Abstract
Apple replant disease (ARD) in production nurseries can negatively impact commercial viability by diminishing tree quality and potentially serving as a source of pathogen inoculum. The current study was carried out to determine the potential for plant genotype to influence anaerobic soil disinfestation (ASD) disease control efficacy. M.9, G.41, and G.935 apple rootstock genotypes were employed. ASD was conducted using orchard grass as the carbon input (10 t ha−1 or 20 t ha −1). Rootstock growth in ASD-treated soils was comparable to that attained in response to soil pasteurization or fumigation with 1,3-dichloropropene/chloropicrin (FUM) in both greenhouse (GT) and nursery field trials (NFT). In GT, Rhizoctonia solani AG-5 root infection and growth performance varied with rootstock genotype and soil treatment. ASD reduced pathogen DNA quantity in roots and improved rootstock growth. Genotype, but not soil treatment, influenced root infestation by Pythium ultimum and R. solani in the NFT. ASD with grass input at 20 t ha −1 improved soil nutrient levels, especially NO3− N, and provided significant weed control in the NFT. Treatments significantly altered composition of the bulk soil and rhizosphere microbiome in GT and these effects were prolonged in ASD-treated soils. In NFT, ASD conducted with orchard grass was uniformly as effective as FUM in the control of ARD and increase in trunk diameter increment, the primary determinant of apple rootstock value. This ASD treatment can be suggested as a potential method for effective control of nursery replant disease across rootstock genotypes varying in disease tolerance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Fruit tree crops such as apple, pear, and cherry are propagated from foundation plants in nurseries and subsequently transplanted to orchards. The process of generating a marketable fruit tree generally involves a three-year-long process that includes growing the rootstock, grafting the scion, and producing the finished tree. Apple scion varieties are bred for characteristics related to consumer preference as well as biotic disease resistance of the fruit and aerial parts of the tree (Harshman and Evans 2015; Igarashi et al. 2016). In contrast, apple rootstocks historically have been bred for features such as dwarfing, tree architecture, precocity, and yield (Fazio and Mazzola 2004). More recently, interest in breeding programs has turned to address biotic disease resistance with an initial emphasis on rootstock resistance to fire blight (Norelli et al. 2003). Although the response of apple rootstock germplasm to certain soil-borne pathogens was intermittently examined, these studies did not emanate from an intentional breeding program, but rather were an opportune assessment of available plant materials. Marker-assisted selection has been utilized of late in the Geneva, New York, USA breeding program to develop new apple rootstocks with a focus on several traditional horticultural traits (e.g. dwarfing), but also directed towards identifying resistance to soil-borne diseases including apple replant disease (ARD; Fazio and Mazzola 2004). Traditional approaches to rootstock breeding and selection have involved a long-term process requiring approximately 30 years to generate a new commercial rootstock genotype (Johnson 2000). With the current ability to impose genetic selection methods on rootstock breeding populations, the time span required to attain such a goal has been significantly reduced.
Apple replant disease is a significant problem in apple-growing areas worldwide resulting from intensive cropping practices. The disease is incited by a biological complex consisting of plant pathogenic fungi and oomycetes, and plant parasitic nematodes (Mazzola and Manici 2012). In Washington State the ARD pathogen complex consists of Rhizoctonia solani anastomosis group (AG)-5, AG-6, binucleate Rhizoctonia sp. AG-G, Ilyonectria spp., Phytophthora spp., Pythium spp. and Pratylenchus penetrans (Mazzola 1998). ARD affects apple crop bearing trees when new orchards are planted on sites previously planted to apple and a similar phenomenon can negatively influence nursery production when soils are repeatedly cropped to rootstocks and nursery stock in the absence of pre-plant disease control measures. While ARD reduces nursery tree quality, trees derived from these sites may also act as a source of pathogen inoculum when planted into commercial orchard soils (Gergerich et al. 2015; Moein et al. 2019). Rotation of the nursery operation to sites not previously planted to fruit trees, and pre-plant soil fumigation are the primary measures employed to manage ARD in nursery crop systems (Ramos 1998). However, new holistic approaches to manage the disease are being investigated (Winkelmann et al. 2019). Soil fumigation is commonly effective in reducing P. penetrans populations during the first orchard growing season; however, re-infestation by this and other pathogens that incite replant disease is common by the end of the second growing season (Mazzola et al. 2015). The failure of soil fumigation to provide extended exclusion of these pathogens from the system is of concern due to the small root systems of this multi-year crop and the potential for movement of associated pathogens from the nursery to the production orchard site. Furthermore, effective fumigation of rootstock stool-beds after establishment is not practical. The lack of post-plant control measures allows for accumulation of potential root pathogens in the stool bed and infection of rooted cuttings by ARD pathogens such as Phytophthora spp. (Tidball 1990).
Commercial apple rootstocks possess varying levels of susceptibility or tolerance to ARD (Deakin et al. 2019; Reim et al. 2019; Robinson et al. 2014). In general, field-based assessment of disease tolerance has relied on growth characteristics without examination of rootstock susceptibility to the causal pathogen complex. Thus, minimal information exists regarding association of the observed plant growth response and relative susceptibility or resistance to the disease causal agents. An exception concerns rootstock susceptibility to infestation by P. penetrans with several studies demonstrating that certain Geneva series rootstocks support lower populations of this nematode relative to the Malling series rootstocks in a manner that corresponded with plant growth (Isutsa and Merwin 2000; Mazzola et al. 2009). Recent studies indicate that apple rootstock germplasm derived from the Geneva breeding program may possess functional resistance to the replant pathogen Pythium ultimum (Shin et al. 2016; Zhu et al. 2016). More broadly, the relative tolerance or resistance of apple to the replant pathogen complex may be a function of the differential composition of the rhizosphere microbiome supported by individual rootstock genotypes (Rumberger et al. 2007). Root exudate metabolite composition may drive selection or recruitment of a unique microbiome that confers tolerance to ARD (Leisso et al. 2017).
Relative to soil fumigation with chemistries that demonstrate replant disease control efficacy, such as 1,3-dichloropropene/chloropicrin, alternative biologically-based control measures have provided prolonged soil-borne pathogen suppression over the 2–3 year nursery production period (Mazzola et al. 2015). Anaerobic soil disinfestation (ASD) has been employed to control soil-borne diseases of annual crops as well as perennial crops such as Norway maple, Southern catalpa (Goud et al. 2004), and walnut (Strauss et al. 2017). In the ASD process, carbon source incorporated into soil induces proliferation of aerobic microorganisms which leads to increased aerobic respiration and consumption of oxygen in soil atmosphere. Inundation and tarping prevent reintroduction of oxygen into soil thereby creating an anoxic environment under which multiple modes of disease suppression take place.
Efficacy of ASD will be dependent, in part, on selection of the appropriate carbon source which influences functional changes in the soil microbiome and corresponding spectrum of biologically active metabolites that are produced (Hewavitharana and Mazzola 2016). Additional factors influencing the carbon source used in ASD include availability, cost and ease of application. In a tree fruit nursery setting in the Northwest United States, trees are established in a weed-free strip while maintaining grass between the tree rows. Hence, mowing grass rows is a routine practice in orchards as well as in nurseries and residues from mowing operations may provide a readily available on-site source of carbon that could be used to conduct ASD.
Optimization of carbon input rate is another important consideration in developing an ASD protocol due to labor costs associated with application and availability of the carbon substrate. In growth chamber trials employing apple seedlings, grass input rate was shown to influence efficacy of ASD for control of P. penetrans but did not affect control of R. solani AG-5 (Hewavitharana and Mazzola 2016). Additional greenhouse and nursery field trial experiments using commercially available apple rootstocks are necessary to confirm the results of these initial studies conducted using apple seedlings. Specifically, assessment of tree caliper (trunk diameter) can be instrumental in determining the value of ASD for control of soil-borne disease in tree fruit nurseries as this is the primary metric used to determine commercial rootstock value.
Although ASD implementation variables and modes of action have been investigated, interaction between ASD and plant genotype in determining the relative efficacy of disease suppression has not been examined extensively. The objectives of this study were to: i) evaluate the efficacy of ASD carried out utilizing orchard grass relative to pre-plant soil fumigation or soil pasteurization for control of nursery replant disease and weed control; ii) assess the rate of grass application used in ASD on disease control attained using commercial rootstocks; and iii) assess whether there is an interaction between rootstock genotype and ASD treatment in determining relative improvement of plant growth in the replant nursery environment.
