neck regions; f Vulval region; g–l Female tail regions. Abbreviations: a anus; gr guiding ring; V vulva. (Scale bars: a = 100 μm; b–l = 10 μm)
Female
The Spanish population of this species is characterised by a coiled body habitus forming a more or less close C when killed with heat, a lip region almost elliptical, expanded and offset from the body by a constriction, female reproductive system amphidelphic with two equally developed genital branches, uterine differentiation absent, vulva slit-like and always posterior to mid-body, vagina occupying about 1/2 corresponding body width, tail conoid with pointed terminus, slightly curved ventrally and bearing two caudal pores on each side (Fig. 7). Male not found.
Examination of five female paratype specimens showed odontostyle with poorly developed basal collar and odontophore with weak flanges. Position of pharyngeal gland nuclei and their associated openings as follows (n = 2): DO = 9.4 ± 1.9 (8.0–10.7); DN = 10.1 ± 0.3 (9.0–10.3); DO-DN = 0.7 ± 0.4 (0.4–1.0); SN = 56.9 ± 1.7 (55.–58.14); SO = 74.4 ± 0.6 (73.9–74.8); Distance SN-SO = 13.6 ± 0.6 (13.1–14.0) μm or 16.8 ± 0.7 (16.3–17.3) % of bulb length; SO = 48.7 ± 2.8 (46.7–50.6) μm anterior to end of platelet region. Neck 294.4 ± 14.5 (284–305) μm long. Glandularium 65.1 ± 3.1 (62–69) μm. Female reproductive system with relatively long unipartite uteri (50 μm) and without sperm; oviduct clearly differentiated, sphincter well-developed. Ovarial sac large, ovaries small and compact, not swollen, without symbiotic bacteria and developing oocytes clearly demarcated. Prerectum indistinct, 6.9–7.2 anal body widths long. Morphology and morphometrics of the Spanish population agree closely with the original description from Portugal by Lamberti et al. (1993) (Table 6), except for a lower L and a ratio in females (av. 1,436 μm, 61.8 vs av.1800 μm, 74.0, respectively), more posterior vulva position (av. 65.1 vs av. 60). Nevertheless, these differences further expand but do not exceed intraspecific variation reported by Lamberti et al. (1994) for others X. duriense populations associated with grapevine and weeds in Central and Northern Portugal. Morphologically and morphometrically this species is close to Xiphinema microstilum Lamberti et al. 1994 and Xiphinema opisthohysterum Siddiqi 1961. It can be differentiated from X. microstilum in having a lower L, a, c and c’ ratios (av. 1,436 μm, 61.8, 47.2, 2.2 vs av. 2,600 μm, 86.0, 74.0, 1.8), and a shorter odontophore and tail (av. 36.6 μm, 30.6 μm vs av. 45.0 μm, 35.0 μm); and from X. opisthohysterum in a higher c’ and V ratios (2.0–2.6, 63–67 vs 1.9–2, 56–59, respectively), longer odontostyle and distance from guiding ring to anterior end (70–74 μm, 53–60 μm vs 62–72 μm, 49–51 μm, respectively). Xiphinema duriense is morphologically similar to other species in the X. pachydermum-group, namely X. brevisicum, X. exile, X. lafoense, X. longistilum, X. mesostilum, X. microstilum and X. pachydermum. It differs from these species by the smaller body size (1.5–2.0 mm compared to a range of 1.9–4.4 mm) and by the absence of males and female gonoducts without sperm present. The presence of X. duriense is the first record for Spain, and the third after the original description from Portugal (Lamberti et al. 1993). These data indicate that this species may be an Iberian endemic species as suggested by Peña-Santiago et al. (2006).
The alpha-numeric codes for X. duriense to be applied to the polytomic identification key for the Xiphinema americanum-group species by Lamberti et al. (2000) are: A 2, B 12, C 1, D 34, E 3, F 21, G 1, H 1, I 23, J 1.
Xiphinema incertum Lamberti et al. 1983 (Fig. 8, Table 6)
Light micrographs of Xiphinema incertum Lamberti et al. 1983 from southern Spain. a Whole female; b–f Anterior regions; g–h Vulval regions; i–n Female tail regions. Abbreviations: a anus; af amphidial fovea. (Scale bars: a = 100 μm; b–d, f–n = 10 μm; e = 5 μm)
Female
The Spanish population of this species was characterised by a body forming a coiled spiral when killed with heat, a lip region slightly expanded and set off by a constriction, less pronounced in some specimens (Fig. 8) separated from the body by a depression, female reproductive system amphidelphic with two equally developed genital branches and absence of uterine differentiation, vulva a transverse slit located posterior to mid-body, vagina occupying about 1/2 of the corresponding body diameter, female tail conoid with narrowly rounded terminus and bearing two pairs of caudal pores (Fig. 8). Males unknown. The morphology and morphometrics of this population closely agree with the original description from several localities in the Balkan region e.g. Bulgaria, Croatia, Serbia (Lamberti et al. 1983; Barsi and Lamberti 2002), except for lower a, c’ and V ratios (49.7, 1.2, 52.4 vs 57, 1.5, 57, respectively) and a higher b ratio (9 vs 6.4). The observed differences extend the known range of the features in X. incertum. Xiphinema incertum in the Balkan region co-occurs with X. pachtaicum and is also morphologically very similar to it except for tail shape, and Barsi and Lamberti (2002) questioned the validity of X. incertum. To avoid inflation of the number of new species, we currently consider the few differences observed as intraspecific until molecular data are available from the populations of the Balkan region. This species is quite similar to Xiphinema franci Heyns and Coomans 1994, X. intermedium and X. tenuicutis, but can be differentiated from X. franci in L, a, b, c, and c’ values (1,800 μm, 49.7, 9.0, 64.5, 1.2 vs 1,430 μm, 40.4, 4.7, 47.5, 1.4, respectively), a shorter hyaline region (6.5 μm vs 12.5 μm) and the tail shape (rounded terminus vs pointed terminus), from X. intermedium in a higher c ratio (64.5 vs 47.0), a longer odontostyle (92.2 μm vs 76.0 μm), a shorter tail and hyaline region (28.6 μm, 6.5 μm vs 33.0 μm, 9.0 μm) and the tail shape (ventrally straight with narrowly rounded terminus vs ventrally curved and concave with pointed terminus), and from X. tenuicutis by the shape of lip region (less expanded and separated from the rest of the body by an obscure depression in X. tenuicutis) and a longer odontostyle and odontophore (92.2 μm, 49.7 μm vs 76.0 μm, 45.0 μm, respectively). The presence of X. incertum is the first record for the Iberian Peninsula and the third from Europe after the original description in Bulgaria and later in Croatia (Lamberti et al. 1983; Barsi 1989).
The alpha-numeric codes for X. incertum to be applied to the polytomic identification key for the Xiphinema americanum-group species by Lamberti et al. (2000) are (in parentheses are exceptions): A 2, B 2(3), C 2, D 2(13), E 3 (12), F 1(2), G 2(1), H 3(2), I 2(1), J 2.
