Abstract
Anthropogenic alterations of river systems may have a profound effect on native fish community and habitat use; however, it’s difficult to understand the extent of these impacts without establishing well-defined habitat preferences. We investigated the Shoal chub, Macrhybopsis hyostoma, a native obligate river species from nine sampling locations in the upper Mississippi River Basin (UMRB). Field surveys demonstrated that overall Shoal chubs preferred tributaries, yet this was statistically significant only for gravid females. Diet analysis and comparative morphology suggested that the Shoal chub is insectivorous and prefer benthic habitats. Our analysis of habitat use suggested that juvenile Shoal chubs preferred sand substrate and adults preferred medium to large gravel. Shoal chubs developed more melanophores as they aged, which is a likely an adaptation to their habitat shifts. The field survey identified possible sites where spawning was occurring and may be important for future conservation efforts for the Shoal chub. In addition, we conducted population genomic analysis of Shoal chub samples collected from the streams in three Midwest states (Illinois, Missouri, and Nebraska) and found low genetic diversity among the chubs that raises a concern in conservation. This preliminary study provides insights into further investigation of the impact caused by stream habitat alteration on native species and into the conservation of Shoal chubs in the UMRB.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Mississippi River watershed, including the Missouri River tributary, has undergone dramatic ecological changes in the past century (Hrabik et al. 2015). The Upper Mississippi River Basin (UMRB) is becoming a highly regulated and degraded ecosystem due to human activities such as channelization, the construction of dams, and the removal of natural formations and agricultural discharge (Weitzell et al. 2003). This high degree of the natural hydrological regime in the UMRB may have negatively affected fish habitat use and population structure.
Macrhybopsis chubs are a representative genus of the chub clade consisting of small-bodied fishes that are typically obligate river species in the Mississippi River (Galat et al. 2005). There are twelve species within this genus, and recent studies have determined that this genera is taxonomically complex (Eisenhour 2004; Gilbert et al. 2017). The most morphologically diverse fish in this genus is the Shoal chub. Shoal chubs are very small minnows with a streamlined body that’s dorsoventrally flattened and a rounded snout overhanging the subterminal mouth. They have small upward gazing eyes with smooth scales and a complete lateral line. The caudal fin possesses a white line along the ventral margin (Fig. 1). Populations in the east of the Mississippi River appear to show little variation, in marked contrast to those in western drainages where a large percentage of individuals possess a secondary pair of maxillary barbels (Eisenhour 2004; Gilbert et al. 2017). In the Arkansas and Red River Basins members of this genus have demonstrated introgressive hybridization with other members of the genus (particularly between M. tetranema and M. hyostoma) which may explain some of the morphological variation. This variation is also hypothesized to be a result of a combination of pre-Pleistocene evolutionary processes, together with subsequent long-term instability and changes in stream-drainage patterns and flow regimes related to periodic advances and retreats of the Pleistocene ice sheets (Mayden 1985; Wiley and Mayden 1985; Cross et al. 1986; Gilbert et al. 2017). This study focused on the upper Mississippi River Basin (Nebraska, Illinois, and Missouri) in order to prevent including hybrids in the analysis.
Shoal chubs at different life-history stages. a. Juvenile collected from Pool 20, Mississippi River, Iowa side north of the Des Moines River confluence (Site I); b. Two year old male collected from Segment 8 of the Missouri River (Site D); c. Two year old female collected from Segment 9 of the Missouri River (Site E); d. Gravid female collected from the Loup River (Site C). Each circle represents a portion of the specimen’s skin magnified 40x so the melanophores can be clearly seen. All adult Shoal chubs, regardless of sex, possess more melanophores than juveniles and their melanophores are larger
Within the upper Mississippi River Basin there are only four species of Macrhybopsis chubs including, the Shoal chub, M. hyostoma, the Sturgeon chub, M. gelida, the Sicklefin chub, M. meeki, and the Silver chub, M. storeriana. Recently, population reductions exceeding 70% for all Macrhybopsis chubs have been observed within the upper Mississippi River Basin (Dynesius and Nilsson 1994; Hesse 1994; Steffensen et al. 2014). Two other Macrhybopsis species, the Sturgeon chub, M. gelida, and the Sicklefin chub, M. meeki, have been listed as threatened or endangered throughout much of their historical range (Rahel and Thel 2004) and are currently petitioned to be listed federally endanged. The Silver chub is currently listed as vulnerable in the upper Mississippi River Basin, and is considered a species of Special Concern throughout parts of Canada (Hesse 1994; Mandrak and Holm 2001; Steffensen et al. 2014). The construction of six dams and reservoirs on the mainstem river converted riverine habitat to lentic systems which has been hypothesized as a potential cause for the dramatic population reductions south of Gavins Point Dam (Service 2001). Population declines in this region may suggest these populations are under strong selection pressures and warrant conservation concern as Macrhybopsis chubs serve as key food chain species during the juvenile and adult stages for the endangered pallid sturgeon Scaphirhynchus albus, particularly the Shoal chub (Gerrity et al. 2006; Herman et al. 2008).
