Abstract
Cladobotryum species and strain diversity of isolates collected from cobweb symptomatic Agaricus bisporus or infected casing soil were investigated due to increased incidences of the disease in the South African mushroom industry. Samples were collected from mushroom farms located in Gauteng, the Western Cape and KwaZulu Natal Provinces of South Africa. Moreover, cobweb disease isolates from the USA and Ireland were included in the study as reference cultures. Isolates were characterised using culture and conidia morphology and were identified as Cladobotryum mycophilum. The isolates were characterised by rapid colony growth between 48 and 72 h on malt extract agar and potato dextrose agar, all but two isolates (GP-15 and KZN-2) produced the pink colour of aurofurasin. All isolates could infect A. bisporus fruiting bodies with varying degrees of aggressiveness. Isolates were sequenced for their ITS, and BLAST analysis showed highest similarity (99–100%) to several ITS sequences of Hypomyces odoratus/C. mycophilum for 35 of the isolates except for one from Ireland that was identified as H. rosellus/C. dendroides. Phylogenetic analysis of the isolates showed South African cobweb disease of mushrooms to be caused by a wide diversity of strains some of which may have originated from elsewhere in the world.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cladobotryum anamorphs of Hypomyces are the cause of cobweb disease of Agaricus bisporus, the most cultivated mushroom species in the world (Grogan and Gaze 2000) and is responsible for significant economic losses in the industry (Grogan and Gaze 2000; Back et al. 2010, 2012). The fungus is a pathogen of mushrooms in the wild, usually associated with woodlands and forests (Rogerson and Samuels 1993, 1994; Tamm and Põldmaa 2013), The pathogen is introduced into mushroom growing rooms through contaminated compost, casing or air in the form of spores or mycelium fragments. Attack on fruiting bodies of A. bisporus can be at any stage during development. However, it is mostly reported in late flushes, especially if infection in early production stages is inadequately controlled (Carrasco et al. 2017a). Cladobotryum species responsible for cobweb disease include C. dendroides, C. mycophilum C. varium, C. multiseptatum, C. verticillatum (Adie et al. 2006; Fletcher and Gaze 2008). The disease is characterised by coarse mycelium growth covering the affected mushroom and casing soil (Fletcher and Gaze 2008). With a few exceptions, all the species cause similar symptoms characterised by rapid colonisation of host basidiomata and casing soil with cotton fluffy, white or greyish mycelia which becomes yellowish or reddish-pink in colour with time, due to the pigment aurofusarin and subsequent decay. (Back et al. 2012; Rogerson and Samuels 1993).
The cobweb disease has been controlled with the use of methyl benzimidazole carbamate (MBC) (Grogan 2006; Carrasco et al. 2017b). However, a number of strains resistant to these fungicides have been reported to be responsible for outbreaks in mushroom farms (Carrasco et al. 2017a). For this reason, the disease continues to be a perennial problem to a number of farmers, and has been reported more frequently in the South African mushroom industry (van Greines, South African Mushroom Growers Association, personal communication). Casing soil, is not readily accessible in South Africa and is therefore, imported as sphagnum peat mainly from Ireland and Australia. Consequently, it is possible that MBC-resistant strains can be brought into the country from these countries that have already reported such strains since the early 1990s (Carrasco et al. 2017b). Moreover, casing soil is prone to secondary contamination by soil and air borne spores if mis-managed in transit or at the destination farm, if it is stored for long periods of time as is often the case on South African farms.
Currently no data are available on the prevalence and diversity of various Cladobotryum spp. in South African mushroom farms. This study uses morphological and molecular data to identify cobweb isolates as well as evaluate their similarity to those available in GenBank (Table 1).