Materials and methods
Assessment of ASD/rootstock genotype interactions for control of Rhizoctonia solani AG-5 in greenhouse trials
Rhizoctonia solani AG-5 (isolate 5–103) inoculum was prepared according to previously described methods (Mazzola 1997; Hewavitharana and Mazzola 2016). Soils used in these trials were obtained from the USDA, ARS/Washington State University Columbia View Research and Demonstration (CV) orchard (Orondo, WA; latitude 47.56235 N, longitude 120.24499 W). The dominant soil type of this site is a silt loam (61% silt, 30% sand, 9% clay; Soiltest Farm Consultants, Moses Lake, WA). Replant disease potential at this site was mild to moderate (Mazzola et al. 2001) and components of the pathogen complex include Rhizoctonia solani AG-5 and AG-6, Pythium ultimum, P. sylvaticum, P. irregulare, P. heterothallicum, Phytophthora cactorum, Ph. cambivora and Ilyonectria spp. (Mazzola and Brown 2010). Soils were collected from the root zone (Mazzola et al. 2015) of established cv. Gala grafted on M.26 trees in spring 2015 for the first trial and spring of 2016 for the second trial, respectively. Soil from this site was used as the ARD components resident to this soil were previously characterized. Soil was transported in 19-L closed containers to the USDA-ARS Tree Fruit Research Laboratory and soils collected from different trees were homogenized in a cement mixer (Kobalt, Mooresville, NC). Large roots, stones, and debris were removed by hand.
A 2.5 kg orchard soil sample was measured out into each of 60, 3.8-L plastic pots. Gravimetric moisture content was approximately 10%. Even though, R. solani AG-5 was one of the previously reported ARD components in the soil, in order to elucidate treatment effects clearly, soil was infested with ground R. solani AG-5 oat grain inoculum (0.5% w/w) and homogeneously mixed into the soil of each pot by hand. The pots were loosely covered with Saranex bags (0.41 m × 0.41 m, Bitran Series “S” bags, Com-Pac International Carbondale, IL) to prevent moisture loss, arranged in randomized complete block design with five biological replicates, and incubated for 1 week in the greenhouse at day and night temperature of 20/15 °C. Treatments applied to the infested soils were ASD with orchard grass (GR) at 10 t ha−1 (7.3 g kg−1 soil; ASD-GR10), 20 t ha−1 (14.6 g kg−1 soil; ASD-GR20), pasteurized control (PC), and no-treatment control (NTC). The orchard grass (Dactylis glomerata) was mowed in autumn 2014 at the CV orchard, air dried, cut into approximately 1 cm length pieces, and incorporated into soil at the rate noted above. Orchard grass applied in the greenhouse trial and in the nursery field trial possessed a C:N ratio of 19:1, pH = 6.3, total C, N, P, K, and S at 42.3, 2.19, 0.24, 1.73, and 0.18%, respectively (Soiltest Farm Consultants, Inc., Moses Lake, WA). After incorporation of grass residues, 600 ml water was added to each ASD treatment pot to attain soil moisture at field capacity after drainage. Pots were placed within two Saranex bags to create a double sealed layer and incubated at 24 °C for 2 weeks. Immediately after completion of the anaerobic phase, headspace O2 and CO2 percentage in the sealed bags was analyzed using a handheld gas analyzer (Dansensor A/S, Ringsted, Denmark). ASD treated soils were aerated for 3 weeks. A phytotoxicity assay was conducted using cress seeds (Lepidium sativum; Aslam and VanderGheynst 2008) to determine sufficiency of the aeration period. The assay demonstrated no decrease in germination in the ASD treatments indicating that the three-week soil aeration period was sufficient to preclude potential phytotoxicity resulting from the ASD treatment. PC treatment was applied by pasteurizing artificially infested soil at 80 °C for 30 min two times with a 2-day interval between pasteurization events. Soil moistened to its field capacity was pasteurized in plastic bags laid out on selves of an oven and distributed evenly to attain a soil depth of 2.5 cm. Upon completion of each treatment, soil samples were obtained for determination of pH and microbiome characterization as described below. The experiment was conducted in 2015 and repeated the following year using soils collected from the same site in the spring 2016.
Characterization of rhizosphere microbial communities in greenhouse trials
In order to assess the effect of soil treatment on rhizosphere bacterial and fungal community composition, DNA samples were used in terminal restriction fragment length polymorphism (T-RFLP) analysis. DNA was extracted using the PowerSoil DNA isolation kit (MO BIO Laboratories Inc.) from two independent 0.25-g rhizosphere soil samples obtained (Mazzola et al. 2015) from each tree at harvest. Fluorescently labeled PCR products of the fungal ITS region were generated using the D4 labeled ITS-1F primer in conjunction with D3 labeled ITS4 primer. D4 labeled 8F and D3 labeled 907R primers were used in amplification of the bacterial 16S rRNA gene. Fungal amplicons were double digested using HaeIII and HhaI, and bacterial amplicons were digested using HaeIII in reaction mixtures containing CutSmart buffer (Biolabs Inc., New England) and nuclease-free water (Ambion®, Life Technologies, Carlsbad, CA). Separation of fragments using the CEQ 8000 Genetic Analysis System (Beckman-Coulter, Brea, CA) was conducted as previously described (Weerakoon et al. 2012).
Assessment of treatment effects on disease suppression and rootstock growth in greenhouse trials
Rootstocks have been classified into resistance categories with relation to ARD tolerance/susceptibility (Auvil et al. 2011). Rootstock genotypes M.9 (susceptible), G.41 (moderately tolerant), and G.935 (tolerant) with a 3/8″ trunk diameter were used in both field and greenhouse trials. In the greenhouse trial, one rootstock was planted per pot and grown for 3.5 months at 24 °C with ambient light supplemented using LED lights (LumiGrow PRO 325, Emeryville, CA) with a 12-h photoperiod. Photosynthetically active radiation (PAR, 400–700 nm) at the level of the plant canopy was 197 μE m−1 s−2. Plants were watered with 250 ml every other day and fertilized with Hoagland’s complete nutrient solution at 20 ml per month during the first month and 40 ml per month for the remainder of the growing period (Hoagland and Aron 1938). Root weight, root volume (Harrington et al. 1994), leader branch length, and trunk diameter were determined at harvest (Online Resource 1). Trunk diameter increment was calculated by deducting the trunk diameter at harvest from the initial trunk diameter. For assessment of R. solani AG-5 root infection, soil was removed from roots by rinsing under tap water prior to excising a representative root sample from each rootstock. Rhizoctonia solani AG-5 infection was determined by isolation from root fragments (Hewavitharana and Mazzola 2016) and real time quantitative PCR (qPCR) as previously described (Mazzola and Zhao 2010). DNA used in qPCR was extracted in duplicate from 50 mg root samples for each plant using PowerPlant Pro® plant DNA isolation kit (MO BIO Inc., Carlsbad, CA).
Nursery field trial
A field trial was conducted at the CV orchard to evaluate the efficacy of ASD for control of nursery replant disease. The site cropping history included a 27-year-old block of cv. Red Delicious on seedling rootstock which was removed in 1997, replanted in 2000 to cv. Gala on M.26 rootstock with spacing of 1 m between trees and 4.6 m between rows. Tree removal was conducted in 2008 and the site remained fallow thereafter, with chemical weed control of the previous orchard tree rows until the current trial was established. The site was chosen based on history of apple planting at the site and availability. The experiment was conducted using a modified split-block design with soil treatments as the whole plots and rootstocks as subplots arranged in six blocks. Subplots are described in detail under ‘site preparation and planting’ below. Soil treatments were pre-plant soil fumigation (FUM), ASD-GR20, and NTC, with six replicates per soil treatment. Eighteen individual tree row nursery field plots each with dimension of 1.5 m wide (centered over previous tree rows) and 9.1 m in length were rotovated and ripped using a single tine chisel plough on 3 September 2014. The buffer grass row width between two tree rows was 4.5 m and buffer width between adjacent treatments in the same row was 2 m. Six randomly selected whole-plots were fumigated at 30.5 cm depth according to industry standards by a commercial applicator using 1,3-dichloropropene/chloropicrin (Telone® C35; Dow AgroSciences, IN) at a rate of 0.02526 L m−2 (252.6 L ha−1) on 4 September 2014. ASD-GR20 was applied to an additional set of six whole plots. Orchard grass was mowed from the alley way between rows at the same nursery premises, air dried and incorporated into soil at a rate of 20 t ha−1 by rotovating to a depth of 15–20 cm on 17 September 2014. All the plots received 8 cm of water overnight (17 h) using a sprinkler irrigation system. ASD plots were covered with a clear plastic tarp (FIN, SNX, 3 mm, Filcon Inc., Clare, MI) that was tightly secured by burying the plastic edges under the soil. ASD treated soils were incubated for 19 days prior to tarp removal. The remaining six whole plots were not treated. Soil samples were obtained from each plot on 6 October 2014. Post-treatment soil sampling was conducted using a ‘Z’ pattern across an individual whole plot with a 2.54 cm diameter probe used to collect soil at a depth of 2–20 cm with 10 soil cores per plot. Soil cores from each individual plot were combined and stored at 4 °C. For nutrient analysis (Cascade Analytical Inc. Wenatchee, WA), a pooled soil sample from each treatment was prepared by mixing sub-samples from each of the six whole plot soil samples. Determination of total metals and trace elements was conducted by using Environmental Protection Agency (EPA)‘s method 200.7 (https://www.epa.gov/esam/method-2007-determination-metals-and-trace-elements-water-and-wastes-inductively-coupled-plasma; Cascade Analytical Inc.). The independent soil samples from each whole plot were used for soil microbiome characterization using T-RFLP analysis and soil pH analysis (Burt 1996) as described above. Soil temperature data were obtained from the AgWeatherNet (weather.wsu.edu), Washington State University’s automated weather station at the CV orchard during soil treatment.