One of the major impediments to conservation efforts has been determining habitat preferences. Previous studies have described a wide variety of habitats that Macrhybopsis chubs can utilize ranging from sandy substrate with clear water with moderate currents (Klutho 1983; Luttrell et al. 2002) to deep turbid water with gravel substrate (Starrett 1950; Jones 1997; Eisenhour 2004; Rahel and Thel 2004). Some of the variation in this literature may be due to these preferences changing in relation to age or gender (Starrett 1950; Jones 1997; Eisenhour 2004; Rahel and Thel 2004). The broad continuum of habitats described in the literature makes identifying critical habitats difficult as well as targeting particularly susceptible populations of these species (Galat et al. 2005). There is an accepted ecological premise that an organism’s habitat provides the template for trait adaptation and over time these organisms evolve under these habitat parameters (Southwood 1977; Townsend and Hildrew 1994). Based on this premise ecologists can use morphological characteristics to infer what types of habitats an organism would utilize (Fulton et al. 2001; Irschick et al. 2005; Collar et al. 2010; Colombo et al. 2016). In order to prevent continuing population declines it is imperative to refine key life history parameters for all Macrhybopsis chubs within the upper Mississippi River Basin, and furthermore, it is particularly crucial to understand how the Shoal chub utilizes particular habitats at different life stages to restore historic population level and because they are a key dietary component of the endangered pallid sturgeon.
The other major impediment for conservation efforts is determining a species’ intrinsic genetic resources. Data from previous studies have demonstrated that populations with reduced genetic diversity often experience reduced growth and increased extinction rates (Keller and Waller 2002). Many genera of native upper Mississippi River Basin fishes, including Macrhybopsis chubs, may possess limited genetic resources due to historic glaciation events or adaptation to historical river conditions, such as those demonstrated in other North American fish (Harris and Taylor 2010; Hrabik et al. 2015). Cost and technological limitations have historically restricted these kinds of assessments, however, such evolutionary consequences can now be addressed at a nuclear genome scale with the advent and advances of next-generation sequencing technology (Luikart et al. 2003; Li et al. 2008; Hohenlohe et al. 2010). Genotyping by sequencing (GBS) is a highly multiplexed, low cost system which requires less handling, fewer PCR and purification steps to generate large numbers of single nucleotide polymorphisms for population studies (Davey and Blaxter 2010; He et al. 2014) which quantifies the degree of genetic diversity within the population.
This study attempts to describe habitat preferences for the Shoal chub using morphological and dietary analysis and explore the population structure of the Shoal chub throughout the upper part of the Mississippi River to determine the number of populations that are present and determine the amount of genetic diversity. The degree of genetic diversity along with morphological and habitat use studies, may offer new management strategies for the Shoal chub.
Methods
Sampling was conducted from September 2013 through August 2015 at nine sites, five tributary and four mainstem sites, throughout the upper Mississippi River Basin (Table 1) based on historical ranges and previous field experience. A total of 234 Shoal chubs were collected from these nine sites.