Xiphinema opisthohysterum Siddiqi 1961 (Fig. 9, Table 6)
Light micrographs of Xiphinema opisthohystherum Siddiqi 1961 from southern Spain. a Whole female; b, c Female neck regions; d–f Anterior regions; g Vulval regions; h–k Female tail regions. Abbreviations: a anus; gr guiding ring; V vulva. (Scale bars: a = 100 μm; b–c, f–k = 10 μm; d, e = 5 μm)
Female
The Spanish population of this species was characterised by a body forming a coiled spiral when killed with heat, lip region widely expanded, of elliptical shape and clearly offset from the body by a constriction, female reproductive system with two equally developed genital branches and absent uterine differentiation, vulva clearly posterior to mid-body, vagina occupying about 1/2 of the corresponding body diameter, female tail conical and elongated with almost pointed terminus and bearing two pairs of caudal pores (Fig. 9). Males not found. Morphology and morphometrics of this population agree closely with the redescription by Lamberti and Bleve-Zacheo 1979 (Table 6), except for L, c’ and V ratio (1,377–1,550 μm, 1.5–1.9, 60–67 vs 1,800–1,850 μm, 1.9–2.0, 56–59, respectively) and a shorter tail (24–28 μm vs 30–36 μm). Nevertheless, these differences may be due to the low number of specimens (two) studied by Lamberti and Bleve-Zacheo (1979), and do not exceed the intraspecific variation of species from the X. americanum group. Also, this species is quite similar to X. duriense and X. tenuicutis. From X. duriense it can be differentiated in lower c’ (1.5–1.9 vs 1.9–2.3) and a shorter odontostyle and distance from guiding ring to anterior end (61.3 μm, 48.6 μm vs 70.0 μm, 61.0 μm, respectively), and from X. tenuicutis by the shape of lip region (separated from the rest of the body by a deep depression vs slight depression), a higher a and V ratio (59.2, 63.8 vs 46, 52.4, respectively), and a shorter odontostyle, odontophore and distance from guiding ring to anterior end (61.3 μm, 36.9 μm, 48.6 μm vs 76.0 μm, 45.0 μm, 70.0 μm, respectively). The presence of X. opisthohysterum it is the first record for Spain, and confirms its occurrence in the Iberian Peninsula, first recorded by Sturhan (1983) in North-East Portugal.
The alpha-numeric codes for X. opisthohysterum to be applied to the polytomic identification key for the Xiphinema americanum-group species by Lamberti et al. (2000) are: A 2, B 12, C 1, D 32, E 3, F 12, G 12, H 1, I 12, J 1.
Xiphinema rivesi Dalmasso 1969 (Figs. 6 and 10 Table 7)
Light micrographs of Xiphinema rivesi Dalmasso 1969 from southern Spain. a Whole female; b Whole male; c Female neck region; d–g Anterior regions; h, i Vulval regions; j–m Female tail regions; n Male region. Abbreviations: a anus; af amphidial fovea; gr guiding ring; sp spicules. (Scale bars: a, b = 100 μm; c–n = 10 μm)
Female
Spanish populations of this species are recognisable by a rounded lip region, continuous with rest of the body, vulva slightly posterior to mid-body, female tail conoid with acute rounded tip (Fig. 10). The morphology and morphometrics of these populations are most similar among them (Table 7) and to other populations of this species from Southern Spain (Gutiérrez-Gutiérrez et al. 2011a), except for a slightly higher c’ ratio and a more narrow lip region in the population from Niebla (Table 7), but do not exceed the intraspecific variations, as confirmed by molecular analyses (see below). Similarly, morphology and morphometrics traits of these three Spanish populations agree very well with the original description of this species (Dalmasso 1969) and detailed study of paratypes (Figs. 6, 10) from USDA Nematode Collection (slide T-3440p) (Fig. 6), except for a slightly higher b ratio in the population from Alcala de Guadaira (7.4 vs 6.2) and narrower lip region in the population from Niebla (7.9 μm vs 10 μm), and some differences in a, c, c’ ratio, odontostyle, distance from guiding ring to anterior end, and tail length, which do not exceed the intraspecific variations as showed for these Spanish population. Also, morphology and morphometrics traits of these three Spanish populations agree very well with the other description of X. rivesi associated to grapevine from Central Portugal (Lamberti et al. 1994, Fig. 6w–y), except for some almost negligible differences in a, b ratio and hyaline region length, which do not exceed either the intraspecific variations.
Only one male was found in this investigation (Fig. 10, Table 7), which is similar to female except for reproductive system, and was mainly characterized by six ventromedian precloacal papillae and anterior to adanal pair, and spicules well developed (Fig. 10, Table 7).
General morphology and morphometrics agree very well with the original description of this species (Dalmasso 1969) and other reports (Urek et al. 2005). This species can be differentiated from X. franci by a longer body (1,886 μm vs 1,430 μm), a shorter hyaline tail region (8.6 μm vs 12.5 μm) and a different shape of the lip region (continuous with rest of the body vs offset from the rest of the body by a depression); from X. luci by the tail shape (shorter and more rounded in X. luci) and the shape of the lip region (rounded vs flattened on top); from Xiphinema occiduum Ebsary et al. 1984 by a lower L, b and c ratios (1,886 μm, 7.4, 53.4 vs 2,300 μm, 9, 70, respectively), a longer odontostyle (92.1 μm vs 75 μm), a different lip region shape (continuous with rest of the body vs offset from the rest of the body by a constriction) and a smaller number of ventromediam supplements (6 vs 9). And from X. oxycaudatum by a higher b ratio (7.4 vs 5.5), a slightly longer odontostyle (92.1 μm vs 82 μm), lip region shape (continuous with rest of the body vs offset from the rest of the body by constriction), and tail shape (with rounded terminus vs pointed terminus).
Xiphinema rivesi, originally described from South-East France (Dalmasso 1969), has been subsequently reported from a great number of localities of European countries including Portugal, Slovenia and Spain (Lamberti et al. 1994; Bello et al. 2005; Urek et al. 2005; Gutiérrez- Gutiérrez et al. 2011a) as well as from North America (Ebsary et al. 1984; Robbins 1993; Mekete et al. 2009), South and Central America (Doucet et al. 1998; Auger et al. 2009), Iran and Pakistan (Nasira and Maqbool 1994; Fadaei et al. 2003) and Western Australia (Sharma et al. 2003). The alpha-numeric codes for X. rivesi to be applied to the polytomic identification key for the Xiphinema americanum-group species by Lamberti et al. (2000) are: A 1, B 2, C 21, D 23, E 23, F 1, G 12, H 23, I 32, J 2.
Although there is a general agreement about the morphology and intraspecific variability of this species, rDNA markers suggest an unresolved complex of cryptic species, since populations from Spain, Pennsylvania and Prosser, Washington (USA) were clearly separated in three different clades (see below). These findings did not support the hypothesis by Lamberti and Ciancio (1993) suggesting that X. rivesi may have been introduced from North America into Europe, since X. rivesi is known as a vector species of CLRV, TomRSV, and TRSV, in North America (Georgi 1988; Robbins and Brown 1991; Brown and Halbrendt 1992) and TomRSV and TRSV in Slovenia. (Širca et al. 2007).