Assessment of weed suppression in nursery field trial
Prior to soil tilling and planting of apple rootstocks, a weed survey and an assessment of weed biomass were conducted on 26 March 2015 at the nursery field trial. A 50 cm × 50 cm quadrant was randomly placed at three points in each plot and all weed shoot biomass was excised and wet aerial weed biomass was determined. Weed identification was conducted according to Gaines and Swan (1972) based primarily on flower and seed characteristics and secondarily on vegetative characteristics.
Site preparation and planting in nursery field trial
On 21 April 2015, the nursery field site was irrigated for 3 h using an impact sprinkler irrigation system. Soils in each plot were subsequently tilled in the plot sequence of FUM, ASD-GR20, and NTC to prevent cross contamination. The same rootstock genotypes with the same trunk diameter grade planted in the greenhouse trial were used in this nursery field study. Five rootstocks from each genotype were planted in a random order in two rows per subplot (one row with seven rootstocks and the adjacent row with eight rootstocks) with 0.3 m between trees and 0.76 m between rows in each plot on 22 April 2015. Plots were irrigated throughout the growing season for 3 h, three times weekly using impact sprinklers. Rootstocks were not fertilized based on soil analysis and nursery recommendation.
Growth measurement, soil sampling, and harvest in nursery field trial
Tree trunk diameter was measured monthly during the growing season and at harvest at a pre-determined marked point 5 cm above the soil line. Average growth rate of the rootstocks was calculated per month. On 3 September 2015, an approximately 100 g root zone soil sample (3-month post-planting root zone soil samples) was obtained from each rootstock and stored at 4 °C until use for DNA extraction described below. A separate bulk soil sample was collected to a 15 cm depth from each plot using a ‘Z’ sampling pattern on 21 February 2016 and used for nutrient analysis. Rootstocks were harvested 21–27 June 2016 (Online Resource 2), the root systems were excised from each plant, and roots were transported to USDA ARS Tree Fruit Research Laboratory, Wenatchee, WA. A rhizosphere soil sample was collected from each plant root (at harvest rhizosphere soil samples) by shaking off soil that was loosely adhered to roots and collecting soil adhered firmly to the fine roots with a sterile spatula (Online Resource 2). A representative sub sample of fine roots was collected from each plant at harvest for use in DNA extraction. Root and soil samples were stored at −80 °C until use.
Characterization of nursery field trial soil microbial communities
DNA was extracted from bulk soil samples collected immediately post-treatment, root zone soil samples collected 3 months post-planting, and rhizosphere soil obtained at harvest. DNA was extracted from 5 g of the former two soil samples using PowerMax soil DNA isolation kit (MO BIO Laboratories Inc.) while DNA was extracted from 0.25 g of the latter sample using the PowerSoil DNA isolation kit (MO BIO Laboratories Inc.). DNA extracts were used in conduct of T-RFLP analysis as described previously to assess the effect of soil treatment on composition of the bacterial and fungal communities.
Effect of soil treatment on R. solani and P. ultimum root infection in nursery field trial
For the nursery field study, presence of R. solani and P. ultimum in fine root tissue of apple rootstocks was assessed by qPCR as described below. DNA was extracted from 50 mg fine roots of the rootstocks from each of two randomly selected rootstocks at harvest per genotype per soil treatment for each block using Powerplant Pro® plant DNA isolation kit (MO BIO Inc.). qPCR was conducted using the primer pair ULT 1F/ULT 4R (Schroeder et al. 2006) and Rhsp1/ITS4B (Bruns et al. 1991; Gardes and Bruns 1993) for quantification of P. ultimum and R. solani, respectively. The reaction mixture consisted of 1.0 μl of a 1:100 dilution of root DNA extract, 0.1 μl of each primer (100 pmol μl−1), 3.0 μl SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK), and 5.8 μl nuclease-free water (Ambion®, Life Technologies, Carlsbad, CA). Standard curves were prepared using purified DNA from P. ultimum (isolate 60–1205) and R. solani AG-5 (isolate 5–103) diluted from 0.1 ng μl−1 to 10 fg μl−1. qPCR was conducted using a StepOnePlus Real Time PCR System (Applied Biosystems, Foster City, CA) with three technical replicates consisting of reactions containing identical components and volumes for each root sample and the no-template control. Amplification of P. ultimum was conducted using the following conditions: 95 °C 10 min, (95 °C 15 s − 62 °C 1 min-72 °C 30 s) × 40 cycles, followed by a melt curve with a 0.3 °C s−1 increase in temperature from 60 °C to 95 °C (Wang and Mazzola 2019a). Reaction conditions for R. solani were as follows: 95 °C 10 min, (95 °C 15 s - 59 °C 0.30 min-72 °C 1.30 min) × 40 cycles, with the melt curve generated using the same conditions as described above.
Data analysis
Statistical analyses were completed using SAS 9.4 software (SAS Institute Inc., Cary, NC). Nonmetric multidimensional scaling analysis (NMDS) and one-way analysis of similarity (ANOSIM) using Dice distance metric of bacterial and fungal T-RFLP data were carried out using PAST software package, version 2.14 (Hammer et al. 2001). Quantitative data such as shoot biomass were analyzed by ANOVA using Proc Mixed or Proc GLM procedures and Tukey’s honest significant test for mean separation with appropriate data transformations to satisfy the model assumptions. Soil pH values were converted to [H+] concentration and analyzed as described above. Percentage data were analyzed using logistic procedure (Proc LOGISTIC).
Results
Changes in soil physicochemical parameters in greenhouse and nursery field trials
Headspace gas analysis at termination of the greenhouse ASD incubation period indicated that anaerobic conditions in response to ASD treatment was established irrespective of GR amendment rate as oxygen was recorded at 0% (n = 15) for both ASD-GR10 and ASD-GR20 treatments whereas that of the ambient air was 20.5% (n = 5). However, GR input rate did affect CO2 concentration at termination of ASD treatment with an odds ratio estimate of 0.634 in trial 1 and 0.691 in trial 2 for ASD-GR10 vs ASD-GR20 (P < 0.0001) indicating greater production of CO2 in ASD-GR20 (n = 15).
Soil pH was significantly affected by soil treatment in both greenhouse trials (trial 1: P = 0.0002; trial 2: P < 0.0001). However, the soil treatment effect on pH was inconsistent between the two experiments as ASD significantly lowered pH in trial 2 but not trial 1. In trial 2, NTC had a soil pH (pH = 5.8) that was significantly lower than all other treatments (PC = 5.9, ASD-GR20 = 6.0, and ASD-GR10 = 6.2). In the nursery field study, pH of ASD-GR20 treated soil (6.1) was lower than that of the FUM (6.3) and NTC (6.8) immediately post-treatment. At the end of the first orchard growing season there was no significant difference in soil pH among treatments (Table 1).
At the time of fumigation treatment of nursery field plots, soil temperature was approximately 23 °C at 0.20-m depth. Soil temperature also was 23 °C at initiation of the ASD treatment, but declined to 19 °C at the end of the treatment period. Post-treatment soil nutrient concentrations in the nursery field study were not statistically analyzed as the assessment was conducted on a single pooled sample from among all blocks for each treatment. After one growing season, soil NO3− N, P, K, organic matter percentage (OM), and electric conductivity (EC) were higher in ASD-GR20 compared to FUM and NTC although NH4+ N was lower in ASD compared to FUM and NTC (Table 1). A higher level of NO3− N and lower level of NH4+ N in the ASD-GR20 treatment compared to FUM and NTC could be attributed to the grass amendment possessing a C:N ratio of 19:1. Soil NO3− N, and EC were higher in FUM compared to NTC, but, P, K, and OM were higher in NTC compared to FUM. Soil nutrient analysis of samples collected from each replicated block after one growing season indicated that NO3− N, Fe, and Mn were significantly higher and S was significantly lower in ASD soils compared to FUM or NTC soils. Soil P, K, and OM were higher in ASD compared to FUM but were not significantly different from that of the NTC.
