Abstract
Policy makers and researchers need to develop long-term priorities using reliable, quantitative tools to assess the risks associated with plant diseases over a range of plant pathogens and over space. EPIWHEAT is a generic simulation model designed to analyse potential disease epidemics in wheat, i.e., epidemics that depend only on the physical environment, and that are not constrained by any disease control. The model is developed on a core structure involving healthy, latent, infectious, and removed sites, and accounts for lesion expansion. It simulates in a simple way host dynamics (growth and senescence). The model involves as few parameters as possible, and a few driving functions. Here, EPIWHEAT is populated with parameters for brown rust (leaf rust; Puccinia triticina) and Septoria tritici blotch (Zymoseptoria tritici). Simulated epidemics are compared to observations at the field, national (France), and European scales. The model appears to represent a sound basis for predicting potential epidemics of wheat foliar diseases at large scales. Areas for model development are documented and discussed. EPIWHEAT appears to provide a simple, generic, transparent, flexible, and reliable platform to modelling potential epidemics caused by leaf pathogens of wheat.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The variability in intensity of plant diseases across locations and seasons is a major obstacle to their strategic management. The large number of factors that may influence the occurrence and the intensity of epidemics is a main difficulty for policy makers and scientists to develop long term control strategies. Yet, only long term strategies are likely to significantly contribute to the sustainability of agricultural systems (Alston et al. 2009). Plant breeders, for example, must identify selection targets, such as disease resistances, decades ahead of the use of breeding products (Duveiller et al. 2007). This is true in particular in the case of wheat, the first food crop worldwide in terms of cultivated area and trade, and the second in terms of human consumption (Shiferaw et al. 2013).
Simulation modelling constitutes a key approach to quantitatively predict the behaviour of biological systems such as plant disease epidemics based on their underpinning processes (Rabbinge et al. 1989). Several process-based simulation models have been developed to predict the dynamics of disease intensity in specific wheat diseases. Examples include models for leaf rust (e.g., Rossi et al. 1997; Räder et al. 2007) and for Septoria blotches (e.g., Djurle and Yuen 1991; El Jarroudi et al. 2009). These models have been mainly developed in order to provide tools to help tactical decisions at the field scale, in particular to help optimizing fungicide use. Most of these models are very detailed in terms of the processes modelled (including, e.g., spore germination or spore dispersal), the canopy dynamics (including leaf layers), and sometimes require hourly weather data as input. Such models can be very useful for tactical decisions, but may be difficult to share, and may lack of transparency, because of their complexity. They are usually specific to a disease, and may address small regional areas.
Developing and deploying long term strategies for disease management first requires considering the set of main pathogens affecting a given crop plant: when considering crop health as a whole, one cannot address one disease in particular, but the entire array of diseases that may cause epidemics. In the case of wheat, one should therefore envision a group of five to 15 diseases, depending on the scale, country or continent, considered (Duveiller et al. 2007). Moreover, long term strategies require considering large spatial scales (Savary et al. 2006), which can be seen as a large (meta-)population of fields, characterized by robust estimates of epidemic patterns. Both requirements can be fulfilled by the use of a simulation model able to handle a wide range of diseases, that would be generic (same structure across diseases), transparent and simple (i.e., easy to share, to test, and to use – with parsimonious parameters and inputs), and that would provide robust estimates of epidemic intensity at large spatial scales. Such a model, EPIRICE, was developed for rice (Savary et al. 2012), whose structure accommodates fungal, bacterial, and viral pathogens, which can develop lesions on small fractions of leaves, entire leaves, entire tillers, or whole plants.
The purpose of EPIWHEAT is to provide a generic, robust modelling structure for a wide array of wheat pathogens that may cause epidemics. The anticipated utility of EPIWHEAT is to produce reasonably accurate predictions of potential epidemics, i.e., of epidemics that would only be driven by climatic conditions, and that would not be constrained or suppressed by any control action. Specifically, the blueprint of EPIWHEAT can be summarized by the following specifications:
-
1.
A simple modelling framework that enables to account for epidemiological processes, especially processes underpinning polycyclic epidemics;
-
2.
A model involving as few parameters as possible, that can be retrieved from the literature;
-
3.
A model driven by as few environmental factors as possible, which can be readily retrieved from local, regional, or international sources;
-
4.
A model that incorporates in a simplified manner crop growth and plant senescence, which are documented major causes for epidemiological variability;
-
5.
A model incorporating crop development and age, in order to account for change in host susceptibility;
-
6.
A model accounting for, if necessary, and in the most simplified way, the effect of aggregation on disease increase;
-
7.
A model that considers a convenient system size – in the case of wheat, 1 m2 of crop;
-
8.
A simple numerical integration: the rectangular (Euler; Leffelaar 1993; Thornley and France 2007) integration method over a time step of 1 day.
This article describes the structure of EPIWHEAT, and its parameterisation, evaluation, and use for two important wheat diseases in temperate areas (Duveiller et al. 2007): brown rust (also known as leaf rust) and Septoria tritici blotch.
Materials and methods
Structure of EPIWHEAT
Modelling structure
The structure of EPIWHEAT is generic, involving the main monocyclic processes underpinning plant disease epidemics. EPIWHEAT is derived in its structure from EPIRICE, a simulation model developed to analyse potential disease epidemics in rice (Savary et al. 2012). The core of this structure is based on the seminal epidemiological model developed by Zadoks (1971), with sites evolving from healthy, to latent, infectious, and removed. The structure used here further incorporates host growth and senescence in a very simplified manner. Lesion expansion, which was not accounted for in EPIRICE, is incorporated in EPIWHEAT. EPIWHEAT is parameterized for host-pathogen epidemiological interactions and crop growth of soft winter or spring wheat, but may be used for durum wheat as well. The time step of EPIWHEAT is 1 day, and the system modelled is 1 m2 of wheat crop.
Modelled processes
Brown rust and Septoria tritici blotch epidemics are modelled in the same fashion, considering four state variables that are expressed as numbers present at any time in the considered system (i.e., densities): healthy, latent, infectious, and removed sites. A site is defined as a fraction of the host tissues where an infection may take place, and where a lesion may develop (Zadoks 1971; Savary et al. 1990, 2012; Djurle and Yuen 1991). Sites becoming infected flow from healthy to latent. Latent sites become infectious at the end of a latency period, p, and infectious sites become removed at the end of an infectious period (i; Fig. 1; Table 1). The rate of infection (number of newly infected sites per unit time) is modelled as a function of the number of infectious sites (INF), a daily multiplication factor (Rc), a correction factor for diseased sites (COFR), and a coefficient for disease aggregation (Agg; Waggoner and Rich 1981):
Simplified representation of EPIWHEAT structure. Only one feed-back loop (linking INF to RI) is indicated, representing the disease cycle, from healthy (HSITE) to diseased and latent (LAT) via the rate of infection (RI), which depends on the amount of infectious (INF). Not all relationships (simple curved arrows) present in the model are indicated. See Table 1 for symbols
Equation 1 implies that the rate of variation of infected-latent sites (LAT, Fig. 1) is proportional to the number of infectious sites (INF), a power function of the proportion COFR of sites that are healthy relative to the total number of sites in the system, and to R c , the basic infection rate corrected for removals (Van der Plank 1963).
Another interpretation of Equation 1 is to consider that the basic reproduction number, R 0 (Madden et al. 2007), is equivalent to the integral over i: ∫ i Rc • dt. Equation (1) therefore amounts to: RI ≈ (R 0 • INF) / i under the following two assumptions: (1) only the first wave of infections occurring in a population of healthy sites is considered, and (2) the quantity: Rc is constant over i (Savary and Willocquet 2014).