Sampling protocol
Water velocity, turbidity, and depth were measured with a water velocity meter (Marsh-McBirney Flo-Mate™, Frederick, MD), a turbidity meter (Hach 2100P Portable Turbidimeter) and a meter stick or boat mount sounder, respectively. Substrate composition samples were collected using a glass jar when the bottom of the sampling site could be reached, or by pipe dredge for water depths exceeding 1.5 m. Particles were classified according to the Wentworth scale (Wentworth 1922).
The type of gear used to collect fish was dictated by accessibility and depth. A 3.66 m wall seine with 6-mm mesh was used to sample the Elkhorn and Loup Rivers, Sites B and C respectively, because sites B and C were shallow enough to allow wading. A benthic 4.9-m otter trawl was actively towed downstream at Sites A, D, and E following the protocols of the Pallid Sturgeon Population Assessment Program by the U.S. Fish and Wildlife Service, Missouri River Fish and Wildlife Conservation Office (Bismarck, ND) and the Nebraska Game and Parks Commission (Welker and Drobish 2010; Welker and Drobish 2011; Steffensen et al. 2014). A bottom trawl was used to sample Sites E-I by the Missouri Department of Conservation, Southeast Regional Office and Jim Lamer at the Kibbe Field Station (Jim Lamer pers. comm.). This bottom trawl consisted of two-seam, 4.8-m slingshot balloon trawls (TRL16BC, Memphis Net and Twine Co., Inc., or the equivalent). The body of the trawl was made of No. 9 nylon with stretch mesh 18 mm in diameter. The cod end was made of No. 18 nylon with stretch mesh 18 mm in diameter. The cod end contained a 1.8-m liner consisting of 3 mm Ace-type nylon mesh. Floats were spaced every 0.91 m along the headrope, and a 4.8-mm steel chain was tied to the footrope. The trawl was equipped with 37-cm-high by 75-cm-long iron “V” doors (otter boards) (Bartels et al. 2003, 2004). Specimens were preserved in 100% ethanol solution for further investigations.
Relative abundance
Catch per unit effort was used to assess the habitat usage. Catch per unit effort was calculated as the total number of fish in relation to the area (length of the gear x number of meters trawled) at each collection site (Hahn et al. 2007; Welker and Drobish 2010). These catch per unit efforts were then standardized using multigear mean standardization (Gibson-Reinemer et al. 2016). Mean standardized catch of species \( \left(\mathrm{i}\right)\left(\mathrm{j}\right)\ \left(\mathrm{MSCij}\right)=\left(\frac{C_{ij}}{\mathrm{e}}\right)/\left(\frac{{\overline{TC}}_j}{\mathrm{e}}\right) \) where (cij/e) is the CPUE of species (i) in observation (j) and \( \overline{TC}j/e \) is the mean total catch per unit effort (Gibson-Reinemer et al. 2016). Chi square analysis was used to determine the statistical significance for standardized catch per unit effort in relation to system type and substrate type.
Morphological analysis
Each specimen was photographed with a Canon EOS Rebel SL1 digital camera with a Canon EFS 60 mm f/2.8 Macro USM. Each melanophore on the lateral side of each specimen was visually counted and measured using GIMP (Gimp 2008). The number of melanophores were visually counted and measured with a microscopic scale. To ensure accuracy, the total number of melanophores was counted until the same number was acquired three separate times for each fish. Melanophores were classified as small (smaller than 0.15 mm in diameter), medium (between 0.15–1.5 mm) or large (greater than 1.5 mm).
Fish were aged to determine how morphological features were affected by age class. Six cycloid scales were removed between the lateral line and the dorsal fin and then placed on a clear plastic slide with ridges down. Each slide was sandwiched between two more pieces of plastic and run through a roller press. Age was determined by counting the number of annuli as described by Schneider (Schneider 2000). Life stage was assigned to each specimen using the scale given in Table 2.
Gut contents are a significant indicator of habitat resource use (Starrett 1950). Gut contents were visually identified to a genus level to determine the individual components of each species’ diet using reference texts (Merritt and Cummins 1978; Wiggins 1977; Borror et al. 1989). The frequency of occurrence for each prey item was calculated as the number of stomachs in which each item occurs and expressed as a total number of stomachs examined using the following equation:
Frequency of Occurrence (Oi) = Ji/P, where Ji is number of fish containing prey i and P is the number of fish with food in their stomach .The frequency of occurrence (%F) of each dietary item provides the most robust and interpretable measure of diet composition (Baker et al. 2014).