Effect of soil treatments on weed suppression in nursery field trial
Fresh aerial weed biomass in the nursery field plots prior to rootstock planting was significantly affected by soil treatment (P = 0.0084). Mean fresh aerial weed biomass in ASD-GR20, NTC, and FUM treatments were 80.06 g−1 m−2, 289.47 g−1 m−2, 277.36 g−1 m−2 respectively. ASD-GR20 soil treatment significantly reduced weed biomass relative to the NTC. There was no significant difference in weed biomass recovered from FUM and NTC plots. Composition of the weed species population was similar among all treatments. The most abundant species across the treatments included Taraxacum officinale (common dandelion), Sisymbrium altissimum (tall tumblemustard), Poa annua (annual meadow grass), Capsella bursa-pastoris (shepherd’s purse), Lamium amplexicaule (henbit dead-nettle), Digitaria sanguinalis (hairy crabgrass), Holosteum umbellatum (jagged chickweed), Polygonum spp. and Malva neglecta (common mallow). Stellaria media (common chicken weed), and Erodium cicutarium (redstem filaree) were only found in NTC plots while Trifolium pratense (red clover) was only found in the fumigated plots.
Effect of soil treatments on rootstock growth greenhouse trials and nursery field trial
Soil treatment significantly affected rootstock root weight (trial 1: P = 0.0006; trial 2: P = 0.0006), leader branch length (trial 1: P < 0.0001; trial 2: P < 0.0001), and root volume (trial 1: P = 0.0005; trial 2: P = 0.0003) in both greenhouse experiments. ASD treatments resulted in significantly greater root weight, leader branch length, and root volume in both trials (Table 2). Root volume and root weight was greater for ASD-GR20 treatment than ASD-GR10 treatment in one of two trials. Rootstock genotype significantly affected root weight (trial 1: P = 0.0272; trial 2: P < 0.0001) and volume (trial 1: P = 0.0172; trial 2: P = < 0.0001), in a manner corresponding to relative rootstock vigor; G.935 > G.41 > M.9. Leader length was not significantly affected by rootstock genotype (Trial 1: P = 0.8988; Trial 2: P = 0.1371). Trunk diameter increment was significantly affected by soil treatment in both greenhouse trials (Trial 1: P = 0.0003; Trial 2: P = 0.006) and rootstock genotype in trial 2 (P = 0.0005). Trunk diameter increment was largest for rootstocks cultivated in ASD-GR20 and ASD-GR10 treated soils, which were not significantly different from each other in both trials (Fig. 1). Trunk diameter increment did not differ between PC and NTC treatments in either trial.
Effect of soil treatment on apple rootstock growth in greenhouse trials. Soil treatments were anaerobic soil disinfestation with grass as the carbon input applied at 20 t ha−1 (ASD-GR20) or at 10 t ha−1 (ASD-GR10), no treatment control (NTC), and pasteurization (PC). Rootstock genotype effect was not significant, hence trunk diameter values across different treatments were averaged. Values represent mean trunk diameter increment with n = 15 and error bars represent standard error of mean. Means designated with the same letter are not significantly different (P > 0.05) as determined by Tukey’s honest significant test
The rate of increase in trunk caliper for all rootstocks at the end of two field seasons (14 months) was significantly greater in response to ASD compared to NTC, while FUM soil treatment improved growth of G.935 (P < 0.0001) and M.9 (P < 0.0001), but not G.41 (P = 0.162), relative to NTC (Fig. 2). Growth rate of all rootstock genotypes was comparable between ASD and FUM treatments.
Effect of soil treatment on apple rootstock trunk diameter growth rate of (a) G935, (b) G41, and (c) M9 rootstocks in a nursery field trial conducted at the Columbia View Research and Demonstration orchard (Orondo, WA; latitude 47.56235 N, longitude 120.24499 W). Soil treatments included anaerobic soil disinfestation with grass as the carbon input applied at 20 t ha−1 (ASD), no treatment control (NTC), and pre-plant soil fumigation with 1,3-dichloropropene/chloropicrin (Telone® C35; FUM). Apple rootstock genotypes included G.935 (tolerant to apple replant disease [ARD]), G.41 (moderately tolerant to ARD), and M.9 (susceptible to ARD). Values represent growth rate per month with n = 30 and error bars represent standard error of mean. Means designated with the same letter are not significantly different (P > 0.05) as determined by Tukey’s honest significant test
Rhizosphere microbiome composition in greenhouse trials
Rhizosphere bacterial communities in ASD-GR10 and ASD-GR20 treated soils were more similar to each other than to the NTC and PC treatments in the greenhouse study (Fig. 3a). A similar trend was observed with respect to rhizosphere fungi where ASD-GR10 and ASD-GR20 treatments possessed fungal communities that were more similar to each other than to NTC and PC fungal communities when assessed across all rootstock genotypes (Fig. 3b). Based on analysis of similarity (ANOSIM), rootstock genotype did not have a significant effect on rhizosphere bacterial community composition. However, ANOSIM indicated that rootstock genotype significantly affected the rhizosphere fungal community composition for all rootstock comparisons (G.935 vs G.41: P = 0.015; G.935 vs M.9: P = 0.029; G.41 vs M.9: P = 0.0223).
Effect of treatment on rhizosphere soil (a) bacterial communities and (b) fungal communities at harvest in the greenhouse study as assessed by nonmetric multi-dimensional scaling of terminal restriction fragment length polymorphism (T-RFLP)-derived data using the Dice similarity index. A total of 30 rhizosphere soil samples were included in T-RFLP analysis of the bacterial and fungal communities. Symbols signify the following soil treatments: □ = NTC (No treatment control), ■ = ASD-GR10 (ASD using 10 t ha−1 rate grass), ♦ = ASD-GR20 (ASD using 20 t ha−1 grass) and ○ = pasteurized control
Bulk soil microbiome composition in the nursery field trial
ASD-GR20 and NTC bulk soil bacterial communities did not differ significantly (P = 0.0688) immediately post-treatment. Bacterial communities in NTC and ASD-GR20 soils differed significantly from the populations in FUM bulk soil (P = 0.0001). At the same sampling period, fungal communities of ASD-GR20, NTC, and FUM treatments were significantly different in composition from each other (ASD-GR20 vs NTC: P = 0.0058; ASDGR20 vs FUM: P = 0.0002; NTC vs FUM: P = 0.0001).
Soil treatment was also observed to influence composition of rhizosphere microbial communities. While there was an apparent block effect, there were clear distinctions among treatments, particularly in blocks 1, 3 and 6 (Fig. 4 a-f), with composition of the rhizosphere bacterial communities from the ASD-GR20 and FUM treatments differing significantly (P < 0.036) from that of the NTC. When examined across all plots, the rhizosphere bacterial community for the ASD treatment was dissimilar from the FUM (P = 0.0259) and NTC (P = 0.0161) treatments, and the communities from the FUM and NTC treatments differed from each other (P = 0.001).
Effect of treatment on rhizosphere soil (a) block 1, (b) block 2, (c) block 3, (d) block 4, (e) block 5, and (f) block 6 bacterial communities at harvest of the nursery field trial conducted at the Columbia View Research and Demonstration orchard (Orondo, WA; latitude 47.56235 N, longitude 120.24499 W). Figures represent community analysis from a single orchard block as assessed by nonmetric multi-dimensional scaling of terminal restriction fragment length polymorphism (T-RFLP)-derived data using the Dice similarity index. A total of 2 rhizosphere soil samples (from two randomly selected rootstocks of each genotype: G.935, G.41, and M.9) per soil treatment per block in the nursery field trial were used for conduct of T-RFLP analysis. Symbols signify the following soil treatments: □ = NTC (No treatment control), ♦ = ASD-GR20 (ASD using 20 t ha−1 grass) and, ○ = FUM (1,3-dichloropropene/chloropicrin [Telone® C35] fumigation)
NMDS analysis of T-RFLP derived data indicated that rhizosphere fungal community composition was highly dissimilar (P < 0.0081) among soil treatments when assessed across all rootstocks in a specific block with apparent block effect (Fig. 5 a-f). When assessed across all blocks and rootstocks, analysis of similarity indicated that the ASD-GR20 treatment resulted in generation of a rhizosphere fungal community that was unique relative to the community detected in the fumigation (P = 0.003) and NTC (P = 0.0075) treatments. Likewise, composition of the rhizosphere fungal community from rootstocks cultivated in fumigated soil was dissimilar from the NTC (P = 0.006). For both bacterial and fungal communities, a significant effect of rootstock genotype was not observed.