Many pathosystems are characterized by the ability of lesions to expand (Berger et al. 1997), providing to some diseases the ability to progress even though new infections cannot occur. Lesion expansion corresponds to the speed at which tissues infected from expansion become diseased. Lesion expansion, as a process, therefore incorporates the lag phase during which tissues are colonized, but (lesion expanded) are not yet infectious. The expanded lesion area is therefore infectious almost immediately (Berger et al. 1997). Lesion expansion translates into a flow of healthy sites becoming directly infectious as lesions expand, healthy site infection thus bypassing the latent stage. Lesion expansion therefore directly depends on the number of infectious sites and (as the infection process), is constrained by the carrying capacity of the host. Lesion expansion is therefore written as:
Where RRLEX is the relative rate of lesion expansion.
Host growth is hypothesized to result from healthy sites only. Sites are defined here as leaf area units corresponding to the size of a lesion. Crop growth is represented as a logistic growth, where a maximum size of the host is considered (SMax). Crop growth is written as:
Where Total_SITES is the total number of sites currently present in the modelled system and RRG is the relative rate of crop growth.
Senescence of healthy sites is hypothesized to depend on both disease and physiological senescence of plant tissues. The two sub-processes are assumed independent and thus additive. Physiological senescence is made proportional to the number of healthy sites, and to a relative rate of senescence which increases as crop develops. Senescence associated to disease is made equal to the flow of sites becoming removed from the epidemic, so as to reflect the effect of disease on the senescence of healthy tissues (Savary et al. 2012).
Effects of crop age, temperature, and rainfall on infection
The effects of crop age, temperature, and rainfall on epidemics are incorporated in EPIWHEAT by altering the daily infection process. Rc is made dependent on RcOpt, which is the optimum value of Rc, and on modifiers (Loomis and Adams 1983) corresponding to three factors:
Where RcA, RcT, and RcW are modifiers of RcOpt for crop age, temperature, and moisture duration, respectively.
Parameterization of EPIWHEAT
EPIWHEAT was parameterized for two important wheat diseases, brown rust (BR; also referred as leaf rust, caused by Puccinia triticina) and Septoria tritici blotch (SB; Zymoseptoria tritici; former Septoria tritici; teleomorph Mycosphaerella graminicola) using published studies (Table 2).
Monocyclic processes
Values of p and i were retrieved from the literature for both diseases (Table 2). These values correspond to optimum conditions for infection, that is, to fully compatible host-pathogen interactions under conducive physical environment.
RcOpt was estimated for each disease following Savary et al. (2012), i.e., from disease progress curves representing epidemics developing under optimum environmental conditions on susceptible varieties. Several approaches to estimate R c have been proposed (Van der Plank 1963; Campbell and Madden 1990; Sun and Zeng 1993; Segarra et al. 2001). In the early stage of an epidemic:
where x 1 and x 2 are diseased fractions at two successive dates t 1 and t 2 , and r l is the apparent rate of disease increase. R c can then be estimated from:
Equation 6 was thus used to estimate RcOpt from r l , estimated from published disease progress curves (Bowen et al. 1984; Coakley et al. 1985; Table 2) under near-optimum conditions for both diseases.
Lesion expansion was documented from the literature as well. Lesion expansion is an important feature of SB. The report by Magboul et al. (1992) allowed deriving an estimate of RRLEX for this disease (Table 2). A non-zero lesion expansion for BR is indicated in Berger et al. (1997), but expansion is comparatively very small. For the sake of simplicity, it was assumed that RRLEX = 0 in the case of BR. Lesion sizes for both BR (Robert et al. 2004) and SB (prior to lesion expansion and coalescence; James 1971; Zhang et al. 2011) were retrieved from literature.
Disease aggregation was a point of specific consideration. The two diseases quite differ in their dispersal (e.g., Bockus et al. 2010), BR having a predominantly wind-driven, dry dispersal mechanism, whereas SB is commonly assumed to have a strong rain splash dispersal component (e.g., Magboul et al. 1992; Bockus et al. 2010). One may therefore expect different levels of overall disease aggregation, which may further be expected to differentially interact with crop growth, so that Agg not only varies over time (Campbell and Madden 1990; McRoberts et al. 1996), but also with crop growth and canopy structure (e.g., Lannou and Savary 1991; Yang and TeBeest 1992; Xu and Madden 2002). For the sake of simplicity again, a neutral hypothesis was made, and Agg was set to 1 for both diseases.
Modifiers
Functional responses of BR to temperature and plant age were derived from De Vallavieille-Pope et al. (1995) and Tomerlin et al. (1984), respectively (Table 2). Wetness was assumed not to influence the monocycle of BR (Bockus et al. 2010). Conversely, functional responses of SB to temperature and wetness were derived from Magboul et al. (1992), and the response to plant age was documented from Wainshilbaum and Lipps (1991). The standard procedure to link driving functions (e.g., temperature) to functional responses (e.g., infection efficiency) was used, that is, through linear interpolation thus producing the desired numerical response to a given running value of the driving function between two experimentally measured (driving function) - (response) couplets (Savary and Willocquet 2014).
Carrying capacities, crop growth and plant development
SMax (Fig. 1, Table 1) represents the maximum number of sites that can be sustained by a canopy covering 1 m2 of a wheat crop, i.e., the disease carrying capacity. SMax values (Table 2) were determined considering (1) an LAI of 3, (2) the maximum number of lesions that can be carried by a leaf given (3) an average lesion size, and (4) an average leaf size (18.27 cm2; Zhang et al. 2011). In the case of BR, an estimated SMax value of 750,000 was derived, assuming a lesion size of 4 mm2 (Robert et al. 2004). In the case of SB, the Smax value was 174,055, considering a maximum possible severity (% leaf area actually covered with lesion) of 25 %, consisting of 106 individual lesions per leaf (James 1971). A maximum SB severity of 25 % concurs with observations (22.87 %) by Eyal and Brown (1976).
The relative rates of growth (Spitters and Kramer 1985) and senescence (Willocquet et al. 2008) of healthy sites were determined from the literature, as well as crop development, which was described as a function of the daily temperature sum using a base temperature of 0 °C (Spitters et al. 1989; Willocquet et al. 2008)
Model initialization
The initialisation of EPIWHEAT was designed so that simulations begin when spring growth starts, i.e., when the sum of temperature accumulated since January 1 has reached 200 °C.day (base 0 °C; Spitters et al. 1989), in the case of winter wheat. This corresponds to a development stage of approximately 0.33 on a 0–2 scale (Penning de Vries et al. 1989). The initial development stage of wheat growth was set to 0.33 at the beginning of each run (day = 0), which corresponds to a sum of temperature of 500 °C.day from seedling emergence. Epidemics are initiated by a single inflow (INOCP, Fig. 1) of infections (10 infections in the case of BR, and 15 in the case of SB) at day = 20. The initial number of sites was set to 250 for both diseases, which is commensurate with the smallest LAI value reported by Groot (1987) at the end of winter.
Driving functions
Daily mean temperature and rainfall (Fig. 1) were used as drivers influencing RcT and RcW (the latter set to a default 1 value for BR). A simple rule was applied to translate rainy days into wetness effects on infection efficiency in SB (Magboul et al. 1992) via the wetness modifier RcW (Fig. 1, Table 1): absence of rainfall over four consecutive days corresponds to a RcW = 0.23, 1 rainy day to RcW = 0.36, and 2, 3, and 4 consecutive rainy days to RcW values of 0.68, 0.93, and 1, respectively (Magboul et al. 1992). Daily mean temperature was also used as a driver to compute daily changes in crop development stage.