Materials and methods
Fungal isolates
A total of 36 isolates previously identified as Cladobotryum spp. used in this study were obtained from mushroom farms in South Africa (28 isolates) as well as from research institutes including those in the USA (6) and Ireland (2) (Table 1). South African isolates were collected from out breaks reported by commercial mushroom farms in the major producing regions including Gauteng (20 isolates), the Western Cape (six) and KwaZulu Natal (two) Provinces of South Africa. Isolates collected from local mushroom farms were either sampled from diseased mushrooms or collected as infected casing, while those from research institutions around the world were received as preserved cultures on agar slants. For each isolate, single spore cultures were prepared for use in the study and preserved in sterile distilled water with 10% glycerol at −75 °C for short- to medium-term storage.
Characterisation of culture- and conidial morphology
Petri dishes (90 mm diameter) were inoculated by placing a 5 mm plug from an actively growing colony approximately 1 cm from the margin of a Petri dish. Malt extract agar (MEA), cornmeal agar (CMA) (BBL™) and potato dextrose agar (PDA) (Merck, Johannesburg) were used. Colony characteristics were evaluated and growth rates were measured after 72 h of incubation at 25 °C. Photographs of cultures on different media were taken with a Nikon DC3100 camera (Nikon, Japan).
Microscopic features were studied with a Zeiss DIC/Phase Contrast (Axiovision release 4.8) microscope (Carl Zeiss, Germany) using cultures grown on malt extract agar (Merck, Johannesburg) for 72 h at 25 °C. A total of 30 conidia per isolate were examined after mounting in 85% lactic acid and the length and width averages, as well as the length/width ratios (Q) calculated.
Colonisation of mushrooms with cobweb isolates’
The ability of Cladobotryum isolates to infect fruiting bodies of A. bisporus was investigated as previously described with slight modifications (Back et al. 2012). Spore suspensions (50 ml, at a concentration of 5 × 106 conidia/ml) were prepared from 10 to 15 day old cultures and spray inoculated onto A. bisporus fruiting bodies. Five fruiting bodies resulting from same flush were used per trial and sterile distilled water was used as a control. The inoculated mushrooms were placed on moistened paper towels in plastic containers which was then covered with glass. The inoculated mushrooms together with the control were incubated for up to 4 days at 20 °C and the Cladobotryum development was observed visually beginning day one after inoculation.
Molecular and phylogenetic analysis
Each culture was derived from a four-day old single spore colony. Fungal mycelia were harvested by scraping from cultures on MEA plates and genomic DNA extracted with a DNeasy Plant Mini kit (QIAGEN), according to manufacturer’s instruction using the bead beating technique. The ITS region for all the 36 isolates were amplified with the following primers: ITS1F (Gardes and Bruns 1993) and ITS4 (White et al. 1990). PCR was performed as previously described (Põldmaa 2011). All samples were sequenced at Inqaba Labs Inc. (Pretoria, South Africa).
DNA Sequence Assembler v4.36.0.2 (2013), (Heracle BioSoft, www.DnaBaser.com) analysis software was used to edit sequences. Sequences were preliminary identified by comparing with those deposited in the GenBank (NCBI) database through megaBLAST searches (Morgulis et al. 2008) (Table 1). Sequences were aligned separately using MAFFT version 6 (Katoh and Toh 2008) and phylogenetic relationships among strains analysed by the software MEGA7 (molecular evolutionary genetics analysis) (Kumar et al. 2016) using the using the Maximum Likelihood method based on the Jukes-Cantor model (Jukes and Kantor 1969). Bootstrap analysis was carried out to examine the reliability of interior branches (Felsenstein 1985). Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method (Kimura 1980) and are in the units of the number of base substitutions per site. Sepedonium sp. (HQ604857) was designated as the outgroup for the phylogenetic analysis.