Xiphinema santos Lamberti et al. 1993 (Figs. 6, 11, Table 8)
Light micrographs of Xiphinema santos Lamberti et al. 1993 from southern Spain. a Whole female; b Female neck region; c–e Anterior regions; f, g Detail of pharyngeal bulb; h, i Vulval regions; j–p Female tail regions. Abbreviations: a anus; gr guiding ring; V vulva. (Scale bars: a = 100 μm; b–l = 10 μm)
Female
The Spanish population of this species was characterised by a body forming a close C-shaped to coiled spiral when killed by heat. Cuticle smooth, 2.1 ± 0.2 (2.0–2.5) μm thick along body but thicker at tail tip (Table 8). Lip region rounded anteriorly, slightly expanded and separated from the body by a depression, 3.3 ± 0.3 (3.0–3.5) μm high. Pharyngeal basal bulb 70.3 ± 8.4 (61.5–84.0) μm long and 19.2 ± 4.6 (15.5–29.5) μm wide, occupying about 1/3 to 1/4 of the total pharyngeal length (Fig. 11f,g). Pharyngeal-intestinal valve inconspicuous, hemispherical. Reproductive system amphidelphic, both branches apparently equally developed, opposed and reflexed, without any uterine differentiation. Ovaries with symbiont bacteria, oviducts relatively long. Vulva slightly post-equatorial, a transverse slit-like; vagina 15.3 ± 1.1 (13.5–17.0) μm long with short distal part and well-developed proximal. Prerectum often indistinct. Rectum 18.8 ± 2.6 (17.5–25.5) μm long, or 1.0–1.2 times the anal body diameter. Tail short, conoid, weakly curved ventrally with conoid-rounded terminus, bearing two pairs of caudal pores (Fig. 11j–p). Two juvenile stages were detected (third- and fourth-stage), which were morphologically similar to females apart from smaller body and longer, more tapered tails (Table 8). The present population of X. santos was found in a sandy soil around the roots of grapevine (Vitis vinifera L.) and Stone pine (Pinus pinea L.) at Rociana del Condado, Huelva province, southern Spain; it is the first record for Spain, and confirms its presence in the Mediterranean Basin, after several reports in Portugal and North Egypt (Lamberti et al. 1994, 1996).
Morphology and morphometrics of this population agree with original description by Lamberti et al. (1993), except for L, a, c’ ratio, and tail length (1,466–1,633 μm, 42.7–48.1, 1.0–1.4, 25.0–34.0 μm vs 1,700–2,000 μm, 54.0–59.0, 1.5–1.8, 32.0–37.0 μm respectively). Nevertheless, these differences are within the range ofintraspecific variation, as showed in different Portuguese populations of this species (Lamberti et al. 1994). This species is also close to X. intermedium. After detailed study of paratypes (Fig. 6) from Rothamsted and USDA Nematode Collection (T-3449p) the Spanish population differs from X. intermedium by female tail (25–34 μm long, short, conoid, weakly curved ventrally with conoid-rounded terminus vs 31–38 μm long, conoid elongated, dorsally convex and ventrally clearly curved and concave with pointed terminus, Fig. 11g–m), and some differences in measurements and ratios, including body length (1,466–1,633 μm vs 1,400–1,900 μm), odontostyle and odontophore length (76–82, 41–49 μm vs 68–80, 39–50 μm, respectively), V (52–55 vs 50–57), a, c, and c’ ratio (43–48, 46–65, 1.0–1.4 vs 38–51, 41–52, 1.3–1.7, respectively).
The alpha-numeric codes for X. santos to be applied to the polytomic identification key for the Xiphinema americanum-group species by Lamberti et al. (2000) are: (A 2, B 12, C 1, D 21, E 21, F 1, G 12, H 2, I 23, J 2).
Phylogenetic relationships of the Xiphinema americanum-group
The amplification of D2-D3 expansion segments of 28S rDNA, ITS1, partial 18S, and COI regions yielded a single fragment of approximately 800 bp, 1,030 bp, 1,600 bp, and 400 bp, respectively, based on gel electrophoresis. Sequences from other species of Xiphinema americanum-group obtained from NCBI were used for further phylogenetic studies. Sequences for X. parabrevicolle n. sp., X. parapachydermum n. sp., X. paratenuicutis n. sp., X. duriense, X. incertum, X. opisthohysterum, X. pachtaicum, X. rivesi, and X. santos were obtained for these species in this study. Sequences for X. pachtaicum and X. rivesi matched well with former sequences deposited in GenBank, except for one isolate of X. rivesi from Pensylvania, USA, extending the molecular diversity of these species to newly studied areas.
Xiphinema parabrevicolle n. sp. (JQ990042) matched well with the D2-D3 sequences of the X. americanum-group deposited in GenBank, being 98 % to 97 % similar to some of them, such as, X. citricolum (DQ299490-DQ299494 and DQ285668), X. santos (AY601587), X. brevicolle (HQ184473), and X. diffusum (AY601600). ITS1 sequence showed similarity with several sequences of X. brevicolle with 85 % similarity (HQ184474, FM211421-25, AY430190), and 84 % with X. diffusum (AY359858). Using partial 18S, many similar sequences were found differing in seven nucleotides with a 99 % similarity. COI sequences from X. parabrevicolle n. sp. did not show any similarity with other sequences from Xiphinema americanum-group or non americanum-group, but showed a 75 % similarity with Longidorus helveticus Lamberti et al. 2001 (EF538747).
Xiphinema parapachydermum n. sp. D2-D3 showed 96 % similarity with X. pachtaicum (HM921393-HM921395), and 88 % with X. pachydermum (AY601608). Intra-specific variation detected among the three studied populations was four nucleotides. Partial 18S also agree with results obtained from D2-D3, the maximum similarities were related to ‘X. pachtaicum’ (AM086682) with 98 % similarity. No similarities in the GenBank were found for ITS1 and COI sequences in this species.
Xiphinema paratenuicutis n. sp. D2-D3 showed 85 % similarity with X. brevisicum (AY601610), and 84 % to 83 % with some species, including X. simile, X. pachydermum, X. pachtaicum, Xiphinema utahense Lamberti and Bleve-Zacheo 1979, Xiphinema thornei Lamberti and Golden 1986 (AY601609, AY601608, AY601607, AY601598, AY601595, respectively). Sequencing of the ITS1, partial 18S and partial COI sequences were not successful despite several attempts.