Starting day 1, running time and driving functions start feeding the computation sequence, and the numerical integrations follow the rectangular (Euler; Leffelaar 1993; Thornley and France 2007) method for each day.
Model evaluation
Given the objectives of EPIWHEAT, model evaluation was conducted not only at the field scale, but also at the regional (country) scale, in order to evaluate the capacity of EPIWHEAT to provide robust estimate of epidemic severity at a spatial scales that are beyond the individual field.
Comparison of simulated and reported disease progress curves
Comparison of observed and simulated epidemics was conducted using published disease progress curves for both diseases, with two steps: (1) visual examination of the dynamics of simulated vs. observed disease severity (Penning de Vries 1977) and (2) regression analyses of simulated over observed values (Teng 1981). Disease progress curves under conditions where disease was not controlled (use of susceptible varieties, no chemical protection) were sought in the literature; disease progress curves reported by Kulkarni et al. (1981) were used for BR, and from Hess and Shaner (1987a) for SB. The corresponding weather data (mean daily temperatures and rainfall) were retrieved from the India Meteorological Department website (http://www.imd.gov.in/) and from the Indiana State Climate Office, Purdue University website (http://www.iclimate.org/data_archive.asp), for BR and SB, respectively.
Comparison of simulated and reported disease occurrence in France
The French ministry for agriculture (Organisation Nationale de la Protection des Végétaux, ONPV) publishes yearly reports summarizing the status of plant health in the past growing season for a number of crops. We used the reports for the period 2005–2010 for wheat. These reports describe the overall levels of the various wheat diseases in the form of maps or tables where the regions of France are displayed and disease intensities are indicated in three classes: “severe”, “average”, and “weak to absent”. Some reports split the “weak to absent” in two. Some regions do not report information in some years, thus leading to missing data. These reports were compiled in yearly tables for each of the two diseases for the period 2005–2010, where only three classes of observations (BRobs and SBobs) were considered: “weak to absent”, “average”, or “severe”, and coded as: ‘0’, ‘1’, and ‘2’, respectively.
The regional crop health assessments provided information on BR and on Septoria blotches. While little doubt exists on the diagnosis of BR in these reports, no distinction is made in the reports between Septoria tritici blotch (Zymoseptoria tritici; teleomorph Mycosphaerella graminicola) and Septoria nodorum blotch (caused by Parastagonospora nodorum; syns. Stagonospora, Septoria, Phaeosphaeria nodorum). Z. tritici is widely predominant in France, and therefore the reported “Septoria” disease was attributed to this disease.
The observed levels were then binarized. Reported BR levels (BRobs) in the regional crop health assessments indicated a majority of non-epidemic region-years (i.e., BRobs = 0), especially before 2007. All region-years with BRobs = 1 or 2 were therefore categorized as ‘epidemic’ (i.e., BRobs = 1). On the contrary, a majority of the reported SB levels (SBobs) indicated occurrence of the disease at “average” level (i.e., SBobs ≠ 0). Only region-years were SB was reported as “severe” were therefore categorized as ‘epidemic’ (i.e., SBobs = 1, and SBobs = 0 otherwise).
Weather data corresponding to French regions and years were retrieved from the MARS (Monitoring Agricultural ResourceS; see details below) meteorological database, enabling simulations for both diseases, and the computation of areas under disease progress curves (AUDPC; Campbell and Madden 1990) for a period of 120 days for each region-year. The simulated AUDPCs (AUBRPC and AUSBPC for BR and SB, respectively) were in turn converted in binary variables (BRsim and SBsim, respectively) using the means and standard deviations of the simulated AUDPCs:
The association between the binarized simulated AUDPCs (BRsim and SBsim) and reported disease intensities (BRobs and SBobs) were tested by means of chi-square tests. Furthermore, logistic regressions involving the binarized reported intensities and the continuous simulated AUDPC variables were then tested:
Logistic regressions and statistical tests associated to them, and to Bayesian statistics in general, provide a powerful and formal, as well as an intuitive way to assess a model during its development phase and to test its ability to predict and to guide for decision (Yuen and Hughes 2002). Such an approach has been used in several instances to develop and assess models for disease management (i.e., Pethybridge et al. 2009; Caffi et al. 2011).
Sensitivity analyses
Sensitivity analyses were performed to assess the model behaviour with varying input parameters, therefore enabling model verification (Teng 1981). Four parameters were addressed: RcOpt, the timing of inflow of primary infections (Day, Fig. 1), temperature (T), and rainfall patterns (rain, only for SB) in simulation runs of 120 days.
The default values of RcOpt were set to 1.472 and 1.17–RRLEX for BR and SB, respectively. For each disease, three values of RcOpt were considered: the default RcOpt value, and the default value + or − 20 %. The default value for Day was set to 20 for both diseases. Three values were again considered: the default Day value and its variation of + or − 20 %. T was made constant over the simulations. Default values of 15 and 22 °C (corresponding to optimum temperatures for infection efficiency for each disease) were set for BR and SB respectively. For each disease, two T values were used: the default value and the default value − 20 %. The default rainfall patterns (rain) considered for SB corresponded to 12 rainy days (chosen at random) out of 30 days. This frequency corresponds to the average number of rainy days per month in France from March to July, when accounting for the range of climatic regions of wheat production. Patterns with an additional 20 %, and a reduced 20 % rainy days were then considered. Two or three additional (or removed) rainy days were thus chosen at random every other month, and added (or removed) to the rainy patterns set in the first stage, in order to consider the 20 % variation above (or below) the default rainy pattern. All combinations of levels of inputs yielded 18 and 54 simulations for BR and SB, respectively. The dynamics of severity were displayed for most combinations for visual examination, and AUDPCs were computed for all combinations.
Simulation of potential epidemics in Europe
Weather data
Daily meteorological data from 1993 to 2012 (20 years) on a 50 × 50 km grid across Europe and its neighbouring countries were retrieved from the MARS (Monitoring Agricultural ResourceS) meteorological database at http://mars.jrc.ec.europa.eu/mars/About-us/AGRI4CAST/Data-distribution/AGRI4CAST-Interpolated-Meteorological-Data. These data are generated from spatial interpolation of weather data collected by meteorological stations. A detailed description of the database is available at: http://marswiki.jrc.ec.europa.eu/agri4castwiki/index.php/Meteorological_data_from_ground_stations. Three daily meteorological variables were used: mean rainfall (rain, mm), and maximum (Tx, °C) and minimum (Tn, °C) daily temperatures.
Simulation runs
EPIWHEAT was developed using STELLA® (2008) and translated to the R language (R Development Core Team, 2014, http://www.r-project.org) for multiple simulations purpose. R is a free, multiplatform, statistical programming environment, which can be linked to database and GIS softwares as was done for rice diseases (Savary et al. 2012). For each disease, the model was run at all locations of the MARS grid, i.e., for all year-site climatic situations using the same initial conditions as indicated above, and the AUDPC of epidemics were computed over 120 days.
Disease maps
Elevation data were retrieved from http://api.geonames.org/astergdem (derived from the Aster Global Digital Elevation Model; http://asterweb.jpl.nasa.gov/gdem.asp). Points of the weather database grid corresponding to an elevation above 1000 m were excluded. The spatial grain chosen to display simulation outputs corresponds to the NUTS (Nomenclature of territorial units for statistics classification) European geographic units. This spatial grain (called level 2, or NUTS2) corresponds to a total of 270 units in Europe (and translates for France in its 22 administrative “Régions”). This spatial grain also allows using base maps from the EUROSTAT website: (http://epp.eurostat.ec.europa.eu/portal/page/portal/nuts_nomenclature/introduction).