Effect of treatment on rhizosphere soil (a) block 1, (b) block 2, (c) block 3, (d) block 4, (e) block 5, and (f) block 6 fungal communities at harvest of the nursery field trial conducted at the Columbia View Research and Demonstration orchard (Orondo, WA; latitude 47.56235 N, longitude 120.24499 W). Figures represent community analysis from a single orchard block as assessed by nonmetric multi-dimensional scaling of terminal restriction fragment length polymorphism (T-RFLP)-derived data using the Dice similarity index. A total of 2 rhizosphere soil samples (from two randomly selected rootstocks of each genotype: G.935, G.41, and M.9) per soil treatment per block in the nursery field trial were used for conduct of T-RFLP analysis. Symbols signify the following soil treatments: □ = NTC (No treatment control), ♦ = ASD-GR20 (ASD using 20 t ha−1 grass) and, ○ = FUM (1,3-dichloropropene/chloropicrin [Telone® C35] fumigation)
Effect of soil treatment and rootstock genotype on R. solani AG-5 root infection in greenhouse trials
Logistic regression analysis of percent root infection based on isolation frequency of the fungus from apple roots indicated that only soil treatment was a significant factor (Trial 1: P = 0.0048; Trial 2: P = 0.0002) in greenhouse trials. Odds ratio estimates of ASD-GR10 vs PC, ASD-GR20 vs PC and NTC vs PC were 0.407, 0.733 and 1.510, respectively, demonstrating that ASD treatments were more robust in reducing root infection compared to PC treatment and that the NTC had a higher incidence of root infection compared to PC. In trial 2, odds ratio estimate of ASD-GR20 vs NTC was greater than 1 indicating higher level of root infection in ASD-GR20. The level of R. solani AG-5 root infection was very low in trial 2, due to low establishment of the pathogen, which may have masked treatment effects. Average percent root infection in the control treatment was 71.4% in trial 1 but less than 1% in trial 2.
DNA extraction from root samples yielded an average 12.58 ng μl−1 of DNA per sample, with a standard error of 0.86 ng μl−1, for use in pPCR quantitation of pathogen concentration in apple roots. In general, results from real-time qPCR supported the findings obtained by root plating in describing treatment effects on R. solani AG-5 root infection. Soil treatment (P < 0.0001), but not rootstock genotype (P = 0.5646), affected quantity of the pathogen detected in apple roots. There was a significant interaction (P = 0.0369) between the soil treatment and rootstock genotype on the abundance of pathogen DNA detected in roots. ASD-GR20, ASD-GR10, and PC treatments significantly reduced the quantity of R. solani AG-5 DNA detected in roots compared to the NTC (Fig. 6). Within a given soil treatment, there was no significant difference among rootstock genotypes in amount of pathogen DNA detected in roots for ASD-GR20, ASD-GR10 and PC treatments. However, in NTC soils, the amount of pathogen DNA detected in apple roots was significantly lower for G.935 rootstock than M.9.
Effect of soil treatment and apple rootstock genotype on quantity of Rhizoctonia solani AG-5 DNA detected in apple roots in the greenhouse trial. Apple rootstock genotypes included G.935 (tolerant to apple replant disease [ARD]), G.41 (moderately tolerant to ARD), and M.9 (susceptible to ARD). Soil Treatment were ASD-GR20 = anaerobic soil disinfestation (ASD) with grass residues applied at 20 t ha−1; ASD-GR10 = ASD with grass residues applied at 10 t ha−1; NTC = No-treatment control and PC = pasteurized control treatment. Values represent log mean pathogen DNA pg g−1 root with n = 5 and error bars represent standard error of mean. Means designated with the same letter are not significantly different (P > 0.05) as determined by Tukey’s honest significant test
Effect of soil treatment and rootstock genotype on R. solani and P. ultimum root infection in nursery field trial
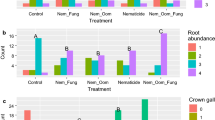
In the nursery field trial, the quantity of P. ultimum and R. solani AG-5 DNA detected in apple roots at the time of plant harvest was significantly affected by rootstock genotype (P. ultimum: P = 0.002; R. solani: P = 0.0273) but not by soil treatment (P. ultimum: P = 0.9456; R. solani: P = 0.2813), and there was no significant interaction between the factors (P. ultimum: P = 0.6997; R. solani: P = 0.2691). Roots of G.41 contained significantly greater amounts of P. ultimum and R. solani DNA compared to M.9 and greater amounts of P. ultimum DNA compared to G.935 (Fig. 7). There was no significant difference between G.935 and M.9 rootstocks in the quantity of DNA for either pathogen detected in roots.
Effect of genotype on quantity of Pythium ultimum and Rhizoctonia solani AG-5 DNA detected in roots of apple rootstocks in the nursery field trial at the Columbia View Research and Demonstration orchard (Orondo, WA; latitude 47.56235 N, longitude 120.24499 W) at the end of the second growing season. Apple rootstock genotypes included G.935 (tolerant to apple replant disease [ARD]), G.41 (moderately tolerant to ARD), and M.9 (susceptible to ARD). Soil treatment was not a significant factor on the quantity of DNA of the pathogens evaluated, hence mean DNA quantities were averaged across different genotypes. Values represent mean pathogen DNA pg g−1 root with n = 36 and error bars represent standard error of mean. Means designated with the same letter are not significantly different (P > 0.05) as determined by Tukey’s honest significant test
Discussion
Apple replant disease constrains successful production of high-quality planting material in apple nurseries. Moreover, effective control of ARD in the nursery can also benefit orchard systems by limiting the potential for transfer of pathogen and parasite inocula to disease free production orchards (Gergerich et al. 2015; Moein et al. 2018). As is the case for soil fumigation (Lembright 1990), numerous environmental variables have potential to modulate the disease control performance of ASD including carbon input type (Hewavitharana and Mazzola 2016), amendment rate (Butler et al. 2014), and soil temperature (Yonemoto et al. 2006). Apple rootstock genotype may interact with soil treatments in a manner that influences resulting disease control and plant growth (Mazzola et al. 2009). Hence, effective use of ASD for control of replant disease will require consideration of these factors in optimization of application protocols.
Carbon source amendment rate can affect the efficacy of ASD in disease suppression as well as plant growth/yield with a general trend of enhanced disease control achieved with higher amendment rates (Shrestha et al. 2016). However, in the current study, GR amendment rates of 10 and 20 t ha−1 resulted in a comparable level of R. solani AG-5 control and plant growth, which confirmed previous findings from studies that employed apple seedlings, rather than rootstocks, in controlled environment assays (Hewavitharana and Mazzola 2016). Reduced carbon input rate will lessen the cost of application as well as reduce residual non-decomposed organic matter at the completion of ASD, which could correspondingly diminish potential saprophytic survival of pathogens when the system is aerated (Hoitink and Boehm 1999; Manici et al. 2004). Pratylenchus penetrans was not identified as a component of the ARD pathogen complex at the orchard site in which the current nursery field trial was performed and ARD is known to be a significant constraint to apple production on sites even where this nematode does not contribute to disease development (Mazzola 1998; Tewoldemedhin et al. 2011). However, previous controlled environment studies indicated that carbon input rate will influence ASD efficacy in control of P. penetrans. Orchard grass at 20 t ha−1 was superior in controlling P. penetrans compared to 5 and 10 t ha−1 rates (Hewavitharana and Mazzola 2016). Thus, field trials are warranted to assess whether effective suppression of P. penetrans can be attained on sites where this nematode contributes significantly to ARD severity.
ASD has provided weed suppression when employed with various carbon inputs at rates greater than 11 t ha−1 (Shrestha et al. 2016). Reports of effective weed suppression include studies in which ASD was implemented using ethanol, manure or agricultural by-products, although weed suppression was not attained when grass was utilized as the input (Shrestha et al. 2016). It also has been reported that effective weed control was only observed when ASD was conducted under high soil temperature conditions (Shrestha et al. 2016). In our nursery field trial, effective suppression of a diverse weed population was obtained when grass was used as the carbon input at a rate of 20 t ha−1. In addition, weed suppression was realized even though ASD was conducted under soil temperature conditions (19–23 ̊ C) that are considered to be moderate (16–35 °C; Shrestha et al. 2016). As weed control is a major impediment to tree performance in newly established orchards (Hoagland et al. 2008), ASD-generated weed suppression will be of significant benefit particularly in organic systems which lack cost-effective weed control strategies in young plantings.