Results
Morphological characterisation of isolates
Mycelia were generally fluffy and grey-white in colour except for isolate IR-2 which had a compact texture. Colony growth was rapid particularly between 48 and 72 h, and reached Petri dish edges in 72–96 h. Colonies initially turned yellowish to pinkish, then purplish after more than 96 h of growth, with the colour changes being more pronounced on the underside while the aerial mycelia remained relatively white. Out of the 35 isolates used in this study, all but two isolates (GP-15 and KZN-2) produced the pink colour of aurofurasin. All isolate produced conidia except for isolate KZN-2. The conidiophores were verticilliately branched, forming subulate phialides tapering at the tips on which conidia were borne. An average of four phialides with a single conidium were formed at branches. Conidia cells had 0–3 septa, were hyaline, ellipsoidal or cylindrical and in some instances ovoid. More than one type of conidia cells could be observed per single isolate. Conidial mean length ranged from 16.41–25.64 μm. and width from 7.65–11.26 μm. The conidial length/width (l/w) ratio (Q-value) ranged from 1.9–3.6 with an average of 2.34 μm, while the majority of isolates had a Q-value measuring between 2.2 and 3.0 μm. All the 35 isolates were identified as C. mycophilum except for one isolate IR-2 which is an H. rosellus of Irish origin.
‘Cobweb growth pattern on infected mushrooms
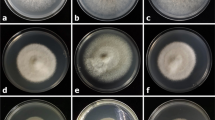
Cobweb disease symptoms developed between 24 and 96 h after spraying mushroom fruitbodies, whereas the control remained healthy as with previous reports (Back et al. 2012; Potočnik et al. 2008). Moreover, Cladobotryum fungi was re-isolated from diseased fruiting bodies but not from the controls. Five distinct symptom groups could be observed (Fig. 1). Nine of the isolates including GP-12, GP-13, GP 17, GP-18, GP-19 and GP-21 isolates grew profusely, covering the entire mushroom cap within 48 h with typical cottony fluffy mycelia growth of C. mycophilum. However, other isolates grew slowly in the 96 h period and included the isolates KZN-1, GP-9 and GP-11. Generally, the mycelia were white and cotton fluffy for most of the isolates and turning pinkish after 72 h. Colony morphology for isolates GP-5 and GP-4 formed a granulated surface as seen in Fig. 1. The slower growing isolates showed white sparse mycelia with no colour change at 96 h. The growth and colony morphology of some isolates observed in culture differed with that seen on mushroom caps. Only a few isolates that showed profuse growth in culture developed poorly on mushroom caps. However, all Cladobotryum isolates that performed poorly on culture media (data not shown) also exhibited the same kind of growth on mushroom caps, similar to previous reports (Potočnik et al. 2008).
Molecular and phylogenetic analysis
The molecular study was carried out to confirm morphological identities and to evaluate the diversity of 36 cobweb isolates collected from diseased A. bisporus or casing in commercial farms in South Africa, Ireland and the USA. Isolates were identified based the internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence (Table 1). BLAST analysis of the isolates revealed highest similarity (99–100%) to several ITS sequences of H. odoratus/C. mycophilum for 35 of the isolates except for one that was identified as H. rosellus/C. dendroides.
Cluster evaluation of the 36 isolates showed four distinct arbitrary groupings which separated the isolates according to their species and strains (Fig. 2). The one isolate (IR-2) identified as C. dendroides/H. rosselus clustered on its own (group A) while all the other isolates identified as C. mycophilum/H. odoratus clustered under one main branch (group B-D). One of the two isolates from Ireland (IR −1) was separately grouped with two isolates from Gauteng (GP-8 and GP-9) in cluster B together with GenBank sequences, MH040808 and KX098648. Cluster C had only one isolate from Gauteng (GP-5) and six sequences from GenBank. The majority of the isolates 80.6% were clustered in a subtree in group D1 and D2. Group D1 was composed of eight isolates from South Africa and five from the USA. The South African isolates in this group were from Gauteng (six isolates) and KZN (2 isolates). Sequences in group C and D2 clustered as previously described for C. mycophilum (Carrasco et al. 2016). ITS sequences in Group C included those from the NCBI with accession numbers JQ004737 (Carrasco et al. 2016) and AB527074 (Back et al. 2010, 2012). All sequences for isolates that were characterised by Profuse growth, of cottony fluffy mycelia-(USA-2, GP-7, GP-17, GP-18 and GP-19 and granulated surface- GP-4 and GP-16 (Fig. 1) were clustered in Group D2 together with GenBank sequences Y17095, and JQ004735 (Carrasco et al. 2016) and JF505112 (Gea et al. 2011). The rest of the observed morphological types were spread among the other cluster groups with no specific pattern. The eighteen isolates in Group D2 included six from the Western Cape Province, 10 from Gauteng and one from the USA (US-2). No isolates from the KZN province were found in this group.