Xiphinema duriense D2-D3 (JQ990032) showed 89 % similarity with X. brevisicum (AY601610), and 88 % to 87 % with X. pachydermum, X. pachtaicum, X. americanum group sp. LZ-2011 (AY601608, AY601607, JN091972, respectively). Partial COI sequence did not show any similarity with sequences deposited in the GenBank. Xiphinema incertum (JQ990031) was closely related in D2-D3 sequence to other species of the Xiphinema americanum-group such as X. pachtaicum (HM921355 and HM921356) with 98 % similarity and ‘X. pachtaicum’ (AY601607) with 93 % similarity, and ITS1 showed 94 % similarity with X. pachtaicum (AY430178, HM921337). However, no similarities in the GenBank were found for COI sequences in this species. D2-D3 region from X. opisthohysterum (JQ990040) showed similarity with X. brevisicum (AY601610) with similarity value of 88 to 87 % with X. brevisicum, X. pachydermum, X. americanum group sp. LZ-2011 (AY601610, AY601608, AY601607, JN091972, respectively); and partial COI sequence did not show any similarity with sequences deposited in the GenBank. D2-D3 and partial COI sequences from X. pachtaicum (JQ990033, JQ990057) matched well with sequences from X. pachtaicum deposited in GenBank, being 99 % similar with all of them (HM921393-HM921395, HM921369-HM921379, respectively). Likewise, X. rivesi D2-D3 region matched closely (99 % similarity) with other species of X. americanum-group such as X. rivesi (AY210845, HM921357 and HM921358) and X. americanum (AY580056). Intra-specific differences were not found among our populations. ITS1 showed even higher similarities with many X. americanum-group species as X. rivesi (HM921338, 100 %), X. inaequale (GQ231530, 98 %), X. thornei (AY430176, 97 %); intra-specific variability within our populations was low (99 %). Partial 18S also showed high similarity with other X. americanum-group species such as X. rivesi (HM921344), 99 % similarity, differing in 5 nucleotides. Some similarity was found with COI sequence, 85 % to 84 % similarity with X. americanum (AY382608) and X. peruvianum (AM086692). Finally, X. santos D2-D3 matched well with the X. americanum-group deposited in GenBank, being identical to X. santos (AY601587), and 99 % similar to some other species, including X. citricolum (DQ299490-DQ299494 and DQ285668), Xiphinema georgianum Lamberti and Bleve-Zacheo 1979 (DQ299495-DQ299502). Similar results were detected for other markers such as ITS1 and partial 18S. ITS1 showed 97 % similarity with several X. americanum-group species such as Xiphinema americanum Cobb 1913 (AF511423, AY430189), Xiphinema laevistriatum Lamberti and Bleve-Zacheo 1979 (DQ299529), and X. georgianum (GQ299521). Also, partial 18S showed a high interspecific similarity, since many sequences differing in only 11 nucleotides (99 % similarity), indicating that this sequence is not adequate for species delimitation. Partial COI sequence showed 86 % similarity with X. peruvianum (AM086692) and 83 % with X. brevicolle (AM086707). Sequencing of the ITS1, partial 18S and partial COI sequences for some species and populations were not successful despite several attempts (Table 1).
Phylogenetic analysis (BI and ML) of X. americanum-group based on D2-D3 expansion segments of 28S rDNA of a multiple edited alignment including 52 sequences and 766 total characters showed two clades clearly separated and supported (Fig. 12). The phylogenetic analysis showed well supported groups at major and terminal clades in both analyses. The phylogenetic tree resolved two major clades: (I) with 37 sequences including X. parabrevicolle n. sp., X. brevicolle, X. santos, X rivesi, and other species, while clade (II) with 14 sequences, includes X. parapachydermum n. sp., X. pachydermum, X. paratenuicutis n. sp., X. duriense, X. incertum, X. opisthohysterum, and X. pachtaicum among others (Fig. 12). In group I, Xiphinema parabrevicolle n. sp. (JQ990042) formed a non well supported clade with X. brevicolle, X. taylori, X. diffusum, X. inaequale, X. lambertii, and X. americanum s. l. from Japan (AB635403) but within the group it is separated from X. brevicolle of which the three sequences (HM163209 HQ184473, and AB635401) do not cluster together. Further there is no support for the sister group composed of three sequences of X. santos (one from Portugal and another from Spain), and two sequences of X. citricolum (from Florida) with no resolution on their relationship. Group I shows a basal group composed of 11 sequences of five species among them X. rivesi from Spain (5 sequences) form a non-supported group separated from X. rivesi (USA); also the two sequences of X. bricolensis do not group together. In group II, Xiphinema parapachydermum n. sp. (JQ990034-JQ990036) formed a well supported clade with X. pachtaicum and X. incertum (JQ990031), as well as with the sister group X. pachydermum (AY601608) (Fig. 12). Similarly, X. duriense (JQ990032) formed other a well supported clade with X. opisthohysterum (JQ990040) as well as X. simile with X. parasimile. The relationship of the three former clades and of X. paratenuicutis n. sp. (JQ990041) within group II is not resolved.
Phylogenetic relationships within Xiphinema americanum-group complex. Bayesian 50 % majority rule consensus trees as inferred from D2 and D3 expansion segments of 28S rRNA sequences alignments under the GTR + G model. Posterior probabilities more than 65 % are given for appropriate clades; bootstrap values greater than 50 % are given on appropriate clades in ML analysis. Newly obtained sequences in this study are underlined. *: species name modified according to Monteiro (2010)
The ITS1 sequences were obtained from five species (Table 1). Phylogenetic analysis (BI and ML) of X. americanum-group based on ITS1 from sequences showing major similarities were included in the analysis, other sequences not included performed weak alignments. Multiple edited alignment including 34 sequences with 842 positions in length showed two clearly separated and supported clades (I and II, Fig. 13). Clade I included 30 sequences and 17 species, separated in two supported sub-clades with 11 and five species, respectively, a) X. georgianum, X. citricolum, X. peruvianum, X. oxycaudatum, X. santos, X. laevistriatum, X. rivesi (JQ990046, JQ990047, HM921338, AY430186, AY430185), X. thornei, X. inaequale, X. californicum, X. brevicolle, X. floridae, and X. tarjanense; and b) X. brevicolle (FM211425, AY359856), X. americanum, X. diffusum, X. incognitum, and X. parabrevicolle n. sp.; the latter subgroup is moderately supported and within it X. brevicolle is paraphyletic. For X. rivesi, the population found in USA (AY430185 and AY430186) is clearly not closely related phylogenetically with European populations. Clade II showed an unresolved relationship with group I. It comprises three species with X. parabrevicolle as a sister group of the well supported clade X. incertum and X. pachtaicum.