The gender of each specimen was determined by macroscopic examination of the gonads under a light microscope at 10x magnification. The gonadosomatic index was used to evaluate sampling sites as potential breeding grounds. Eggs were removed using forceps and individually counted under a light microscope. Each gravid female was weighed on a digital scale before and after egg removal and egg weight was calculated from subtracting the fish’s weight without eggs from the total fish’s weight, including eggs. The gonadosomatic index was calculated by dividing the weight of the eggs by the total weight of the fish (Devlaming et al. 1982; Hassanin et al. 2002).
Melanophore analyses
To better predict how total melanophore abundance fluctuates in relation to age and environmental variables, fourteen linear models were constructed which explored the effects of total length, sex, age class, turbidity, current velocity, water temperature and water depth with the total number of melanophores that were present, including a global model with all the covariates. Models were constructed based on the four continuous covariates for evidence of collinearity with pairwise Pearson’s correlation coefficients. If two variables had ∣r ∣ ≥ 0.75 they were not included in the same model. The total number of melanophores, number of small melanophores, number of medium melanophores and the number of large melanophores were logarithmically transformed based on the pairs plot. Total length was standardized using the formula: standardized total length = (total length-mean total length)/(standard deviation of the total length). AICc and calculated Akaike weights were used to compare models and determine the optimal model. All analyses were carried out in R version 3.1.3 (Team 2014). Non-linear models were run using the package mgcv (Wood and Wood 2015). Graphs were created using ggplot2 (Wickham et al. 2013). Regression analysis was conducted using the package rpart (Therneau et al. 2010).
Population genetic structure
Nuclear genomic DNA of forty-eight Shoal chubs were extracted and purified from fin tissue using the Qiagen DNeasy Blood and Tissue Kit for Genotyping by Sequencing (GBS). PCR free libraries were constructed with a custom Illumina protocol by performing a double digest of 100 ng DNA with PstI-HF and MspI. The sheared DNA was isolated with magnetic beads and re-quantified. The barcoded libraries were constructed from 1 ng of DNA and sequenced on an Illumina NextSeq500 at the USGS Leetown Science Facility. Four libraries with twelve individuals from Sites (C, D, G and I; Loup, Missouri River-Segment 8, MO-Marquette Island Side Channel, IL respectively) per library were constructed. Pooled individuals were identified with unique 9-bp barcodes. All specimens used for genetic analysis are part of the ichthyology collection at the University of Nebraska State Museum (Z-2019-02).
Reads were trimmed and aligned using CLC Genomics Workbench (CLC bio) where only one ambiguous base was allowed. Before trimming, quality scores are converted to an error probability (p = 10(−Q/10), Q is quality score) and during trimming the error rate had to be smaller 0.03 to maintain high quality within the reads. Reads that were shorter than 40 bp were discarded. Over 65% of all the reads had a Phred score of 35 and over 50% of the total reads had a GC content between 40 and 50%.
Single nucleotide polymorphisms (SNPs) were identified using the Stacks software, which utilizes a maximum likelihood statistical model to identify loci de novo (Catchen et al. 2011). The Populations program within Stacks was used to calculate population genetic statistics, including genetic diversity, heterozygosity and FST.
Three locations were selected to determine the genetic structure of the Shoal chub within the upper MRB. One location, from Segment 8 of the Missouri River, had to be removed due to a low number of reads following sequencing. The first population contained eleven samples from the sample site in Illinois (Site I), the second population contained twelve specimens from the sample site from the Loup River in Nebraska (Site C) and the third population was defined as twelve specimens collected in Missouri (Site G). A total of 2,696,647 reads from thirty-five specimens were used for downstream analysis. All raw reads were deposited at the National Center for Biotechnology Information (BioProject ID PRJNA516905).