Discussion
Morphological characterisation
Morphological characterisation of fungal isolates collected from cobweb disease in South African mushroom farms identified them as C. mycophilum. The reference isolates were all identified as C. mycophilum, with the exception of one isolate from Ireland which identified as H. rosellus (IR 2). The C. mycophilum isolates were characterised by verticillate branched conidiophores, forming subulate phialides without evident rachis and with tapering tips on which 0- to 3-septate conidia were borne (Gams and Hoozeman 1970; Rogerson and Samuels 1993, 1994; Grogan 2006; Carrasco et al. 2016). Species morphological identities were further confirmed with the implementation of phylogenetic analyses of the internal transcribed spacer (ITS) region of ribosomal DNA (rDNA) (Tamm and Põldmaa 2013; Zuo et al. 2016; Carrasco et al. 2016). The identification of C. mycophilum as the causal agent of cobweb disease in South African mushroom farms confirms previous reports that it is the most cited causal agent of cobweb disease of mushrooms (Back et al. 2010, 2012; Carrasco et al. 2017a; Potočnik et al. 2008). Studies in Africa, Asia and Europe have reported the fungi to parasitize different fungi, including A. bisporus, Pleurotus eryngii and Ganoderma lucidum (Back et al. 2010; Chakwiya et al. 2015; Carrasco et al. 2016; Zuo et al. 2016; Gea et al. 2017).
The variation in growth and conidia production among Cladobotryum isolates may be attributed to the formation of morphological variants. Formation of distinct morphological variants or sectors in culture is often observed when maintained on artificial media. The type and frequency of sectoring varies among different fungal species and strains have often been attributed to mutation, transposons, double-stranded RNA myco-viruses and genomic rearrangements (Becker et al. 2003). This may explain why the isolates in this study showed different growth patterns (morphologies) and aggressiveness when grown on both mushroom pilei and artificial media such as MEA (data not shown). When grown on MEA, the evidence of formation of sectors was observed, though on a few isolates. Some studies suggest that sectors arise as a result of cultural degeneration caused either by the age of culture, method of propagation, or nature of the culture medium (Booth 1975).
Although the causal agents of cobweb disease in the cultivated mushroom (A. bisporus) has been associated with several Cladobotryum species (Carrasco et al. 2017a), findings from the molecular analyses showed the South African isolates in this study to belong to H. odoratus/C. mycophilum. This confirms C. mycophilum as the main causal agent of cobweb disease in commercial mushroom production systems in South Africa, similar to previous reports in other countries (Tamm and Põldmaa 2013). Consequently, Tamm and Põldmaa (2013) went on to suggest that a large part of the earlier identifications of the causal agent of cobweb disease described as H. rosellus to be incorrect.