The means and standard deviations over years of AUDPCs in each NUTS were computed and partitioned in quintiles corresponding to different shades. Maps displaying these means and standard deviations of potential BR and SB epidemics were developed.
Evaluation of simulated maps for potential epidemics
A third phase of evaluation consisted in assessing the simulated potential epidemic maps. A key reference for this purpose is the Atlas of Cereal Diseases and Pests in Europe (Zadoks and Rijsdijk 1984). It is important to note that the Atlas (1) was often based on ‘soft’ rather than on ‘hard’ evidence, (2) was developed using information that pertains to the period 1965–70, and (3) did not refer to epidemics, but rather to crop losses. Comparison of simulated outputs with maps of the Atlas should therefore be done carefully, considering broad patterns rather than specific geographical details.
Results
Model evaluation - disease progress curves
Figure 2a indicates a good agreement of observed (symbols) vs. brown rust (BR) simulated severities (curve). EPIWHEAT tends to slightly overestimate severities in the beginning of epidemic, but as the epidemic amplifies, the adjustment of simulated and observed severities seems good. A regression of simulated on observed BR severities (R 2 = 0.979; regression: F = 187.69, P < 0.001) yielded a non-significant intercept (b = 0.067, SE = 0.041, P = 0.180) and a significant slope (a = 0.849, SE = 0.062, P = 0.0002). t-tests of the null hypotheses that b equals 0 and a equals 1 yielded P values of 0.18 and 0.07, respectively.
The simulated progress of Septoria tritici blotch (SB) seems to produce a better fit than that of BR in the beginning of the observed epidemic (Fig. 2b), but as the epidemic intensifies, EPIWHEAT tends to progressively overestimate severity. A regression of simulated on observed SB severities (R 2 = 0.991; regression: F = 289.85, P < 0.001) yielded a non-significant intercept (b = −0.021, SE = 0.033, P = 0.554) and a significant slope (a = 1.211, SE = 0.071, P < 0.001). t-tests of the null hypotheses that a equals 1 and b equals 0 yielded P values of 0.55 and 0.03, respectively.
Model evaluation - disease variation in French regions
Data retrieved from the Organisation Nationale de la Protection des Végétaux (ONPV) indicated relatively regular variation in SB over the six considered years (2005–2010), but a sharp increase of BR in 2007. That year actually corresponds to the emergence of new BR pathotypes, associated with the widespread deployment of resistance genes Lr10, Lr13, and Lr37 (Huerta-Espino et al. 2011; Goyeau and Lannou 2011), as well as Lr14a and Lr23 in durum wheat (Goyeau et al. 2012). Important changes in the ranking of host plant resistances to BR were therefore expected to be reflected in the data after 2006, and therefore our analyses were limited for this disease to the years 2005 and 2006.
Building a contingency table for observed and simulated BR levels, there was one false negative (BRsim = 0 whereas BRobs = 1), 4 false positives (BRsim = 1 whereas BRobs = 0), 12 true negatives (BRsim = BRobs = 0), and 24 true positives (BRsim = BRobs = 1), totalling 41 cases (Table 3). This yielded a chi-square value of 22.7 allowing rejection (P < 0.001) of the null hypothesis that BRobs and BRsim are independently distributed. Analysis of binarized information on SB yielded 0 false positives and negatives, 71 true negatives, and 23 true positives, and a chi-square value of 94.0, leading to rejection (P < 0.001) of independence of binarized distributions of SBsim and SBobs (Table 4).
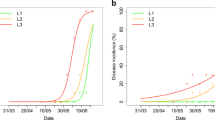
A plot comparing the three classes of reported disease levels (“weak to absent”, “average”, and “severe”) with the simulated areas under disease progress curves (Fig. 3) indicates that EPIWHEAT was unable to distinguish “severe” from “average” BR levels, but was however a reasonable predictor of “weak to absent” vs. other levels (Fig. 3a). EPIWHEAT performed better for SB (Fig. 3b), with gradual distinction between the three levels, despite some overlap.
Comparisons between disease intensity reported in wheat health annual reports (BRPV and SBPV, abscissa) and areas under simulated disease progress curves (AUDPC, ordinates) in French regions. a: leaf rust (2005 and 2006 only, see text); b: septoria blotch (2005–2010). Abscissas: reported disease levels. Ordinates: areas under simulated disease progress curves for brown rust (BR) and septoria blotch (SB). Box plots indicate medians between first and third quartiles (boxes) and further interquartile ranges (bars). Insert in each box plots are the ROC curves corresponding to the simulated area progress curves as predictors of binarized (see text) reported disease intensities in logistic regressions. The logistic models (Table 3) are of the shape: ln [Bin(xONPV)/ (1 - (Bin(xONPV))] = α + β * AUxPC. where Bin (xONPV) refers to binarized (see text) reported disease (x) levels
Further analysis of observed (ONPV) and simulated epidemics involved the binarized version of the observed epidemics (i.e., ‘epidemic’ vs. ‘non-epidemic’) and the numerical values of the areas under the simulated disease progress curves (AUBRPC and AUSBPC for BR and SB, respectively).
Both logistic models were significant (P < 0.01, Table 5), with likelihood ratios of 21.2 and 10.6 for BR and SB, respectively. Both models yielded significant (P < 0.01, Table 5) and positive estimates of slope parameters. These logistic models correspond to areas under the receiver operating characteristic curves (ROC) of 0.87 for BR and 0.72 for SB. ROC curve displays the relationship between the sensitivity (proportion of true positive; TPP) and 1 - specificity (proportion of false positive; FPP) of the model. ROC curves with shapes corresponding to TPP close to 1 and FPP close to zero correspond to a model with good performances, i.e., enabling a good discrimination of epidemics vs. non epidemics (Hughes et al. 1999).The shapes of the ROC curves (Fig. 3) show the differences in logistic predictions, largely binary (“weak to absent” vs. “average” or “severe”) in the case of BR, and more gradual for SB.
Sensitivity analyses
Sensitivity analysis of EPIWHEAT for BR indicates a large effect of Rc (Fig. 4, A1), with epidemics achieving faster saturation of the host population when Rc is high. Comparing disease progression curves may sometimes be misleading. The distribution of simulated areas under progress curves (Fig. 4, A4) confirms the overall effect of variations in Rc: each of the 3rd areas are larger than the two preceding ones among the parameter combinations tested. A similar conclusion is reached with the day of epidemic initiation day (Fig. 4, A2); however, the effect of earlier epidemic initiation seems much stronger than that of a later one (Fig. 4, A4). Temperature T (Fig. 4, A3) has also a very strong effect on epidemics, the higher temperatures leading to stronger epidemics (Fig. 4, A4).
Sensitivity analyses of EPIWHEAT. a. Brown rust simulations. 1, 2, 3, disease progress curves at varying values of Rc, day, and T, respectively. 4, corresponding areas under disease progress curves represented as bars. The reference for Rc, day, and T is shown in dark. b. Septoria tritici blotch simulations. 1, 2, 3, 4, disease progress curves at varying values of Rc, day, T, and rain, respectively. 5, corresponding areas under disease progress curves represented as bars. The reference for Rc, day, T, and rain is shown in dark. See text for details
SB epidemics, too, appear very sensitive to Rc (Fig. 4, B1); as for BR epidemics, each of the 3rd areas under progress curve is always higher than the preceding two (Fig. 4, B5). The effect of day (Fig. 4, B2) seems higher in this case, with regular decreases from early, to medium, and to late onsets (Fig. 4, B5). One of the strongest effects seems to be that of T (Fig. 4, B3), a reduction in T leading to a strong reduction of epidemic. By contrast, the effect of rain seems moderate, with disease curves largely overlapping (Fig. 4, B4), and this is confirmed by the comparisons of disease progress curves (Fig. 4, B5). Simulations conducted with a number of rainy days reduced by 20 % as compared to the default value yielded to simulated epidemics with reduced speed, and to reduced AUDPC (results not shown). The range of reduction (in epidemic speed and AUDPC) was similar to the range of increase obtained from sensitivity analyses with a number of rainy days increased by 20 % (corresponding to Fig. 4).