The POPULATIONS program in Stacks was used to analyse the organization of the populations using multilocus genotypic information using output SNP data from across all GBS sites into a STRUCTURE-format file (Pritchard et al. 2000; Hubisz et al. 2009; Catchen et al. 2013). Due to computational limitations of handling many more than this number of loci in the current STRUCTURE application, we implemented a custom Perl script to randomly choose 10,000 of these SNPs. STRUCTURE 2.3.4 (Pritchard et al. 2000; Hubisz et al. 2009; Catchen et al. 2013) was used to infer historical lineages through clustering of similar genotypes. The admixture model of STRUCTURE and the option of correlated allele frequencies between populations were used. For the entire population set K ranged from 1 to 3. The optimal K was determined using the deltaK method and visual inspection of the change in the Ln P(D) of each model (Evanno et al. 2005). A burn-in of 100,000 steps followed by 1000,000 additional Markov Chain Monte Carlo iterations were performed.
This same set of 10,000 SNPs from 251 nuclear loci created by Stacks was downloaded into GenoDive which calculated pairwise FST values for all population pairs (Meirmans and Van Tienderen 2004; Meirmans 2009). This was accompanied by 1000 randomization tests to determine if each FST value is different from zero utilizing a strict Bonferroni correction due to the multiple comparisons (Rice 1989; Catchen et al. 2013).
Results
Overall, Shoal chubs preferred tributaries (p value = 0.002), particularly those with moderate current velocities (0.38–0.57 m/s) and relatively shallow water depth (1.04–2.69 m). The total lengths of specimens ranged from 27 to 57 mm. The site with the highest relative abundance was the Loup River (MSC 17.65, Table 3). Although Shoal chubs preferred tributaries, different life stages had varying habitat preferences, particularly substrate preferences.
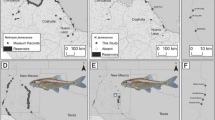
The total lengths of juvenile Shoal chubs were less than 37 mm (Table 2). The area with the highest mean standard catch (MSC) of juvenile Shoal chubs was Pool 20 of the Mississippi River in Illinois (MSC 0.61, Table 3). Overall, juvenile Shoal chubs did not demonstrate a statistically significant system preference (p = 0.210) (Fig. 2). Regardless of system type, however, juvenile Shoal chubs demonstrated a statistically significant preference for sand substrate (p = 0.003) (Fig. 3).
The mean standardized catch of Shoal (MSCS) chubs at various life-history stages in relation to the system types. See Table 1 for system delineations and locations. Asterisk denotes significant differences
Specimens were considered ‘adults’ if their total length exceeded 37 mm, their age corresponded to a year class 1+ and had reproductive organs that could be visually identified as male or female (Table 2). Overall adult Shoal chubs preferred tributaries (p = 0.001). This preference may be skewed by gravid female’s significant preference for tributaries (p = 0.010) (Fig. 2). Neither adult males nor females demonstrated a significant preference for system type when their life stages were solely considered. All adult Shoal chubs preferred medium gravel significantly more (p < 0.001) (Fig. 3). Gravid females had total lengths ranging from 45 to 56 mm. The locations with the highest relative abundance of gravid female Shoal chubs (~80%) were collected in early June from the Loup River (MSC 17.65), suggesting Shoal chubs may use this area as a spawning site. Gravid females were also found in the Elkhorn in late September but only had a gonadosomatic index of 6–7%, indicating the specimens were collected at the end of the spawning season or display a bimodal or multi-modal spawning cycle. One other gravid female was collected from Segment 8 of the Missouri River in early July; however, it also had a relatively low gonadosomatic index (9%.) This may indicate that this portion of the mainstem river was unsuitable for spawning or spawning had already completed.
Chi square analysis suggested that adults preferred larger substrate than juveniles. The best linear model based on Akaike information criterion that explored the effects of total length, sex, age class, turbidity, current velocity, temperature and depth with the total number of melanophores that were present determined that there was an additive effect amongst total length and age classes in relation to total melanophores present, AICc 267.60 (Table 4). The older Shoal chubs had more melanophores and a greater number of larger melanophores.
Shoal chubs ate primarily small dietary items, namely Chironomidae larvae (90%), regardless of age or collection site. The rest of the Shoal chub’s diet consisted of various dipteran body parts consisted of the remaining 10%. These body parts were too small for further identification.