Previous studies by Chakwiya et al. (2015) on C. mycophilum in South Africa reported reduced sensitivity to prochloraz manganese and carbendazim on some of the isolates collected in that study. The resistance to fungicides in H. odoratus has been cited as the likely factor in the efficient spread of C. mycophilum in mushroom farms worldwide (Tamm and Põldmaa 2013). A number of hypothesis on the spread of cobweb disease are plausible including, long distance conidia dispersal, imported casing soil, international movement of personnel and equipment and accidental release from imported cultures. In South Africa the use of local peat as casing soil was banned in 2007, as it was listed as a threatened natural resource (Richardson 2010). As a result, farmers rely on imported casing soil from Ireland and Australia, hence, it is probable that new strains of C. mycophilum may be introduced from imported peat. However, the worldwide Agaricus bisporus mushroom industry is effectively a monoculture with similar production systems providing, the same selective pressure to local populations to become resistant. Research on H. odoratus worldwide have shown very low genetic diversity hence firm conclusions on the origin of MBC resistant isolates are difficult to make without detailed studies on a much larger collection of isolates. Moreover, local populations of MBC-sensitive isolates of H. odoratus on mushroom farms have been reported to be linked to their local wild populations (Tamm and Põldmaa 2013). The report on MBC sensitive isolates from previous studies in the country support the hypothesis that H. odoratus also occurs within the South African natural environment (Chakwiya et al. 2015). In the USA and Europe, the reported MBC-resistant clade of isolates has been shown to be weakly supported by multigene analysis. Consequently, the geographic origin of fungicide resistance remains unknown as it occurs due to a single point mutation (McKay et al. 1998). Hence, there is the potential for ‘spontaneous emergence and independent spread.
Previous studies have reported nearly identical isolates from different countries including Serbia, Ireland, Hungary, USA and the UK. While isolates from the UK had already been established to be strongly resistant to thiabendazole and weakly resistant to carbendazim, a similar observation was made with the isolates from Serbian which were shown to be sensitive to carbendazim (Grogan and Gaze 2000; Potočnik et al. 2009). Regarding resistance to thiabendazole and carbendazim, a recent study has indicated metrafenone use as an alternative effective control for which resistance has not as yet been reported and has no phytotoxic effects on mushroom. Moreover, it has been shown to be better than prochloraz-Mn as it was shown to be totally inhibiting to cobweb disease development (Carrasco et al. 2017b).
However, Resistance to pesticide including benzimidazoles can occur relatively quickly when products are used continuously, as would have occurred in many mushroom growing countries, including South Africa. It is likely that resistance has emerged independently in each mushroom growing country, as has happened with another mushroom pathogen, Lecanicillium fungicola (syn. Verticillium fungicola). Hence, in the context of spread of isolates from one country to another there is need for detailed analysis of more isolates to resolve the phylogeny of H. odoratus from mushroom farms and their relationship to the background H. odoratus populations in each country as well as to populations found on mushroom farms around the world. This is particularly important given that infraspecific variation within this species is considerably lower than any other Hypomyces/Cladobotryum taxa studied including isolates collected between 1940–1980s. These originated from diverse substrates, and were not benzimidazole resistant (Tamm and Põldmaa 2013),
The distribution of H. odoratus strains in South Africa shows the presence of unique strains in particular regions as well as strains common to all the provinces sampled. Moreover, some isolates including both unique and common strains have been reported to have reduced sensitivity to MBC and prochloraz manganese fungicides (Chakwiya et al. 2015). Origins of fungicide resistance in H. odoratus generally remains unknown, and it is probable that divergent mutations led to MBC resistance, and it occurred independently in different regions as has been reported for the Verticillium mushroom pathogens (Tamm and Põldmaa 2013; Gea et al. 2005). More evidence is however needed to establish the idea of spontaneous emergence and independent spread of MBC-resistant mutants of H. odoratus. The concurrent spread of genetically homogenous MBC-resistant strains in mushroom farms has been suggested to result from importation of contaminated A. bisporus spawn (Carrasco et al. 2017a, b). However, given the high quality standards in the spawn chain and rapid growth by the pathogen, it is highly unlikely.
The success, dominance and persistence of H. odoratus in mushroom farms in various parts of the world has been suggested to be a result of its clonality in nature (Tamm and Põldmaa 2013). Fungicide resistance that occur as a result of single point mutations can easily be spread in the absence of recombination during reproduction. This is particularly important given that evidence from molecular markers suggest the prevalence of clonal reproduction in H. odoratus (Tamm and Põldmaa 2013). Furthermore, the presence of H. odoratus, in natural environments may serve as a source of inoculum providing air borne conidia that may infect casing and carpophores, resulting in disease development and if not properly contained, an outbreak (Carrasco et al. 2017a).