Phylogenetic relationships within Xiphinema americanum-group complex. Bayesian 50 % majority rule consensus trees as inferred from ITS1 rRNA gene sequence alignment under the TMP3uf + G model. Posterior probabilities more than 65 % are given for appropriate clades (in bold letters); bootstrap values greater than 50 % are given on appropriate clades in ML analysis. Newly obtained sequences in this study are underlined
Phylogenetic analysis (BI and ML) of X. americanum-group based on the partial COI of a multiple edited alignment including 40 sequences with 383 positions in length showed six moderate to highly supported clades (Fig. 14). Clade I included 11 species in two sub-clades, a) X. santos, X. parabrevicolle n. sp., X. peruvianum (USA, AM086692), X. citricolum, X. cf. americanum RN-2005 (USA), and X. americanum s.str.; and b) X. rivesi (Arkansas,AM086697), X. floridae, X. georgianum, and X. tarjanense. Clade II included four species X. duriense, X. opisthohysterum, X. peruvianum (Brazil, AM086712), and X. rivesi (Spain, JQ990060). Clade III included 10 species in two sub-clades: a) Xiphinema sp. JPN-HS-09, X. inaequale, X. cf. americanum RN-2005 (Nepal), X. lambertii, X. incognitum; and b) X. diffusum, X. brevicolle (Japan, AB604337), X. taylori, and X. brevicolle (Czech Republic, HM163206). Clade IV included three species X. pachtaicum, X. parapachydermum n. sp., and X. incertum. Clade V included two species X. pachtaicum (Czech Republic, GU222424) and X. simile (Slovakia, AM086708). Finally, clade VI included three species X. cf. americanum RN-2005 (Portugal), X. simile (Czech Republic, GU222427) and X. simile (Serbia, AM086711).
Phylogenetic relationships within Xiphinema americanum-group complex. Bayesian 50 % majority rule consensus trees as inferred from COI gene sequence alignment under the TVM + I + G model. Posterior probabilities more than 65 % are given for appropriate clades (in bold letters); bootstrap values greater than 50 % are given on appropriate clades in ML analysis. Newly obtained sequences in this study are underlined
Discussion
The primary objective of this study was to identify and molecularly characterize species belonging to the Xiphinema americanum-group in cultivated and natural environments in southern Spain and one sample in Italy. Our results demonstrate that the application of rDNA and mtDNA molecular markers integrated with morphological studies revealed that some long-assumed single species are in fact cryptic species. Results of this study suggest that species from Xiphinema americanum-group show a high diversity in the southern part of Iberian Peninsula, and demonstrate that intensive surveys within a region may still increase substantially our knowledge of nematode diversity. However, the main contribution of this work in the X. americanum-group is the holistic approach of diagnosis, that also has critical phytopathological implications, since some species are virus vectors or are listed as A1 (X. americanum, X. californicum, X. bricolensis) and A2 (X. rivesi) quarantine organisms by the EPPO.
Phylogenetic analysis of D2-D3 of this and previous studies demonstrate that X. brevicolle comprises a complex of species with similar morphological traits but differing molecularly as shown in Figs. 12 and 13, implying the need for additional integrative studies for clarification. Current results agree with Lamberti et al. (2000) which discussed the synonymies proposed by Luc et al. (1998) within the X. brevicolle species complex (i.e. X. diffusum, X. parvum, X. pseudoguirani, X. taylori), as it was demonstrated later for some of these species by using ITS (Chen et al. 2005; Oliveira et al. 2005). Similarly, on the basis on D2-D3 and ITS1 phylogeny, X. rivesi appears to be also a species complex, since isolates from Spain were related to an isolate from Prosser, USA (AY210845), but separated from another isolate from Pennsylvania, USA (AY601589, AY430185, AY430186) (Figs. 12, 13). These differences were also confirmed in the phylogeny of partial COI, which clearly separate the Spanish isolates from an isolate from Arkansas, USA (AM086697) (Fig. 14). Similar differences were detected by Vrain (1993) studying several American isolates of X. rivesi using of restriction fragments of amplified internal transcribed spacer regions. Nevertheless, this situation needs to be clarified with additional integrative studies and using other molecular markers (i. e. ITS1 and COI) for those isolates, and also checking possible misidentification of some of these populations.
D2-D3 expansion segments of 28S rDNA and ITS1 showed congruent phylogenetic information. Xiphinema parabrevicolle n. sp. is well associated in the same clade as X. brevicolle, X. diffusum and X. americanum. Similar results were obtained for X. rivesi, in which one population found in USA (AY430185 and AY430186) is clearly not closely related phylogenetically with European populations. Xiphinema incertum showed a congruent position as D2-D3 expansion segments of 28S rDNA with X. pachtaicum (HM921337). The position of X. santos is completely different, showing a well supported clade with X. laevistratum Lamberti and Bleve-Zacheo 1979 (DQ299531) and X. inaequale (DQ299530). However, the position of X. santos in D2-D3 analysis is not supported.
The phylogenetic relationships inferred in this study based on the D2-D3 of 28S rRNA, the ITS1 of rRNA gene, and partial COI sequences mostly agree with the morphological grouping of species obtained by the cluster analysis carried out by Coomans et al. (2001) based on key diagnostic characters of the genus Xiphinema. In comparison with a cluster analysis based on morphometrics that resulted in the subdivision of the X. americanum-group into several subgroups (Lamberti and Ciancio 1993), our results did not support these subgroups apart from the X. pachtaicum group (D2-D3 analysis), since species of these subgroups clustered in several clades (Figs. 12, 13, and 14). Nevertheless, as stated by Coomans et al. (2001), molecular and morphological evolution may differ because of e.g. proceeding at different speed as a result of different mechanisms. The present study also rejects the hypothesis that X. americanum-group represents numerous morphotypes with large inter-and intra- population variability resulting from environmentally driven morphometric plasticity, based on the phylogeny of the 18S rRNA gene (Oliveira et al. 2004). Phylogenetic analyses of D2-D3 and partial COI regions in this study were congruent supporting the clade including X. brevicolle, X. diffusum, and X. taylori (Figs 12, 14), whereas X. parabrevicolle n. sp. clustered with X. santos and outside this group in the partial COI tree (Fig. 14). For ITS1, X. parabrevicolle n. sp. clustered in the sub-clade b) with X. incognitum, X. diffusum, X. americanum, and X. brevicolle ((AY359856, FM211426). Phylogenetic relationships based on the D2-D3 and ITS1 agree with those by He et al. (2005) and partially support the hypothesis by Luc et al. (1998) and Coomans et al. (2001), that the X. pachydermum-subgroup (including X. brevisicum, X. exile, X. lafoense, X. longistilum, X. mesostilum, X. microstilum, X. pachydermum) comprises a subgroup outside the X. americanum-group, as well as some other species belonging to other X. americanum subgroups, viz. X. brevisicum, X. pachtaicum, X. incertum, X. duriense, X. opisthohysterum, X. simile and X. paratenuicutis n. sp. (Figs. 12, 13). Nevertheless, the partial COI reject this hypothesis, since X. parapachydermum n. sp., X. pachtaicum from Spain and X. incertum clustered together and clearly within the X. americanum-group species (Fig. 14). These results agree with those by He et al. (2005) based on D2-D3 region which differentiate a group with X. brevicolle and X. diffusum (and other species), as well as the X. pachtaicum group including X. pachydermum, X. brevisicum and X. simile. Consequently the relationships of this subgroup remain unclear and need the availability of additional ITS sequences for a larger analysis and other molecular markers which may allow the construction of a more complete and precise phylogeny in this group. Surprisingly, the partial COI for X. duriense (JQ990053) and X. opistohysterum (JQ990053) showed 100 % similarity and 97 % alignment coverage from the query sequence, but were clearly separated by D2-D3 (96 % similarity and 99 % alignment coverage from the query sequence), which may suggests a recent speciation event in this gene and maybe only mutations with amino acid changes occurred and they have been not kept in the species separation.