Changes in composition of the soil microbiome in response to induction of ASD have been extensively studied (Messiha et al. 2007; van Agtmaal et al. 2015; Runia et al. 2014; Streminska et al. 2014; Hewavitharana and Mazzola 2016). ASD conducted using grass or potato haulms as the carbon source induced distinct shifts in bacterial community composition relative to the control as assessed using PCR-DGGE analyses of 16S rDNA (Messiha et al. 2007). These findings also were observed in our previous studies where ASD treatment with grass resulted in significant compositional changes in soil bacterial and fungal communities (Hewavitharana and Mazzola 2016). Van Agtmaal et al. (2015) reported that ASD conducted with a defined protein-rich vegetal by-product of the food processing industry, reduced operational taxonomic unit (OTU) richness at 3 months post-treatment, but by 15 months post-treatment OTU richness was restored in ASD treated soil. In the current nursery field study, it was evident that ASD treatment with grass as the carbon source had a prolonged effect on composition of the soil fungal and bacterial communities. In the greenhouse study, when evaluated four months post-treatment, rhizosphere microbial communities in ASD-GR treated soils were distinct from that of the PC and NTC treatments. In the nursery field trial, unlike the fungal community, differences in bulk soil bacterial community composition between the control and ASD treatments were not apparent immediately post-treatment (19 days), potentially due to a rapid reversion in composition of the bacterial population after the anaerobic phase. Although ASD consistently stimulates shifts in composition of the soil microbiome, documentation of these changes may be regulated by various factors including soil type, carbon input, temperature and time of sampling (Runia et al. 2014; Hewavitharana et al. 2019). When examined at 14 months post-treatment, rhizosphere soil bacterial and fungal communities in the ASD treatment were distinct from the control, but somewhat more similar to the fumigation treatment. In a study that investigated the application of a B. juncea-S. alba seed meal soil-amendment for control of ARD, trees cultivated in seed meal amended soils possessed a rhizosphere microbiome that was distinct from that of trees grown in non-treated or fumigated orchard soil after two growing seasons (Mazzola et al. 2015). These documented changes were associated with prolonged soil-borne pathogen suppression.
The apparent block effect in soil microbial community composition observed in the study may have stemmed from several edaphic factors. The nursery field site consisted of rolling topography across the north-south direction that may have caused water retention for prolonged period in some plots relative to others. Although we did not assess Phytophthora spp. as a significant component of the ARD pathogen complex affecting rootstock growth in this trial, Phytophthora cambivora was shown to contribute to replant disease development at a previous trial conducted at this location (Mazzola and Brown 2010). It could be assumed that water retention in those plots may have incited proliferation of Phytophthora spp. The field site also exhibited east-west effect from the sunlight due to slope, with soils in west side plots likely remaining cooler than those of plots in the east side of the study site.
Certain apple rootstock genotypes demonstrate superior growth performance when planted on replant orchard sites (Auvil et al. 2011; Bezuidenhout et al. 2014; Kviklys et al. 2016). Field level tolerance has been demonstrated across sites, however the relative performance of an individual rootstock has demonstrated significant site-to-site variability. Thus, rootstocks reported to be tolerant on one site have been described as susceptible on others (Auvil et al. 2011; St. Laurent et al. 2010). Assessments of tree performance in the field have consistently been based upon horticultural characteristics and have not documented rootstock genotype differences relative to root infection by ARD pathogens. Recent findings have indicated that functional resistance towards certain replant pathogens, including P. ultimum, exists within apple rootstock germplasm including the rootstock G.935 (Zhu et al. 2016). However, expression of this resistance in the field has not been explored. In the current field study, no significant difference in root infestation by P. ultimum or R. solani was observed among rootstocks previously described as tolerant (G.935) or susceptible (M.9) to replant disease. When cultivated in the field, the moderately tolerant rootstock G.41 possessed significantly higher quantities of DNA for both pathogens than did the susceptible rootstock M.9. This observation was consistent with a study conducted at the same experimental orchard where Pythium spp. in NTC and fumigation treatments were significantly higher in Gala/G.41 trees than in Gala/M.26 trees (Wang and Mazzola 2019b). This finding suggests that superior growth performance of rootstocks previously described as tolerant to ARD may depend upon factors other than resistance mechanisms as suggested by Atucha et al. (2014). For example, apple rootstock root architecture has been identified as a heritable trait (Fazio et al. 2013). ARD tolerant rootstocks, such as G.210, possess finer root branching structure with roots of smaller diameter and a thinner cortex compared to the susceptible rootstock M.26 (Emmett et al. 2014). In addition, relative to M.26, G.210 possessed greater root biomass and a higher ratio of second-to-first order roots, but roots had lower N concentration and shorter lifespan (Atucha et al. 2014). Based on these findings, authors surmised that ARD tolerant rootstocks invest fewer resources in development of individual roots (Atucha et al. 2014).
In our study, ASD soil treatment had a greater influence on resulting composition of the rhizosphere microbiome than did rootstock genotype. A distinction in composition of the rhizosphere microbiome, and in particular the fungal community, was detected among different soil treatments both in field and greenhouse experiments. Although a significant rootstock genotype effect was observed for composition of the rhizosphere fungal community in studies conducted in the greenhouse environment, such a response was not in the nursery field trial. Genotype effect on rhizosphere bacterial communities was not prominent in both the greenhouse and nursery field trials. A similar phenomenon was observed in relation to B. juncea-S. alba seed meal soil-amendment where rootstock genotype effects were observed in greenhouse trials conducted at low seed meal amendment rates (Wang and Mazzola 2019a), but were not observed in field trials at a higher seed meal amendment rate (Mazzola et al. 2015). In contrast, previous reports indicated that rhizosphere bacterial and fungal communities were influenced by rootstock genotype and orchard planting position (Rumberger et al. 2007) but not by pre-plant compost or fumigation treatments (Rumberger et al. 2004). In concert these findings indicate that certain soil treatments that have stronger effects on directing composition of the soil microbiome may mask the overall effect of rootstock genotype on the rhizosphere microbiome.
Changes in the microbiome could not be directly linked to disease control in the nursery field study as there was no significant relationship between soil treatment and relative root infestation at the end of the second growing season by the pathogens monitored in this study. We hypothesize that even in the presence of ARD pathogens, specific transformations in the rhizosphere microbiome may have taken place in response to ASD in the context of unique root architectural differences among tolerant and susceptible genotypes, which may have functioned to improve growth of the rootstocks. For instance, some rhizosphere inhabitants have been identified as plant growth promoting bacteria (PGPR) that are functionally important to plant growth when acting in roles such as biofertilizers and phytostimulators (Somers et al. 2004). Moreover, tripartite associations among plant roots, fungi, and endobacteria may interact to directly promote plant growth (Sharma et al. 2008). Despite differential growth rates corresponding to relative rootstock vigor, growth rates of G.935 and M.9 genotypes were enhanced in the ASD treatment in a manner similar to that observed for the FUM treatment. Although growth rate of G.41 was not improved with FUM in comparison to NTC, it was superior with ASD treatment compared to NTC. Hence, ASD treatment may have resulted in an additive effect on tolerant rootstock genotypes and a complementary effect on moderately tolerant and susceptible rootstock genotypes through selection of a rhizosphere microbiome that is beneficial for growth promotion. Apart from soil microbial changes that occur in response to ASD, a myriad of microbe generated soil metabolites that persist longer than volatiles produced during the ASD process may contribute to pathogen suppression (Hewavitharana et al. 2019) as well. Further research is needed to validate these hypotheses.
Studies have shown that ASD affects soil nutrient levels. When a variety of warm season cover crops including cowpea, sunn hemp, pearl millet, and sorghum-Sudan grass were used as ASD carbon sources, it was found that post-treatment soil N form was primarily NH4+-N which was hypothesized to be due to poorly aerated conditions (Butler et al. 2012). Furthermore, NH4+-N level was found to be lower in soils where ASD was conducted using a grass cover crop amendment relative to soils receiving ASD treatment with all other cover crop inputs. This finding was attributed to the high C:N ratio of the amendment, however the values were not presented relative to a control treatment (Butler et al. 2012). In contrast to Butler et al. (2012), in the current study NH4+-N was lower in ASD-GR20 soil and NO3− N was the primary N form detected following ASD-GR20 treatment indicating that nitrification occurred towards the end of the incubation period probably due to high C:N ratio in the grass amendment. Relative to the FUM and NTC soils, increased levels of NO3− N, Fe and Mn were maintained in ASD-GR20 treated soils throughout the initial field growing season, and the improved nutrient availability may have contributed to enhanced tree growth.