Potential wheat epidemics in Europe
The simulated potential epidemics generated by EPIWHEAT, expressed as areas under disease progress curves (AUDPCs), are mapped in Fig. 5.
EPIWHEAT simulated distribution of potential brown rust (a and b) and Septoria tritici blotch (c and d) in Europe using daily meteorological data from 1993 to 2012 (20 years) on a 50 × 50 km grid across Europe. Maps indicate quintiles of the distributions of means (a and c) and of standard deviations (b and d) of the areas under simulated disease progress curve over 120 days
Potential BR epidemics (mean AUBRPC, Fig. 5a) are expected to be on average stronger in the north-eastern parts of Europe, owing to abrupt springs and relatively warm summers. They also are predicted to be strong in the Southern part of the Nordic countries, as a result of cool springs and relatively warm summers as well. They are predicted to be strong in the southern part of the Iberian Peninsula too (as well as in Sicily), which reflects a rapid crop growth and development in the spring, and warm, but not hot, periods when the crops approach maturity. Western Europe, with mild weather in the spring (allowing rapid crop development, but not necessarily favouring BR multiplication) and cool summer (that are sub-optimal for rust) are not mapped as conducive to strong epidemics. Central Eastern Europe is predicted unfavourable too, reflecting surging temperatures when the crop approaches maturity. The map for standard deviations in mean values of areas under progress curves of potential BR (Fig. 5b) indicates a strong West–east gradient, variability increasing when moving East, where temperature variability in the end of the cropping season increases.
The mean values of potential SB (Fig. 5c) epidemics indicate by contrast a clear West–east decreasing gradient of intensity. This spatial pattern may be attributed to the combined effects of temperature and rainfall patterns. The more humid, cooler parts of Europe (including parts of the Alps where wheat cultivation below 1000 m of elevation is considered in these simulations) are predicted to be more strongly affected by the disease. The distribution of standard deviation of AUSBPC (Fig. 5d) appears in agreement with this pattern, as the strongest variances are found in the western part (where summers may vary in wetness and temperatures) and the eastern part (where summers may be more rainy in some years) of Europe.
Discussion
Modelling objectives
The objective of this work was to develop and test the generic value of a modelling structure that can mobilize existing quantitative information on some key wheat diseases, and generate robust estimates of patterns of potential epidemics at spatial scales higher than the individual field. This structure was designed to be as simple, involving as few parameters, as possible, in order to be easily tested, improved, shared, and used. In the present work, the modelling structure has been populated with processes and parameters pertaining to two important wheat diseases, but it is flexible enough to be modified so as to address other wheat diseases. The geographical domain addressed here is Europe and France, but the model structure should enable applications in any environment where wheat is grown in the world.
Specifically, we made use of (1) a well-established modelling framework that has been applied in a number of pathosystems, temperate or not, on cereals or on other crops, and (2) a set of published studies directly providing, or enabling easy calculation of, the required parameters. The modelling structure is based on the design of Zadoks (1971) as a numerical integration of the integro-differential equation 8.3 of Van Der Plank (1963), which Madden et al. (2007, Chap. 5) discuss in detail. A number of epidemiological simulation models using this design have been developed successfully, for cereal diseases (e.g., Teng et al. 1980; Djurle and Yuen 1991; Rossi et al. 1997) or dicotyledonous crops (e.g., Savary et al. 1990; Berger et al. 1995). The epidemiological characteristics of the two wheat diseases considered here, on the other hand, are quite well documented, so that parameterization of EPIWHEAT using reliable data was made possible (Table 2).
Key hypotheses
The specifications of EPIWHEAT listed in the Introduction are driven by the overall objective of modelling patterns of potential epidemics, that is, of epidemics that only depend on physical environmental conditions, and therefore that are not constrained by any disease management actions, whether direct or indirect (i.e., host plant resistance, chemicals, crop management, landscape and timing of crop establishment).
The structure of EPIWHEAT is congruent with our modelling objectives and with the specifications listed in its blueprint. These specifications have generated a series of key modelling hypotheses, which we can list as follow:
-
a.
Specification 1 (a simple model structure) implies that the spatial properties and attributes of epidemics are not addressed in EPIWHEAT, except for the inclusion of an aggregation coefficient (specification 6). The model therefore implicitly assumes random distribution of diseased sites and uniform vulnerability (equation 1) of healthy sites. This hypothesis corresponds to the implicit assumption that any healthy site may become infected, and derives from the integro-differential model of Van der Plank (1963). Such a simplifying hypothesis is shared by a number of epidemiological models, which nevertheless have proven to be acceptable research tools (Zadoks and Schein 1979). This simplifying hypothesis does not appear to hamper strongly EPIWHEAT as a model for potential epidemics; this is further addressed below.
-
b.
Specifications 1, 2 (as few processes and parameters as possible) and 3 (few environmental factors considered) lead to the use of modifiers that reflect daily changes in temperature and (for SB) in wetness. This modelling hypothesis leads to processing environmental information in a most simplified manner, which may entail risks. The use of mean daily temperature, for instance, leads to erasing daily temperature variation that may have important consequences on epidemiological processes (e.g., Scherm and Van Bruggen 1994). Similarly, the representation of canopy moisture is very crude. While improvements of these aspects should be sought, it seems that the resulting outputs can provide a reasonably good working basis.
-
c.
Specifications 1 and 4 (inclusion of crop growth and plant senescence) lead to using a simple logistic function to account for crop growth, and to make the rate of senescence an additive function of (exponential) senescence and of (disease-induced) removal (Fig. 1, Table 1). The modelling of crop dynamics provides much more realism to the simulations. Incorporation of crop growth in an epidemiological simulation model does represent a considerable improvement of its own (Campbell and Madden 1990; Savary et al. 2012).
-
d.
Specification 5 (effect of crop development on host susceptibility) corresponds to a very simple function (RcA, Table 2) representing physiological age; this constitutes the only element to characterize the quality of healthy sites in the model. Under this hypothesis, a simple, tabular representation (RcA, Table 2) of change in site susceptibility is supposed adequate. Our simulation outputs suggest that this seems to be the case.
-
e.
Specification 6 (accounting for aggregation) provides some flexibility in the capacity of EPIWHEAT to reflect some spatial structure, although in a very simple manner (which was not actually put into use in the two pathosystems considered here). Further, the combination of site age (RcA, above), aggregation (current hypothesis), and crop growth (modelling hypothesis c) can produce an implicit representation of disease in time and space.
-
f.
Specifications 7 (system’s dimension of 1 m2) and 8 (1-day time-step, Euler integration method) further emphasize the design of a very simple model structure, which assumes a system in a steady state of propagule inflow and outflow (specification 6), and implies that the model must involve processes with time characteristics matching this time-step. This modelling hypothesis implies that processes underpinning epidemics may be accounted for in such a small system size, and with a 1 day time step. This hypothesis is commonly used in epidemiological systems (e.g., Teng 1985; Rossi et al. 1997; Savary et al. 2006). It entails explicit and implicit assumptions which deserve further discussion, even for a generic model of potential epidemics.