The Shoal chub’s recent population declines, and dietary status make it an ideal representative of this genus to begin to explore population structure. Next generation sequencing yielded 105,433,437 nuclear sequences. The sequence lengths varied from 31 to 142 bp with a GC content of 42%. A total of 109,108,396 remained after CLC Genomics Workbench preliminary processed and trimmed the total number of reads. Over 50% of the total reads had a GC content between 40 and 50%. Ustacks utilized a total of 2,696,647 sequences to form 122,382 stacks. The Stacks program utilizes short-read sequences to assign identical short read sequences to a unique stack which is equivalent to a nuclear locus. The mean merged coverage depth was 21.2 and the maximum number of nuclear sequences present in a stack was 14,576. The minimum depth of coverage used to create a stack was three and the maximum distanced allowed between stacks was 2. The standard deviation for the coverage depth was 74.2.
For all loci that were polymorphic in at least one population in the entire data set, the average major allele frequency (P) ranged from 0.79 to 0.84 and the average observed heterozygosity ranged from 0.30 to 0.38. When considering all nucleotide positions, the values increase to 0.999 for the major allele frequency (P), and the observed average heterozygosity decreased to a range from 3.07e−4-3.37e−4. Genetic diversity is the best method to assess a species’ capacity to respond to disturbance. Nucleotide diversity was 0.31, 0.39 and 0.38 with a standard error of 0.001 for all three populations, respectively. This reduction in genetic variation is particularly evident in the percentage of loci that are polymorphic at all loci which was 0.05 at Illinois (Site I) and the Marquette Island Side Channel (Site G) and 0.06 for the Loup River (Site C). The mean inbreeding coefficient (FIS) was 0.003, 0.02 and 0.06 for samples from the site in Illinois (Site I), the Loup River (Site C) and the Marquette Island Side Channel (Site G), respectively (Table 5).
Pairwise comparisons of FST amongst these three populations reveals exceptionally low FST values at all populations indicating a high degree of gene flow amongst populations in the upper MRB. Using a numerical randomization approach of 10,000 randomly chosen SNPs the only pairwise FST values that were significantly different were the Illinois and Loup River populations at the alpha = 0.05 level (Table 6). Although not statistically significant, the pairwise FST is lower for the Loup River and Missouri Marquette Island Side Channel collection sites (0.001) compared to the pairwise FST between Illinois and the Missouri Marquette Island Side Channel (0.013) collection sites.
As a further test of potential population structure, we analysed 10,000 randomly chosen SNPs using STRUCTURE. By examining the change in Ln P(D) and using the deltaK approach of Evanno et al. (2005), we found that a model with K = 2 best fits the data which was supported by STRUCTURE’s plot of posterior probabilities.
Pairwise FST and STRUCTURE analysis support the presence of two possible populations of Shoal chubs within the upper Mississippi River Basin. One population consists of the Loup River (Site C) and the Missouri Marquette Island Side Channel (Site G) collection sites and the other population consists of the Illinois (Site I) collection site.
Discussion
This is the first study that demonstrated that Shoal chubs utilize various types of habitats, including different types of substrates, at different life-history stages. Overall, Shoal chubs preferred tributaries, but this was highly affected by the preference of gravid females. Juvenile Shoal chubs were prevalent in areas with sand substrate, and adults preferred medium gravel substrate. In addition, gravid females were more frequently found in sites with medium gravel. Melanophores may be one morphological feature that facilitates these fluctuating habitat preferences and may allow the Shoal chub to take advantage of a variety of habitats, particularly different types of substrate throughout their life history. Age classes and the total lengths of the Shoal chubs have an additive effect on the number of melanophores. As they age, the Shoal chubs can use larger substrate particles. Our results resolved a previous confusion about the habitat parameters described for the Shoal chub that is likely associated with the change of habitat preferences in different life-history stages.