ITS1 diversity was higher than that for D2-D3 for differentiating species of the X. americanum-group and should be considered as a very useful marker for the identification of species in this group (Chen et al. 2005; Meza et al. 2011). Furthermore, our results on mtDNA did not support the hypothesis by Lazarova et al. (2006) on the apparent dichotomy existing among X. americanum-group populations from North America and those from Asia, South America and Oceania. On the contrary, as shown by the phylogenetic tree of partial COI sequence, the Italian and Spanish populations of the X. americanum-group clustered with populations from North America, South Africa, or Brazil (Fig. 14).
In summary, present study establishes the importance of using polyphasic identification highlighting the time consuming aspect and difficulty of a correct identification at species level within the Xiphinema americanum group. This study also provides molecular markers for precise and unequivocal diagnosis of some species of the X. americanum-group in order to differentiate virus vector or quarantine species, since the morphology is quite similar among them and mixed populations of the X. americanum-group in the same soil sample are quite frequent all over the world.
Notes
The species epithet refers to Gr. prep. para, alongside of, resembling; N.L. masc. n. brevicolle, because of its close resemblance to Xiphinema brevicolle
The species epithet refers to Gr. prep. para, alongside of, resembling; N.L. masc. n. pachydermum, because of its close resemblance to Xiphinema pachydermum
The species epithet refers to Gr. prep. para, alongside of, resembling; N.L. masc. n. tenuicutis, because of its close resemblance to Xiphinema tenuicutis
References
Auger, J., Leal, G., Magunacelaya, J. C., & Esterio, M. (2009). Xiphinema rivesi from Chile transmits tomato ringspot virus to cucumber. Plant Disease, 93, 971.
Barsi, L. (1989). The longidoridae (Nematoda: dorylaimida) in Yugoslavia. I. Nemtologia Mediterranea, 17, 97–108.
Barsi, L., & De Luca, F. (2008). Morphological and molecular characterisation of two putative Xiphinema americanum-group species, X. parasimile and X. simile (Nematoda: Dorylaimida) from Serbia. Nematology, 10, 15–25.
Barsi, L., & Lamberti, F. (2002). Morphometrics of three putative species of the Xiphinema americanum group (Nematoda: Dorylaimida) from the territory of the former Yugoslavia. Nematologia Mediterranea, 30, 59–72.
Bello, A., Robertson, L., Díez-Rojo, M. A., & Arias, M. (2005). A re-evaluation of the geographical distribution of quarantine nematodes reported in Spain. Nematologia Mediterranea, 33, 209–216.
Brown, D. J. F., & Halbrendt, J. M. (1992). The virus vector potential of Xiphinema americanum and related species. Journal of Nematology, 24, 584.
Castillo, P., Vovlas, N., Subbotin, S., & Troccoli, A. (2003). A new root-knot nematode, Meloidogyne baetica n. sp. (Nematoda: Heteroderidae), parasitizing wild olive in Southern Spain. Phytopathology, 93, 1093–1102.
Chen, D. Y., Fang, H., Yen, J. H., Cheng, Y. H., & Tsay, T. T. (2005). Differentiation of the Xiphinema americanum-group nematodes X. brevicollum, X. incognitum, X. diffusum and X. oxycaudatum in Taiwan by morphometrics and nuclear ribosomal DNA sequences. Nematology, 7, 713–725.
Cobb, N. A. (1913). New nematode genera found inhabiting fresh water and non-brackish soils. Journal of the Washington Academy of Science, 3, 432–444.
Coolen, W. A. (1979). Methods for extraction of Meloidogyne spp. and other nematodes from roots and soil. In F. Lamberti & C. E. Taylor (Eds.), Root-knot nematodes (Meloidogyne species). Systematics, biology and control (pp. 317–329). New York: Academic.
Coomans, A., Huys, R., Heyns, J., & Luc, M. (2001). Character analysis, phylogeny, and biogeography of the genus Xiphinema Cobb, 1973 (Nematoda, Longidoridae). Vol. 287, Annales du Musée Royal de l’Afrique Centrale (Zoologie), Tervuren, Belgique.
Dalmasso, A. (1969). Étude anatomique et taxonomique des genres Xiphinema, Longidorus et Paralongidorus (Nematoda: Dorylaimida). Mémoires du Muséum National d’Histoire Naturelle, Nouvelle Série, A. Zoologie, 61, 34–82.
Decraemer, W., & Robbins, R. T. (2007). The who, what and where of Longidoridae and Trichodoridae. Journal of Nematology, 39, 295–297.
Doucet, M. E., Ferraz, L. C. C. B., Magunacelaya, J. C., & Brown, D. J. F. (1998). The occurrence and distribution of longidorid nematodes in Latin America. Russian Journal of Nematology, 6, 111–128.
Ebsary, R. A., Potter, J. W., & Allen, W. R. (1984). Redescription of Xiphinema rivesi Dalmasso, 1969 and X. americanum Cobb, 1913 in Canada with a description of Xiphinema occiduum n. sp. (Nematoda, Longidoridae). Canadian Journal of Zoology, 62, 1696–1702.
Ebsary, R. A., Vrain, T. C., & Graham, M. B. (1989). Two new species of Xiphinema (Nematoda: Longidoridae) from British Columbia vineyards. Canadian Journal of Zoology, 67, 801–804.
ECBOL web (European Consortium for the Barcode of Life). http://www.ecbol.org/index.php?/pa/ge/welcome
Fadaei, A. A., Coomans, A., & Kheiri, A. (2003). Three species of the Xiphinema americanum lineage (Nematoda: Longidoridae) from Iran. Nematology, 5, 453–461.
Flegg, J. J. M. (1967). Extraction of Xiphinema and Longidorus species from soil by a modification of Cobb’s decanting and sieving technique. Annals of Applied Biology, 60, 429–437.
Georgi, L. L. (1988). Transmission of tomato ringspot virus by Xiphinema americanum and X. rivesi from New York apple orchards. Journal of Nematology, 20, 304–308.
Goodey, T. (1936). A new dorylaimid nematode Xiphinema radicicola, n. sp. Journal of Helminthology, 14, 69–72.
Gozel, U., Lamberti, F., Duncan, L., Agostinelli, A., Rosso, L., Nguyen, K., et al. (2006). Molecular and morphological consilience in the characterisation and delimitation of five nematode species from Florida belonging to the Xiphinema americanum-group. Nematology, 8, 521–532.
Gutiérrez-Gutiérrez, C., Palomares-Rius, J. E., Cantalapiedra-Navarrete, C., Landa, B. B., Esmenjaud, D., & Castillo, P. (2010). Molecular analysis and comparative morphology to resolve a complex of cryptic Xiphinema species. Zoologica Scripta, 39, 483–498.