Evaluation of hypotheses
EPIWHEAT entails a number of simplifying hypotheses, some of which are inherent to the class of epidemiological models it belongs to, and others that result from its particular features. We shall not dwell on the former, which have been discussed in detail (Zadoks 1971; Teng et al. 1980; Teng 1985; Madden et al. 2007; Segarra et al. 2001; Savary and Willocquet 2014), and rather emphasize the latter.
An important hypothesis is related to the use of an integration time step of 1 day. This choice amounts to consider that all the processes considered in the system have time characteristics such that their variation can accurately be modelled within a 1 day time-step. The hypothesis can be summarized in saying that variation of each of the six processes (i.e., their six rates) shown in Fig. 1 – for example the rate of infection (RI) – can be reliably modelled using a 1-day interval. This has implications in terms of reaction time of the system modelled (Leffelaar 1993). Many epidemiological mechanisms involved in the infection process actually have time constants that are quite smaller than 1 day: for instance, spores may be liberated within seconds through, for example, the ‘tap and puff’ phenomenon (Hirst and Stedman 1963); spore deposition may be slow, or quite fast, for instance in the case of dry spore deposition on wet canopies (Chamberlain 1967; Savary et al. 1990); and the sequence from propagule germination to infection initiation may take place at different periods of the climatic day, sometimes favourable and consecutive (and thus, within a few hours), or sometimes unfavourable, causing delays of several days (Zadoks and Schein 1979). These infection sub-processes, and many others, each may take place in less than 1 day, leading to the biological possibility of infection itself taking less than a day.
Rapilly (1983) referred to the concatenation of sub-processes as ‘epidemiological sequences’, which constitute the biological heart of an epidemic. In detailed simulation models, one approach is to consider daily weather patterns where the relative rate of each sub-process is assigned a value, given the current state of a range of environmental variables represented by driving functions (e.g., Savary et al. 1990). Despite their importance, sub-processes cannot be explicitly incorporated in simple modelling structures: they must be summarized. In a very simple manner, EPIWHEAT uses daily weather patterns which translate into changing values of modifiers (RcT and RcW, Table 1).
EPIWHEAT involves only two climate driving functions, daily rainfall and temperature ― far fewer than most mechanistic epidemiological simulation models (e.g., Rossi et al. 1997). This is a considerable advantage in terms of portability of the structure: most global climate data provide this basic climate information. However, this ignores the considerable knowledge accumulated on the strong effects of moisture on Septoria blotches (e.g., Hess and Shaner 1987b; Magboul et al. 1992). Djurle and Yuen (1991) for instance compute leaf moisture on 10 canopy layers in their detailed model for Septoria nodorum blotch. Another example is the effect of leaf moisture on the infection efficiency of P. triticina (Rossi et al. 1997), which was not incorporated here. Further evaluation of EPIWHEAT, especially when additional diseases are considered, should further assess the need of incorporating additional driving climate functions.
A common limitation of simple epidemiological models such as EPIWHEAT is the lack of account for the spatial disease aggregation, which can have important consequences on the speed of epidemics (Madden and Hugues 1995). The aggregation coefficient introduced in the model may help overcoming this limitation. Campbell and Madden (1990) however pointed to issues with the approach of Waggoner and Rich (1981), which we used: disease aggregation is dynamic, too, and cannot be constant throughout an epidemic. Additional work is necessary on this aspect in two directions, first a further evaluation of the use of an aggregation coefficient, and second, the implementation of a dynamic aggregation.
In EPIWHEAT, disease-induced senescence is made equal to the rate of site removal. The underlying assumption is that the number of sites senescing from disease corresponds to the number of sites becoming removed from the epidemiological process. While this may be an acceptable simplification in the case of necrotrophic pathogens, it may be questioned for biotrophic pathogens. This is however rendered suitable by the very long infectious period of BR (31 days, Table 2).
The simulations with EPIWHEAT used fixed values of p and i for the two diseases. Both parameters are well known to vary, especially with temperature, in the course of an epidemic. Further, variation of p and i do affect very strongly the speed of epidemics (Zadoks and Schein 1979; Savary and Willocquet 2014). Further improvements of EPIWHEAT will have to incorporate variable latency and infectious periods, which are well documented in the literature.
The modelled pattern of epidemic initiation is another element which entails hypotheses. Epidemics have been initiated as single impulses of inoculum (INOCP) injected in the system at a given date (START, Fig. 1, Table 2). The analysis of epidemiological importance of the primary inoculum made by Bergamin Filho and Amorim (1996) underlines the very large differences in epidemics resulting from the temporal shape of primary inoculum inflow. This concurs with simulation results obtained by Savary et al. (1990). Further, there is accumulated knowledge on wheat diseases (e.g., SB; Suffert et al. 2011) to indicate that complex patterns of inflows of primary inoculum may occur. New research (Duvivier et al. 2013) suggests that disease spread in septoria tritici blotch may owe much to dispersal of ascospores, rather than pycnidiospores. This has potentially strong implications on (1) the amount of primary inoculum that initiates epidemics, (2) the pattern of initial inoculum inflow, and (3) secondary infections throughout entire SB epidemics. Better representation of epidemic onset, through (1) the initial influx of infections (INOCP), and (2) its pattern of distribution over time (START), possibly represents the single most important change to incorporate in the modelling structure, and warrants the need for further improvements.
Model evaluations
The evaluation of EPIWHEAT was conducted in successive phases. The first phase entails the conventional comparison of simulated vs. actually observed disease progress curves. The overall shapes of disease progress curves (Penning de Vries et al. 1989) for both BR and SB correspond well to the observed patterns for the two diseases. Visual examination of simulations vs. observed data suggest that EPIWHEAT provides a good account of the early progress of SB epidemics, which frequently show a lag phase in its early increase (e.g., Thomas et al. 1989), compared to that of typical rusts such as P. triticina. From a formal statistical standpoint, the comparison between observed and simulated severities suggests that EPIWHEAT is correctly simulating BR epidemics, whereas SB epidemics are over-estimated by the model at the end of epidemics (Fig. 2b). Overall, the performances of EPIWHEAT nevertheless appear to be acceptable with respect to this first evaluation phase.
Comparison of EPIWHEAT outputs with qualitative data retrieved from reports of the Organisation Nationale de la Protection des Végétaux (ONPV) allowed assessing the model in a range of French regions over two (2005–2006) or six (2005–2010) years for BR and SB, respectively, i.e., at a spatial scale beyond the individual field. In a first stage, chi-square tests allowed to reject (P < 0.001) the null hypothesis that the distribution of binarized simulation outputs (‘epidemic’ vs. ‘non-epidemic’) and the binarized reported (ONPV) levels of disease were independently distributed for both diseases. In a second stage, logistic regressions of the likelihood of strong epidemics (“average” or “severe”, and “severe”, for BR and SB, respectively) being observed were tested, using the simulated areas under (120-day) disease progress curves as predictors. Both logistic models were significant, with positive, significant coefficients. This second phase of evaluation suggests that EPIWHEAT is sufficiently robust to account for epidemics occurring in different locations and climatic conditions.
A third phase of evaluation consists in comparing the simulated potential epidemic maps with maps of the Atlas of Cereal Diseases and Pests in Europe (Zadoks and Rijsdijk 1984), bearing in mind the differences and reservations indicated earlier in Materials and Methods. As the Atlas does, the EPIWHEAT-generated map for BR shows an increasing disease gradient from West to East, with a maximum level in Central Europe (Fig. 5a). But unlike the Atlas, the simulated map shows a decline east of the Carpathian mountains; and unlike the Atlas, the simulated map shows no BR decline in Northern Germany, Poland, and the Nordic countries. The simulated variance however increases in Northern Europe, and even more so, in Eastern Europe, while the Atlas (by construction) provides no indication of variability. The simulated outputs therefore do indicate that strong epidemics can occur in Eastern Europe, and are weaker in Northern Europe, as the Atlas indicates. Simulated maps for SB (Fig. 5c and d) concur with the Atlas, with strong epidemics occurring in the British Isles and Northern France, and a steady decline towards the south-east of Europe. Overall, the mapped outputs from EPIWHEAT can therefore be considered satisfactory.