This study confirms the prevalence of cobweb disease in the South African mushroom industry to be caused by diverse C. mycophilum strains that may potentially have varied aggressiveness and pesticide resistance profiles. This data is important for further research and evaluation of alternative pesticides and cultural practices critical in the control of cobweb disease in South Africa.
References
Adie, B., Grogan, H., Archer, S., & Mills, P. (2006). Temporal and spatial dispersal of Cladobotryum conidia in the controlled environment of a mushroom growing room. Applied and Environmental Microbiology, 72, 7212–7217.
Back, C. G., Kim, Y. H., Jo W. S., Chung, H. & Jung, H, Y. (2010). Cobweb disease on Agaricus bisporus caused by Cladobotryum mycophilum in Korea. Journal of General Plant Pathology, 76, 232–235.
Back, C. G., Lee, C. Y., Seo, G. S., & Jung, H. C. (2012). Characterisation of species of Cladobotryum which cause cobweb disease of edible mushrooms grown in Korea. Mycobiology, 40(3), 189–194.
Becker, T. C. A., Chiuchetta, S. J. R., Baptista, F., & de Castro-Padro, M. A. A. (2003). Increase in mitotic recombination in diploid cells of Aspergillus nidulans in response to ethidium bromide. Genetics and Molecular Biology, 26(3), 381–385.
Booth, C. (1975). The present status of Fusarium taxonomy. Annual Review of Phytopathology, 13, 83–93.
Carrasco, J., Navarro, M. J., Santos, M., Diánez, F., & Gea, F. J. (2016). Incidence, identification and pathogenicity of Cladobotryum mycophilum, causal agent of cobweb disease on Agaricus bisporus mushroom crops in Spain. Annals of Applied Biology, 168(2), 214–224.
Carrasco, J., Navarro, M. J., & Gea, F. J. (2017a). Cobweb, a serious pathology in mushroom crops: A review. Spanish Journal of Agricultural Research, 15(2), e10R01.
Carrasco, J, Navarro, M. J., Santos, M. & Gea, F. J, (2017b). Effect of five fungicides with different modes of action on cobweb disease (Cladobotryum mycophilum) and mushroom yield. Annals of applied biology, https://doi.org/10.1111/aab.12352.
Chakwiya, A., Van der Linde, E. J., & Korsten, L. (2015). In vitro sensitivity testing of Cladobotryum mycophilum to carbendazim and prochloraz manganese. South African Journal of Science, Volume 111. https://doi.org/10.17159/sajs.2015/20140408.
Felsenstein, J. (1985). Phylogenies and the Comparative Method. The American Naturalist, 125(1), 1–15.
Fletcher, J. T., & Gaze, R. H. (2008). Mushroom pest and disease control – A colour handbook. London: Manson Publishing Ltd..
Gams, W., & Hoozeman, A. C. M. (1970). Cladobotryum-konidienformen von Hypomyces-Arten. Persoonia, 6, 95–110.
Gardes, M., & Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecology, 2(2), 113–118.
Gea, F. J., Navarro, M. J., & Tello, J. C. (2005). Reduced sensitivity of the mushroom pathogen Verticillium fungicola to prochloraz-manganese in vitro. Mycological Research, 109, 741–745.
Gea, F. J., Navarro, M. J., & Suz, L. M. (2011). First report of Cladobotryum mycophilum causing cobweb on cultivated king oyster mushroom in Spain. Plant Disease, 95(8), 1030.
Gea, F. J., Carrasco, J., Suz, L. M., & Navarro, M. J. (2017). Characterization and pathogenicity of Cladobotryum mycophilum in Spanish Pleurotus eryngii mushroom crops and its sensitivity to fungicides. European Journal of Plant Pathology, 147(1), 129–139.
Grogan, H. M. (2006). Fungicide control of mushroom cobweb disease caused by Cladobotryum strains with different benzimidazole resistance profiles. Pest Management Science, 62, 153–161.