Gutiérrez-Gutiérrez, C., Palomares-Rius, J. E., Cantalapiedra-Navarrete, C., Landa, B. B., & Castillo, P. (2011). Prevalence, polyphasic identification, and molecular phylogeny of dagger and needle nematodes infesting vineyards in southern Spain. European Journal of Plant Pathology, 129, 427–453.
Gutiérrez-Gutiérrez, C., Castillo, P., Cantalapiedra-Navarrete, C., Landa, B. B., Derycke, S., & Palomares-Rius, J. E. (2011). Genetic structure of Xiphinema pachtaicum and X. index populations based on mitochondrial DNA variation. Phytopathology, 101, 1168–1175.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98.
He, Y., Subbotin, S., Rubtsova, T. V., Lamberti, F., Brown, D. J. F., & Moens, M. (2005). A molecular phylogenetic approach to Longidoridae (Nematoda: Dorylaimida). Nematology, 7, 111–124.
Heyns, J., & Coomans, A. (1994). Four species of the Xiphinema americanum group (Nematoda: Longidoridae) from Islands in the Western Indian Ocean. Nematologica, 40, 12–24.
Huelsenbeck, J. P., & Ronquist, F. (2001). MrBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17, 754–755.
Jairajpuri, M. S., & Ahmad, W. (1992). Dorylaimida. Freeliving, predaceous and plant-parasitic nematodes. New Delhi: Oxford & IBH Publishing Co. 458 pp.
Kirjanova, E.S. (1951). Soil nematodes in cotton fields and in virgin soil of Golodnaya Steppe (Uzbekistan). [In Russian]. Trudy Zoologicheskogo Instituta Akademiya Nauk SSSR, 9, 625–657.
Kumari, S., Decraemer, W., De Luca, F., & Tiefenbrunner, W. (2010). Cytochrome c oxidase subunit 1 analysis of Xiphinema diversicaudatum, X. pachtaicum, X. simile and X. vuittenezi (Nematoda, Dorylaimida). European Journal of Plant Pathology, 127, 493–499.
Lamberti, F., & Bleve-Zacheo, T. (1979). Studies on Xiphinema americanum sensu lato with descriptions of fifteen new species (Nematoda, Longidoridae). Nematologia Mediterranea, 7, 51–106.
Lamberti, F., & Golden, A. M. (1986). On the identity of Xiphinema americanum sensu lato in the nematode collection of Gerald Thorne with description of X. thornei sp.n. Nematologia mediterranea, 14, 163–171.
Lamberti, F., & Ciancio, A. (1993). Diversity of Xiphinema americanum-group species and hierarchical cluster analysis of morphometrics. Journal of Nematology, 25, 332–343.
Lamberti, F., Choleva, B., & Agostinelli, A. (1983). Longidoridae from Bulgaria (Nematoda, Dorylaimida) with description of three new species of Longidorus and two new species of Xiphinema. Nematologia Mediterranea, 11, 49–72.
Lamberti, F., Ciancio, A., Agostinelli, A., & Coiro, M. I. (1991). Relationship between Xiphinema brevicolle and X. diffusum with a redescription of X. brevicolle and descriptions of three new species of Xiphinema (Nematoda, Dorylaimida). Nematologia mediterranea, 19, 311–326.
Lamberti, F., Lemos, R. M., Agostinelli, A., & D’Addabo, T. (1993). The Xiphinema americanumgroup in the vineyards of the Dão and Douro regions (Portugal) with description of two new species (Nematoda, Dorylaimida). Nematologia Mediterranea, 21, 215–225.
Lamberti, F., Bravo, M. A., Agostinelli, A., & Lemos, R. M. (1994). The Xiphinema americanum-group in Portugal with descriptions of four new species (Nematoda, Dorylaimida). Nematologia Mediterranea, 22, 189–218.
Lamberti, F., Agostinelli, A., & Radicci, V. (1996). Longidorid nematodes from Northern Egypt. Nematologia Mediterranea, 24, 307–339.
Lamberti, F., Molinari, S., Moens, M., & Brown, D. J. F. (2000). The Xiphinema americanum group. I. Putative species, their geographical occurrence and distribution, and regional polytomous identification keys for the group. Russian Journal of Nematology, 8, 65–84.
Lamberti, F., Kunz, P., Grunder, J., Molinari, S., De Luca, F., Agostinelli, A., & Radicci, V. (2001). Molecular characterization of six Longidorus species from switzerland with the description of Longidorus helveticus sp. n. (Nematoda, Dorylaimida). Nematologia Mediterranea, 29, 181–205.
Lazarova, S. S., Malloch, G., Oliveira, C. M. G., Hübschen, J., & Neilson, R. (2006). Ribosomal and mitochondrial DNA analyses of Xiphinema americanum-group populations. Journal of Nematology, 38, 404–410.
Loof, P. A. A., & Luc, M. (1990). A revised polytomous key for the identification of species of the genus Xiphinema Cobb, 1913 (Nematoda: Longidoridae) with exclusion of the X. americanum-group. Systematic Parasitology, 16, 36–66.
Loof, P. A. A., Luc, M., & Baujard, P. (1996). A revised polytomous key for the identification of species of the genus Xiphinema Cobb, 1913 (Nematoda: Longidoridae) with exclusion of the X. americanum-group: Supplement 2. Systematic Parasitology, 33, 23–29.
Lordello, L. G. E., & Costa, C. P. (1961). A new nematode parasite of coffee roots in Brazil. Revista Brasileira de Biologia, 21, 363–366.
Luc, M., Coomans, A., Loof, P. A. A., & Baujard, P. (1998). The Xiphinema americanum group (Nematoda: Longidoridae). 2. Observations on Xiphinema brevicollum Lordello & da Costa, 1961 and comments on the group. Fundamental and Applied Nematology, 21, 475–490.
Mekete, T., Gray, M. E., & Niblack, T. L. (2009). Distribution, morphological description, and molecular characterization of Xiphinema and Longidorous spp. associated with plants (Miscanthus spp. and Panicum virgatum) used for biofuels. Global Change Biology Bioenergy, 1, 257–266.
Meza, P., Aballay, E., & Hinrichsen, P. (2011). Molecular and morphological characterisation of species within the Xiphinema americanum-group (Dorylaimida: Longidoridae) from the central valley of Chile. Nematology, 13, 295–306.
Monteiro, A. R. (2010). Nomenclatorial note on Xiphinema brevicolle Lordello & Costa, 1961 (Nematoda: Longidoridae). Nematologia Brasileira, 34, 1–2.
Nasira, K., & Maqbool, M. A. (1994). Occurrence of virus vector nematodes in Pakistan. Pakistan Journal of Nematology, 12, 79–85.
Oliveira, C. M., Hubschen, J., Brown, D. J., Ferraz, L. C. C., Wright, F., & Neilson, R. (2004). Phylogenetic relationships among Xiphinema and Xiphidorus nematode species from Brazil inferred from 18S rDNA sequences. Journal of Nematology, 36, 153–159.