Overall, our findings demonstrate that ASD treatment with orchard grass residues as the carbon source may be an effective means to control nursery ARD. Trunk diameter increment is a primary determinant of tree value in a nursery setting, and our results show that use of ASD to control ARD may result in greater overall returns to nursery growers due to the resulting increase in tree caliper size. Although the ASD-GR20 treatment effectively controlled disease and improved tree growth in the nursery field trial, greenhouse studies suggest that a similar effect could be achieved using an ASD grass amendment rate of 10 t ha−1. Improved tree growth was obtained independent of apple rootstock genotype, and 14-month growth increments attained in ASD treated soil were similar to that achieved in fumigated soils for each genotype examined. Increased tree growth realized through ASD was multifaceted most likely resulting from proliferation of beneficial microorganisms in the rhizosphere, suppression of soil-borne plant pathogens as well as increased availability of nitrate nitrogen.
References
Aslam, D. N., & VanderGheynst, J. S. (2008). Predicting phytotoxicity of compost-amended soil from compost stability measurements. Environmental Engineering Science, 25, 72–81.
Atucha, A., Emmett, B., & Bauerle, T. L. (2014). Growth rate of fine root systems influences rootstock tolerance to replant disease. Plant and Soil, 376, 337–346.
Auvil, T. D., Schmidt, T. R., Hanrahan, I., Castillo, F., McFerson, J. R., & Fazio, G. (2011). Evaluation of dwarfing rootstocks in Washingotn apple replant sites. Acta Horticulturae, 903, 265–271.
Bezuidenhout, C. M., van Schoor, L., & Cook, N. C. (2014). Rootstocks evaluated for apple replant disease tolerance. Acta Horticulturae, 1058, 553–558.
Bruns, T. D., White, T. J., & Taylor, W. J. (1991). Fungal molecular systematics. Annual Review of Ecology and Systematics, 22, 525–564.
Burt, R. (1996). Soil survey laboratory methods manual. National Soil Survey Center, Natural Resources Conservation Service, United States Department of Agriculture. https://www.nrcs.usda.gov/Internet/FSE_DOCUMENTS/nrcseprd1026806.pdf
Butler, D. M., Rosskopf, E. N., Kokalis-Burelle, N., Albano, J. P., Muramoto, J., & Shennan, C. (2012). Exploring warm-season cover crops as carbon sources for anaerobic soil disinfestation (ASD). Plant and Soil, 355, 149–165.
Butler, D. M., Ownley, B. H., Dee, M. E., Eichler Inwood, S. E., McCarty, D. G., Shrestha, U., et al. (2014). Low carbon amendment rates during anaerobic soil disinfestation (ASD) at moderate soil temperatures do not decrease viability of Sclerotinia sclerotiorum sclerotia or Fusarium root rot of common bean. Acta Horticulturae, 1044, 203–208.
Deakin G., Fernández-Fernández, F., Bennett, J., Passey, T., Harrison, N., Tilston, E. L., et al. (2019). The effect of rotating apple rootstock genotypes on apple replant disease and rhizosphere microbiome. Phytobiomes, doi.org/10.1094/PBIOMES-03-19-0018-R.
Emmett, B., Nelson, E. B., Kessler, A., & Bauerle, T. L. (2014). Fine-root system development and susceptibility to pathogen colonization. Planta, 239, 325–340.
Fazio, G., & Mazzola, M. (2004). Target traits for the development of marker assisted selection of apple rootstocks-prospects and benefits. Acta Horticulturae, 663, 823–828.
Fazio, G., Aldwinckle, H., & Robinson, T. (2013). Unique characteristics of Geneva® apple rootstocks. New York Fruit Quarterly, 21, 25–28.
Gaines, X. M., & Swan, D. G. (1972). Weeds of eastern Washington and adjacent areas (349 pp). Davenport, WA: Camp-Na-Bor-Lee Association.
Gardes, M., & Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecology, 2, 113–118.
Gergerich, R. C., Welliver, R. A., Gettys, S., Osterbauer, N. K., Kamenidou, S., Martin, R. R., et al. (2015). Safeguarding fruit crops in the age of agricultural globalization. Plant Disease, 99, 176–187.
Goud, J. C., Termorshuizen, A. J., Blok, W. J., & van Bruggen, A. H. C. (2004). Long-term effect of biological soil disinfestation on Verticillium wilt. Plant Disease, 88, 688–694.
Hammer, Ø., Harper, D. A. T., & Ryan, P. D. (2001). PAST:Paleontological statistics software package for education and data analysis. Palaeontologia Electronica, 4, 1–9.
Harrington, J. T., Mexal, J. G., & Fisher, J. T. (1994). Volume displacement provides a quick and accurate way to quantify new root production. Tree Planters’ Notes, 45, 121–124.
Harshman, J. M., & Evans, K. M. (2015). Survey of moldy core incidence in germplasm from three U.S. apple breeding programs. Journal of the American Pomological Society, 69, 51–57.
Hewavitharana, S. S., & Mazzola, M. (2016). Carbon source-dependent effects of anaerobic soil disinfestation on soil microbiome suppression of Rhizoctonia solani AG-5 and Pratylenchus penetrans. Phytopathology, 106, 1015–1028.
Hewavitharana, S. S., Klarer, E., Reed, A. J., Leisso, R., Poirier, B., Honaas, L., Rudell, D. R., & Mazzola, M. (2019). Temporal dynamics of the soil metabolome and microbiome in response to anaerobic soil disinfestation. Frontiers in Microbiology, 10, 2365.
Hoagland, D.R. & Aron, D.I. (1938). The water-culture method for growing plants without soil. Circular (California Agricultural Experiment Station), 347. eds. Berkeley, California, University of California, College of Agriculture, Agricultural Experiment Station.
Hoagland, L., Carpenter-Boggs, L., Granatstein, D., Mazzola, M., Smith, J., Peryea, F., & Reganold, J. P. (2008). Orchard floor management effects on nitrogen fertility and soil biological activity in a newly established apple orchard. Biology and Fertility of Soils, 45, 11–18.
Hoitink, H. A. J., & Boehm, M. J. (1999). Biocontrol within the context of soil microbial communities: A substrate-dependent phenomenon. Annual Review of Phytopathology, 37, 427–446.
Igarashi, M., Hatsuyama, Y., Harada, T., & Fukasawa-Akada, T. (2016). Biotechnology and apple breeding in Japan. Breeding Science, 66, 18–33.
Isutsa, D. K., & Merwin, I. A. (2000). Malus germplasm varies in resistance or tolerance to apple replant disease in a mixture of New York orchard soils. HortScience, 35, 262–268.
Johnson, W. C. (2000). Methods and results of screening for disease- and insect-resistant apple rootstocks. Compact Fruit Tree, 33, 108–111.
Kviklys, D., Robinson, T. L., & Fazio, G. (2016). Apple rootstock evaluation for apple replant disease. Acta Horticulturae, 1130, 425–430.
Leisso, R., Rudell, D., & Mazzola, M. (2017). Metabolic composition of apple rootstock rhizodeposits differs in a genotype-specific manner and affects growth of subsequent plantings. Soil Biology and Biochemistry, 113, 201–214.
Lembright, H. W. (1990). Soil fumigation: Principles and application technology. Journal of Nematology, 22, 632–644.
Manici, L. M., Caputo, F., & Bambini, V. (2004). Effect of green manure on Pythium spp. population and microbial communities in intensive cropping systems. Plant and Soil, 263, 133–142.
Mazzola, M. (1997). Identification and pathogenicity of Rhizoctonia spp. isolated from apple roots and orchard soils. Phytopathology, 87, 582–587.
Mazzola, M. (1998). Elucidation of the microbial complex having a causal role in the development of apple replant disease in Washington. Phytopathology, 88, 930–938.
Mazzola, M., & Brown, J. (2010). Efficacy of Brassicaceous seed meal formulations for the control of apple replant disease in conventional and organic production systems. Plant Disease, 94, 835–842.
Mazzola, M., & Manici, L. M. (2012). Apple replant disease: Role of microbial ecology in cause and control. Annual Review of Phytopathology, 50, 45–65.
Mazzola, M., & Zhao, X. (2010). Brassica juncea seed meal particle size influences chemistry but not soil biology-based suppression of individual agents inciting apple replant disease. Plant and Soil, 337, 313–324.
Mazzola, M., Granatstein, D. M., Elfving, D. C., & Mullinix, K. (2001). Suppression of specific apple root pathogens by Brassica napus seed meal amendment regardless of glucosinolate content. Phytopathology, 91, 673–679.