Grogan, H. M., & Gaze, R. H. (2000). Fungicide resistance among Cladobotryum spp. - causal agents of cobweb disease of the edible mushroom Agaricus bisporus. Mycological Research, 104(3), 357–364.
Jukes, T. H., & Kantor, C. R. (1969). Evolution of protein molecules. In H. N. Munro (Ed.), Mammalian protein metabolism (Vol. II, pp. 21–123). New York: Academic Press.
Katoh, K., & Toh, H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics, 9(4), 286–298.
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16(2), 111–120.
Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874.
McKay, G. J., Egan, D., Morris, E., Scott, C., & Brown, A. E. (1998). Identification of benzimidazole resisitance in Cladobotryum dendroides using a PCR- based method. Mycological Research, 102, 671–676.
McKay, G. J., Egan, D., Morris, E., Scott, C., & Brown, A. E. (1999). Genetic and morphological characterization of Cladobotryum species causing cobweb disease of mushrooms. Applied and Environmental Microbiology, 65(2), 606–610.
Morgulis, A., Coulouris, G., Raytselis, Y., Madden, T. L., Agarwala, R., & Schäffer, A. A. (2008). Database indexing for production MegaBLAST searches. Bioinformatics, 24(16), 1757–1764.
Põldmaa, K. (2011). Tropical species of Cladobotryum and Hypomyces producing red pigments. Studies in Mycology, 68, 1–34.
Potočnik, I., Recanovick, E., Milijašević, S., Todorović, B., & Stepanović, M. (2008). Morphological and pathogenic characteristics of the fungus Cladobotryum dendroides, the causative agent of cobweb disease of the cultivated mushroom Agaricus bisporus in Serbia. Pesticide and Phytomedicine, 23, 175–181.
Potocnik, I., Vukojevic, J., Stajić, M., Rekanović, E., Milijasević, S., Todorović, B., & Stepanović, M. (2009). In vitro toxicity of selected fungicides from the groups of benzimidazoles and demethylation inhibitors to Cladobotryum dendroides and Agaricus bisporus. Journal of Environmental Science and Health B, 44, 364–370.
Richardson, R. (2010). SAMFA Chairman’s report. The Spawn Run, 1, 2–4.
Rogerson, C. T., & Samuels, G. J. (1993). Polyporicolous species of Hypomyces. Mycologia, 85(2), 231–276.
Rogerson, C. T., & Samuels, G. J. (1994). Agaricolous species of Hypomyces. Mycologia, 86(6), 839–866.
Tamm, H., & Põldmaa, K. (2013). Diversity, host associations, and phylogeography of temperate aurofusarin-producing Hypomyces/Cladobotryum including causal agents of cobweb disease of cultivated mushrooms. Fungal Biology, 117, 348–367.
White, T. J., Bruns, T., Lee, S., & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, & T. J. White (Eds.), PCR protocols. A guide to methods and applications (pp. 315–322). San Diego: Academic Press.
Zuo, B., Lu, B. H., Liu, X. L., Wang, Y., Ma, G. L., & Gao, J. (2016). First report of Cladobotryum mycophilum causing cobweb on Ganoderma lucidum cultivated in Jilin province, China. Plant Disease, 100(6), 1239.
Funding
Funding for this research was provided by the South African Mushroom Association (SAMFA), Technology and Human Resources for Industry Programme (THRIP) and the University of Pretoria. Alinesi Chakwiya is a recipient of a postgraduate fellowship from SIDA through the Organisation for Women in Science for the Developing World (OWSD), formerly TWOWS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Chakwiya, A., Van der Linde, E.J., Chidamba, L. et al. Diversity of Cladobotryum mycophilum isolates associated with cobweb disease of Agaricus bisporus in the south African mushroom industry. Eur J Plant Pathol 154, 767–776 (2019). https://doi.org/10.1007/s10658-019-01700-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-019-01700-7