Oliveira, C. M. G., Fenton, B., Malloch, G., Brown, D. J. F., & Neilson, R. (2005). Development of species-specific primers for the ectoparasitic nematodes species X. brevicolle, X. diffusum, X. elongatum, X. ifacolum and X. longicaudatum (Nematoda: Longidoridae) based on ribosomal DNA sequences. Annals of Applied Biology, 146, 281–288.
Oliveira, C. M. G., Ferraz, L. C. C. B., & Neilson, R. (2006). Xiphinema krugi, species complex or complex of cryptic species? Journal of Nematology, 38, 418–428.
Page, R. D. (1996). TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences, 12, 357–358.
Palomares-Rius, J. E., Subbotin, S. A., Landa, B. B., Vovlas, N., & Castillo, P. (2008). Description and molecular characterisation of Paralongidorus litoralis sp. n. and P. paramaximus Heyns, 1965 (Nematoda: Longidoridae) from Spain. Nematology, 10, 87–101.
Palomares-Rius, J. E., Subbotin, S. A., Liébanas, G., Landa, B. B., & Castillo, P. (2009). Eutylenchus excretorius Ebsary Eveleigh, 1981 (Nematoda: Tylodorinae) from Spain with approaches to molecular phylogeny of related genera. Nematology, 11, 343–354.
Peña-Santiago, R., Abolafia, J., Guerrero, P., Liébanas, G., & Peralta, M. (2006). Soil and freshwater nematodes of the Iberian fauna: a synthesis. Graellsia, 62, 179–98.
Posada, D. (2008). jModelTest: phylogenetic model averaging. Molecular Biology and Evolution, 25, 1253–1256.
Robbins, R. T. (1993). Distribution of Xiphinema americanum and related species in North America. Journal of Nematology, 25, 344–348.
Robbins, R. T., & Brown, D. J. F. (1991). Comments on the taxonomy, occurrence and distribution of Longidoridae (Nematoda) in North America. Nematologica, 37, 395–419.
Romanenko, N. D. (1981). Discovery of a new species of nematode Xiphinema paramonovi n. sp. (Nematoda: Longidoridae) in the territory of the Soviet Union. [1st Conference (9th meeting) on nematodes of plants and insects in soil and water] Tashkent, September 16th-18th, (pp. 68–69).
Sakai, H., Takeda, A., & Mizukubo, T. (2011). First report of Xiphinema brevicolle Lordello et Costa, 1961 (Nematoda, Longidoridae) in Japan. ZooKeys, 135, 21–40.
Seinhorst, J. W. (1966). Killing nematodes for taxonomic study with hot f.a. 4:1. Nematologica, 12, 178.
Sharma, S. B., McKirdy, S., Mackie, A., & Lamberti, F. (2003). First record of Xiphinema rivesi associated with grape vines in Western Australia. Nematologia Mediterranea, 31, 87.
Siddiqi, M. R. (1961). On Xiphinema opisthohysterum, n. sp., and X. pratense Loos, 1949, two Dorylaimid nematodes attacking fruit trees in India. Zeitschrift für Parasitenkunde, 20, 457-465.
Širca, S., Stare, B. G., Plesko, I. M., Marn, M. V., Urek, G., & Javornik, B. (2007). Xiphinema rivesi from Slovenia transmit Tobacco ringspot virus and Tomato ringspot virus to cucumber bait plants. Plant Disease, 91, 770–770.
Sturhan, D. (1983). Description of two new Xiphinema species from Portugal, with notes on X. pachtaicum and X. opisthohysterum (Nematoda, Longidoridae). Nematologica, 29, 270–283.
Swofford, D. L. (2003). PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4.0b 10. Sunderland: Sinauer Associates.
Tarjan, A. C. (1969). Variation within the Xiphinema americanum group (Nematoda: Longidoridae). Nematologica, 15, 241–252.
Taylor, C. E., & Brown, D. J. F. (1997). Nematode vectors of plant viruses. Wallingford: CABI. 296 pp.
Téliz, D., Landa, B. B., Rapoport, H. F., Pérez Camacho, F., Jiménez-Díaz, R. M., & Castillo, P. (2007). Plant-parasitic nematodes infecting grapevine in southern Spain and susceptible reaction to root-knot nematodes of rootstocks reported as moderately resistant. Plant Disease, 91, 1147–1154.
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Research, 22, 4673–4680.
Tulaganov, A. T. (1938). The fauna of nematodes of cotton and surrounding soil in Katta-Kurgan distric of Uzbek SSR. Uzbekskogo Gosundarstvennogo Universiteta, 12, 1–25.
Urek, G., Širca, S., & Karssen, G. (2005). Morphometrics of Xiphinema rivesi Dalmasso, 1969 (Nematoda: Dorylaimida) from Slovenia. Russian Journal of Nematology, 13, 13–17.
Vrain, T. C. (1993). Restriction fragment length polymorphism separates species of the Xiphinema americanum group. Journal of Nematology, 25, 361.364.
Wang, X., Bosselut, N., Castagnone, C., Voisin, R., Abad, P., & Esmenjaud, D. (2003). Multiplex polymerase chain reaction identification of single individuals of the Longidorid nematodes Xiphinema index, X. diversicaudatum, X. vuittenezi, and X. italiae using specific primers from ribosomal genes. Phytopathology, 93, 160–166.
Wang, S., & Wu, X. (1992). Two species of Xiphinema Cobb, 1913 (Dorylaimida: Longidoridae) from around grape roots in China. Acta Phytopathologica Sinica, 22, 117–123.
Wu, Y., Zheng, J., & Robbins, R. T. (2007). Molecular characterization of a Xiphinema hunaniense population with morphometric data of all four juvenile stages. Journal of Nematology, 39, 37–42.
Ye, W., Szalanski, A. L., & Robbins, R. T. (2004). Phylogenetic relationships and genetic variation in Longidorus and Xiphinema species (Nematoda: Longidoridae) using ITS1 sequences of nuclear ribosomal DNA. Journal of Nematology, 36, 14–19.
Acknowledgements
This research was supported by grant AGL2009-06955 from ‘Ministerio de Ciencia e Innovación’ of Spain, grant AGR-136 from ‘Consejería de Economía, Innvovación y Ciencia’ from Junta de Andalucía, and the European Social Fund. The authors thank J. Martín Barbarroja and G. León Ropero from IAS-CSIC for the excellent technical assistance, and B.B. Landa and J.A. Navas-Cortés (IAS-CSIC) for their critically reading of the manuscript prior to submission.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gutiérrez-Gutiérrez, C., Cantalapiedra-Navarrete, C., Decraemer, W. et al. Phylogeny, diversity, and species delimitation in some species of the Xiphinema americanum-group complex (Nematoda: Longidoridae), as inferred from nuclear and mitochondrial DNA sequences and morphology. Eur J Plant Pathol 134, 561–597 (2012). https://doi.org/10.1007/s10658-012-0039-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-012-0039-9