Mazzola, M., Brown, J., Zhao, Z., & Izzo, A. (2009). Interaction of Brassicaceous seed meal and apple rootstock on recovery of Pythium spp. and Pratylenchus penetrans from roots grown in replant soils. Plant Disease, 93, 51–57.
Mazzola, M., Hewavitharana, S. S., & Strauss, S. L. (2015). Brassica seed meal soil amendments transform the rhizosphere microbiome and improve apple production through resistance to pathogen reinfestation. Phytopathology, 105, 460–469.
Messiha, N., van Diepeningen, A., Wenneker, M., van Beuhingen, A., Janse, J., Coenen, T., et al. (2007). Biological soil disinfestation (BSD), a new control method for potato brown rot, caused by Ralstonia solanacearum race 3 biovar 2. European Journal of Plant Pathology, 117, 403–415.
Moein, S., Mazzola, M., Ntushelo, N. S., & McLeod, A. (2019). Apple nursery trees and irrigation water as potential external inoculum sources of apple replant disease in South Africa. European Journal of Plant Pathology, 153, 1131–1147.
Norelli, J. L., Holleran, H. T., Johnson, W. C., Robinson, T. L., & Aldwinckle, H. S. (2003). Resistance of Geneva and other apple rootstocks to Erwinia amylovora. Plant Disease, 87, 26–32.
Ramos, D. E. (1998). Walnut production manual publication 3373. Oakland: University of California Division of Agriculture and Natural Resources.
Reim, S., Siewert, C., Winkelmann, T., Wöhner, T, Hanke, M., & Flachowsky, H. (2019). Evaluation of Malus genetic resources for tolerance to apple replant disease (ARD). Scientia Horticulturae, https://doi.org/10.1016/j.scienta.2019.05.044.
Robinson, T., Fazio, G., & Aldwinckle, H. (2014). Characteristics and performance of four new apple rootstock from the Cornell-USDA apple rootstock breeding program. Acta Horticulturae, 1058, 651–656.
Rumberger, A., Shengrui, Y., Merwin, I. A., Nelson, E. B., & Thies, E. J. (2004). Rootstock genotype and orchard re-plant position rather than soil fumigation or compost amendment determine tree growth and rhizosphere bacterial community composition in an apple replant soil. Plant and Soil, 264, 247–260.
Rumberger, A., Merwin, I., & Thies, J. E. (2007). Microbial community development in the rhizosphere of apple trees at a replant disease site. Soil Biology and Biochemistry, 39, 1645–1654.
Runia, W. T., Thoden, T. C., Molendijk, L. P. G., van den Berg, W., Termorshuizen, A. J., Streminska, M. A., et al. (2014). Unravelling the mechanism of pathogen inactivation during anaerobic soil disinfestation. Acta Horticulturae, 1044, 177–193.
Schroeder, K. L., Okubara, P. A., Tambong, J. T., Lévesque, C. A., & Paulitz, T. C. (2006). Identification and quantification of pathogenic Pythium spp. from soils in eastern Washington using real-time polymerase chain reaction. Phytopathology, 96, 637–647.
Sharma, M., Schmid, M., Rothballer, M., Hause, G., Zuccaro, A., Imani, J., Kämpfer, P., Domann, E., Schäfer, P., Hartmann, A., & Kogel, K. H. (2008). Detection and identification of bacteria intimately associated with fungi of the order Sebacinales. Cellular Microbiology, 10, 2235–2246.
Shin, S. B., Zheng, P., Fazio, G., Mazzola, M., Main, D., & Zhu, Y. (2016). Transcriptome changes specifically associated with apple (Malus domestica) root defense response during Pythium ultimum infection. Physiological and Molecular Plant Pathology, 94, 16–26.
Shrestha, U., Auge, R., & Butler, D. (2016). A meta-analysis of the impact of anaerobic soil disinfestation on pest suppression yield of horticultural crops. Frontiers in Plant Science. https://doi.org/10.3389/fpls.2016.01254.
Somers, E., Vanderleyden, J., & Srinivasan, M. (2004). Rhizosphere bacterial signaling: A love parade beneath our feet. Critical Reviews in Microbiology, 30, 205–240.
St. Laurent, A., Merwin, I. A., Fazio, G., Thies, J. E., & Brown, M. G. (2010). Rootstock genotype succession influences apple replant disease and root zone microbial community composition in an orchard soil. Plant and Soil, 337, 259–272.
Strauss, S. L., Greenhut, R. F., McClean, A. E., & Kluepfel, D. A. (2017). Effect of anaerobic soil disinfestation on the bacterial community and key soil-borne phytopathogenic agents under walnut tree-crop nursery conditions. Plant and Soil, 415, 493–506. https://doi.org/10.1007/s11104-016-3126-4.
Streminska, M. A., van der Wurff, A. W. G., Runia, W. T., Thoden, T. C., Termorshuizen, A. J., & Feil, H. (2014). Changes in bacterial and fungal abundance in the soil during the process of anaerobic soil disinfestation. Acta Horticulturae, 1041, 95–102.
Tewoldemedhin, Y. T., Mazzola, M., Labuschagne, I., & McLeod, A. (2011). A multi-phasic approach reveals that apple replant disease is caused by multiple biological agents, with some agents acting synergistically. Soil Biology and Biochemistry, 43, 1917–1927.
Tidball, C. J. (1990). Phytophthora root and stem rot of apple rootstocks from stool beds. Plant Disease, 74, 141–146.
van Agtmaal, M., van Os, G. J., Hol, W. H. G., Hundscheid, M. P. J., Runia, W. T., Hordijk, C. A., et al. (2015). Legacy effects of anaerobic soil disinfestation on soil bacterial community composition and production of pathogen-suppressing volatiles. Frontiers in Microbiology. https://doi.org/10.3389/fmicb.2015.00701.
Wang, L., & Mazzola, M. (2019a). Interaction of Brassicaceae seed meal soil amendment and apple rootstock genotype on microbiome structure and replant disease suppression. Phytopathology, 109, 607–614.
Wang, L., & Mazzola, M. (2019b). Field evaluation of reduced rate Brassicaceae seed meal amendment and rootstock genotype on the microbiome and control of apple replant disease. Phytopathology, 109, 1378–1391.
Weerakoon, D. M. N., Reardon, C. L., Paulitz, T. C., Izzo, A. D., & Mazzola, M. (2012). Long-term suppression of Pythium abappressorium induced by Brassica juncea seed meal amendment is biologically mediated. Soil Biology and Biochemistry, 51, 44–52.
Winkelmann, T., Smalla, K., Amelung, W., Baab, G., Grunewaldt-Stöcker, G., Kanfra, X., Meyhöfer, R., Reim, S., Schmitz, M., Vetterlein, D., Wrede, A., Zühlke, S., Grunewaldt, J., Weiß, S., & Schloter, M. (2019). Apple replant disease: Causes and mitigation strategies. Current Issues in Molecular Biology, 30, 89–106.
Yonemoto, K., Hirota, K., Mizuguchi, S., & K. Sakaguchi. (2006). Utilization of the sterilization by soil reduction in an open air field its efficacy against Fusarium wilt of strawberry. Pages 15–24 in: Proceedings of Association of Plant. Protection, Shikoku.
Zhu, Y., Shin, S., & Mazzola, M. (2016). Genotype responses of two apple rootstocks to infection by Pythium ultimum causing apple replant disease. Canadian Journal of Plant Pathology, 38, 483–491.
Acknowledgements
This work was funded by the National Institute of Food and Agriculture, United States Department of Agriculture, under award number 2016 -51102-25815. Authors acknowledge Trident AG Products Inc. (Woodland, WA) for supporting this research by generously conducting soil fumigation. We also thank Danielle Graham for taxonomic identification of weed species, and Sheila K. Ivanov, Xiaowen Xiao, Kevin Hansen, Edward Valdez, Drew Reed, Parama Sikdar, Emmi Klarer, Cyrus Desmaris, Joy Lawless and Steven Lawless for their excellent technical support in this study.
Funding
As stated in acknowledgements, this work was funded by the National Institute of Food and Agriculture, United States Department of Agriculture, under award number 2016–51102-25,815.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflicts of interest in the reporting of this work.
Research involving human participants and/or animals
No human participants nor animals were involved in the conduct of this research.
Informed consent
Not relevant to this work.
Rights and permissions
About this article
Cite this article
Hewavitharana, S.S., Mazzola, M. Influence of rootstock genotype on efficacy of anaerobic soil disinfestation for control of apple nursery replant disease. Eur J Plant Pathol 157, 39–57 (2020). https://doi.org/10.1007/s10658-020-01977-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-020-01977-z