In spite of several shortcomings that are mostly reflecting its simplicity, and which can in many cases be addressed, EPIWHEAT therefore appears to represent a sound basis for predicting potential epidemics of wheat foliar diseases. Further efforts will focus on the improvement of the predicting ability of the model from the careful addition of biological characteristics of brown rust and Septoria tritici blotch, and on the expansion of EPIWHEAT to a range of wheat diseases.
References
Alston, J. M., Beddow, J. M., & Pardey, P. G. (2009). Agricultural research, productivity, and food prices in the long run. Science, 325, 1209–1210.
Bergamin Filho, A., & Amorim, L. (1996). Doenças de plantas tropicais: epidemiologia e controle econômico. Sao Paulo: Editora Agronômica Ceres Lda.
Berger, R. D., Hau, B., Weber, G. E., Bacchi, L. M. A., Bergamin Filho, A., & Amorim, L. (1995). A simulation model to describe epidemics of rust of Phaseolus beans: I. Development of the model and sensitivity analysis. Phytopathology, 85, 715–721.
Berger, R. D., Bergamin Filho, A., & Amorim, L. (1997). Lesion expansion as an epidemic component. Phytopathology, 87, 1005–1013.
Bockus, W., Bowden, R., Hunger, R., Morrill, W., Murray, T., & Smiley, R. (Eds.). (2010). Compendium of wheat diseases and pests (3rd ed.). St Paul: APS.
Bowen, K. L., Teng, P. S., & Roelfs, A. P. (1984). Negative interplot interference in field experiments with leaf rust of wheat. Phytopathology, 74, 1157–1161.
Caffi, T., Rossi, V., Legler, S. E., & Bugiani, R. (2011). A mechanistic model simulating ascosporic infections by Erysiphe necator, the powdery mildew fungus of grapevine. Plant Pathology, 60, 522–531.
Campbell, L. C., & Madden, L. V. (1990). Introduction to plant disease epidemiology. New York: Wiley.
Chamberlain, A. C. (1967). Deposition of particles to natural surfaces. In P. H. Gregory & J. L. Monteith (Eds.), Airborne microbes (pp. 138–164). Cambridge: University Press.
Coakley, S. M., McDaniel, L. R., & Shaner, G. (1985). Model for predicting severity of Septoria tritici blotch on winter wheat. Phytopathology, 75, 1245–1251.
De Vallavieille-Pope, C., Huber, L., Leconte, M., & Goyeau, H. (1995). Comparative effects of temperature and interrupted wet periods on germination, penetration, and infection of Puccinia recondita f.sp. tritici and P. striiformis on wheat seedlings. Phytopathology, 85, 409–415.
Djurle, A., & Yuen, J. E. (1991). A simulation model for Septoria nodorum in winter wheat. Agricultural Systems, 37, 193–218.
Duveiller, E., Singh, R. P., & Nicol, J. M. (2007). The challenges of maintaining wheat productivity: pests, diseases, and potential epidemics. Euphytica, 157, 417–430.
Duvivier, M., Dedeurwaerder, G., De Proft, M., Moreau, J. M., & Legrève, A. (2013). Real-time PCR quantification and spatio-temporal distribution of airborne inoculum of Mycosphaerella graminicola in Belgium. European Journal of Plant Pathology, 137, 325–341.
El Jarroudi, M., Delfosse, P., Maraite, H., Hoffmann, L., & Tychon, B. (2009). Assessing the accuracy of simulation model for Septoria leaf blotch disease progress on winter wheat. Plant Disease, 93, 983–992.
Eyal, Z., & Brown, M. B. (1976). A quantitative method for estimating density of Septoria tritici pycnidia on wheat leaves. Phytopathology, 66, 11–14.
Goyeau, H., & Lannou, C. (2011). Specific resistance to leaf rust expressed at the seedling stage in cultivars grown in France from 1983 to 2007. Euphytica, 178, 45–62.
Goyeau, H., Berder, J., Czerepak, C., Lanen, C., & Lannou, C. (2012). Low diversity and fast evolution in the population of Puccinia triticina causing durum wheat leaf rust in France from 1999 to 2009, as revealed by an adapted differential set. Plant Pathology, 61, 761–772.
Groot, J. J. R. (1987). Simulation of nitrogen balance in a system of winter wheat and soil. Simulation Report CABO-TT nr. 13, Wageningen.
Hess, D. E., & Shaner, G. (1987a). Effect of moisture on Septoria tritici blotch development in the field. Phytopathology, 77, 220–226.
Hess, D. E., & Shaner, G. (1987b). Effect of moisture and temperature on development of Septoria tritici blotch in wheat. Phytopathology, 77, 215–219.
Hirst, J. M., & Stedman, O. J. (1963). Dry liberation of fungus spores by rain drops. Journal of General Microbiology, 33, 335–344.
Huerta-Espino, J., Singh, R. P., German, S., McCallum, B. D., Park, R. F., Chen, W. Q., et al. (2011). Global status of wheat leaf rust caused by Puccinia triticina. Euphytica, 179, 143–160.
Hughes, G., McRoberts, N., & Burnett, F. J. (1999). Decision-making and diagnosis in disease management. Plant Pathology, 48, 147–153.
James, C. (1971). A manual of assessment keys for plant diseases. St. Paul: The American Phytopathological Society.
Kulkarni, R. N., Chopra, V. L., & Singh, D. (1981). Observed and hypothetical leaf rust progress curves of some genotypes of wheat. Theoretical and Applied Genetics, 60, 85–88.
Lannou, C., & Savary, S. (1991). The spatial structure of spontaneous epidemics of different diseases in a groundnut plot. Netherlands Journal of Plant Patholology, 97, 355–367.
Leffelaar, P. A. (1993). Basic elements of dynamic simulation. In On systems analysis and simulation of ecological processes (pp. 11–27). Dordrecht: Kluwer Academic Publishers.
Loomis, R. S., & Adams, S. S. (1983). Integrative analysis of host-pathogen relations. Annual Review of Plant Pathology, 21, 341–362.
Madden, L. V., & Hugues, G. (1995). Plant disease incidence: distributions, heterogeneity, and temporal analysis. Annual Review of Phytopathology, 33, 529–564.
Madden, L. V., Hughes, G., & Van den Bosch, F. (2007). The Study of plant disease epidemics. St. Paul: The American Phytopathological Society.
Magboul, A. M., Geng, S., Gilchrist, D. G., & Jackson, L. F. (1992). Environmental influence on the infection of wheat by Mycosphaerella graminicola. Phytopathology, 82, 1407–1413.
McRoberts, N., Hughes, G., & Madden, L. V. (1996). Incorporating spatial variability into simple disease progress models for crop pathogens. Aspects of Applied Biology, 46, 1–8.
Penning de Vries, F. W. T. (1977). Evaluation of simulation models in agriculture and biology: conclusions of a workshop. Agricultural Systems, 2, 99–107.
Penning de Vries, F. W. T., Jansen, D. M., ten Berge, H. F. M., & Bakema, A. (Eds.) (1989). Simulation of ecophysiological processes of growth in several annual crops. Los Baños: IRRI and Wageningen: Pudoc.
Pethybridge, S. J., Gent, D. H., Esker, P. D., Turechek, W. W., Hay, F. S., & Nutter, F. W., Jr. (2009). Site-specific risk factors for ray blight in Tasmanian pyrethrum fields. Plant Disease, 93, 229–237.
Rabbinge, R., Ward, S. A., & Van Laar, H. H. (Eds.). (1989). Simulation and systems management in crop protection. Wageningen: Pudoc.
Räder, T., Racca, P., Jörg, E., & Hau, B. (2007). PUCREC/PUCTRI—a decision support system for the control of leaf rust of winter wheat and winter rye. EPPO Bulletin, 37, 378–382.
Rapilly, F. (1983). Effets de quelques facteurs physiques du climat sur diverses séquences épidémiques. EPPO Bulletin, 13, 63–68.
Robert, C., Bancal, M. O., & Lannou, C. (2004). Wheat leaf rust uredospore production on adult plants: influence of leaf nitrogen content and Septoria tritici blotch. Phytopathology, 94, 712–721.
Rossi, V., Racca, P., Jiosuè, S., Pancaldi, D., & Alberti, I. (1997). A simulation model for the development of brown rust epidemics in winter wheat. European Journal of Plant Pathology, 103, 453–465.
Savary, S., & Willocquet, L. (2014). Simulation modeling in botanical epidemiology and crop loss analysis. APSnet The Plant Health Instructor. The American Phytopathological Society. http://www.apsnet.org/edcenter/advanced/topics/Pages/default.aspx.
Savary, S., De Jong, P. D., Rabbinge, R., & Zadoks, J. C. (1990). Dynamic simulation of groundnut rust, a preliminary model. Agricultural Systems, 32, 113–141.
Savary, S., Teng, P. S., Willocquet, L., & Nutter, F. W., Jr. (2006). Quantification and modeling of crop losses: a review of purposes. Annual Review of Phytopathology, 44, 89–112.
Savary, S., Nelson, A., Willocquet, L., Pangga, I., & Aunario, J. (2012). Modelling and mapping potential epidemics of rice diseases globally. Crop Protection, 34, 6–17.
Scherm, H., & Van Bruggen, A. (1994). Effects of fluctuating temperatures on the latent period of downy mildew (Bremia lactucae). Phytopathology, 84, 853–859.
Segarra, J., Jeger, M. J., & Van den Bosch, F. (2001). Epidemic dynamics and patterns of plant diseases. Phytopathology, 91, 1001–1010.
Shiferaw, B., Smale, M., Braun, H. J., Duveiller, E., Reynolds, M., & Muricho, G. (2013). Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Security, 5, 291–317.
Spitters, C. J. T., & Kramer, T. (1985). Changes in relative growth rate with plant ontogeny in spring wheat genotypes grown as isolated plants. Euphytica, 34, 833–847.
Spitters, C. J. T., van Keulen, H., & van Kraalingen, D. W. G. (1989). A simple and universal crop growth simulator: SUCROS87. In R. Rabbinge, S. A. Ward, & H. H. van Laar (Eds.), Simulation and systems management in crop protection (pp. 147–181). Wageningen: Pudoc.
STELLA® v.9.1, (2008). isee systems, Inc. Wheelock Office Park, 31 Old Etna Road, Suite 7 N, Lebanon, NH 03766 USA, http://www.iseesystems.com/.
Suffert, F., Sache, I., & Lannou, C. (2011). Early stages of septoria tritici blotch epidemics of winter wheat: build-up, overseasoning, and release of primary inoculum. Plant Pathology, 60, 166–177.
Sun, P., & Zeng, S. (1993). On the measurement of the corrected basic infection rate. Zeitschrift fur Pflanzenkrankheiten und Pflanzenschutz, 101, 297–302.
Teng, P. S. (1981). Validation of computer models of plant disease epidemics: a review of philosophy and methodology. Zeitschrift fur Pflanzenkrankheiten und Pflanzenschutz, 88, 49–63.
Teng, P. S. (1985). A comparison of simulation approaches to epidemic modelling. Annual Review of Phytopathology, 23, 351–379.
Teng, P. S., Blackie, M. J., & Close, R. C. (1980). Simulation of the barley leaf rust epidemic: structure and evaluation of BARSIM. Agricultural Systems, 5, 85–103.
Thomas, M. R., Cook, M. J., & King, J. E. (1989). Factors affecting the development of Septoria tritici in winter wheat and its effect on yield. Plant Pathology, 38, 246–257.
Thornley, J. H. M., & France, J. (2007). Mathematical models in agriculture. Oxon, UK and Cambridge: CABI.
Tomerlin, J. R., Eversmeyer, M. G., Kramer, C. L., & Browder, L. E. (1983). Temperature and host effects on latent and infectious periods and on urediniospore production of Puccinia recondita f. sp. tritici. Phytopathology, 73, 414–419.
Tomerlin, J. R., Eversmeyer, M. G., Kramer, C. L., & Browder, L. E. (1984). Environmental and host effects on colony development on Puccinia recondita f. sp. tritici. Phytopathology, 74, 225–229.
Van der Plank, J. E. (1963). Plant diseases epidemics and control. New York: Academic.
Waggoner, P. E., & Rich, S. (1981). Lesion distribution, multiple infection, and the logistic increase of plant disease. Proceedings of the National Academy of Sciences USA, 78, 3292–3295.
Wainshilbaum, S. J., & Lipps, P. E. (1991). Effect of temperature and growth stage of wheat on development of leaf and glume blotch caused by Septoria tritici and S. nodorum. Plant Disease, 75, 993–998.
Willocquet, L., Aubertot, J. N., Lebard, S., Robert, C., Lannou, C., & Savary, S. (2008). Simulating multiple pest damage in varying winter wheat production situations. Field Crops Research, 107, 12–28.
Xu, X. M., & Madden, L. V. (2002). Incidence and density relationships of powdery mildew on apple. Phytopathology, 92, 1005–1014.
Yang, X. B., & TeBeest, D. O. (1992). Dynamic pathogen distribution and logistic increase of plant disease. Phytopathology, 82, 380–383.
Yuen, J. E., & Hughes, G. (2002). Bayesian analysis of plant disease prediction. Plant Pathology, 51, 407–412.
Zadoks, J. C. (1971). Systems analysis and the dynamics of epidemics. Phytopathology, 61, 600–610.
Zadoks, J. C., & Rijsdijk, F. H. (1984). Atlas of cereal diseases and pests in Europe. Wageningen: Pudoc.
Zadoks, J. C., & Schein, R. D. (1979). Epidemiology and plant disease management. New York: Oxford University Press.
Zhang, Y., Zhang, Y., Wang, Z., & Wang, Z. (2011). Characteristics of canopy structure and contributions of non-leaf organs to yield in winter wheat under different irrigated conditions. Field Crops Research, 123, 187–195.
Acknowledgments
We acknowledge the Joint Research Centre Monitoring Agricultural ResourceS of the European Commission for providing Interpolated meteorological data at the European scale (JRC-MARS - Meteorological Data Base), and the French ministry for agriculture and fisheries and its partners (ONPV) for accessing the yearly crop health reports for wheat, 2005–2010. We are grateful to anonymous reviewers for their comments which have helped improving the article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Savary, S., Stetkiewicz, S., Brun, F. et al. Modelling and mapping potential epidemics of wheat diseases—examples on leaf rust and Septoria tritici blotch using EPIWHEAT. Eur J Plant Pathol 142, 771–790 (2015). https://doi.org/10.1007/s10658-015-0650-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-015-0650-7