At times large aggregations of individual Shoal chubs were found and such aggregations may serve as a means to avoid predation for juveniles or to maximize reproductive potential (Pitcher and Parrish 1993). One aggregation of juvenile Shoal chubs was collected from Pool 20 of the Mississippi River in Illinois. We found a proportion of the juvenile fish collected from Pool 20 in Illinois (~19%) infected with Uvulifer spp. ectoparasite. Previous studies have shown that anthropogenic disturbances such as dams may indirectly facilitate parasitism (Hernandez et al. 2007). Pool 20 had extensive damming which may increase the likelihood of secondary fish host infection. The reduction of water velocity or the increase of the intermediate snail host as a result of damming could increase the probability that Shoal chubs would be exposed to Uvulifer spp.. Consequently, this parasite may increase the mortality of Shoal chubs by depleting their nutritional reserves and increasing the probability of being consumed by a predator or reducing the probability of survival during stressful periods (Barber et al. 2000; Pracheil and Muzzall 2010; Ferguson et al. 2011; Markle et al. 2014). Even a single ectoparasite may increase the mortality for larval or juvenile Shoal chub significantly (Grutter et al. 2010).
Life cycles including spawning migrations or the act of spawning rely on natural flow peaks by bringing reproductive adults together and maximizing habitat conditions for larval fish survival (Thomas 1988; Næsje et al. 1995; Hooper et al. 2005). Water redirection has previously affected native minnows by altering flow peaks and may affect spawning potential for the Shoal chub (O'Connor 2002; District and HDR Engineering 2015). We found a second aggregation of Shoal chubs at the Loup River, where 80% of the fish collected from the Loup River were gravid females. This section of the Loup River is under annual water redirection efforts. The water redirection that occurs along this stretch will lower water levels by as much as 75% which may remove valuable spawning areas or hinder recruitment potential for Shoal chubs.
Population genomic analysis revealed low heterozygosity within northern populations of the Shoal chubs. The low degree of heterozygosity displayed amongst upper Mississippi River Basin Shoal chub populations potentially reduces this species capacity to respond to anthropogenic disturbances, which may be responsible for the recent population reductions (Hoffmann and Hercus 2000). The population declines seen in the Shoal chub may eventually lead to inbreeding which would lower resistance to disease and environmental stress and could exacerbate the potential for extinction (Keller and Waller 2002). Southern populations of Shoal chubs have been observed hybridizing with M. tetranema, a southern sister species. These hybridization events may overcome the issues caused by low heterozygosity that may also explain why southern populations of the Shoal chub are not experiencing such dramatic population declines (Underwood et al. 2003). Future genetic studies should investigate the degree of divergence and genetic similarity between northern and southern populations and explore how hybridization has impacted southern populations.
This study refined habitat preferences for Shoal chubs at various life-history stages, explored how melanophores may be related to habitat use, and identified two possible sites where large aggregations of Shoal chubs were present. Future studies should focus on the impact of these sites with juvenile survival, particularly in relation to infection rates of Uvulifer sp., and how water redirection affects reproductive efforts for this species in the Loup River. Future conservation efforts should focus on minimizing stressful environments, such as those disrupted by anthropogenic disturbances, which will likely decrease mean fitness that may continue to compromise the upper Mississippi River Basin’s populations persistence (Kinnison and Hairston 2007; DiBattista et al. 2011).
References
Baker R, Buckland A, Sheaves M (2014) Fish gut content analysis: robust measures of diet composition. Fish Fish 15:170–177
Barber I, Hoare D, Krause J (2000) Effects of parasites on fish behaviour: a review and evolutionary perspective. Rev Fish Biol Fish 10:131–165
Bartels A et al. (2003) Annual status report: a summary of fish data in six reaches of the upper Mississippi River system
Bartels A et al. (2004) 2004 Annual Status Report: A Summary of Fish Data in Six Reaches of the Upper Mississippi River System. Accessed 2/27/2016 2016
Borror DJ, Triplehorn CA, Johnson NF (1989) An introduction to the study of insects, vol Ed. 6. Saunders College Publishing, Philadelphia
Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH (2011) Stacks: building and genotyping loci de novo from short-read sequences G3 (Bethesda, Md) 1:171–182 https://doi.org/10.1534/g3.111.000240
Catchen J, Bassham S, Wilson T, Currey M, O'Brien C, Yeates Q, Cresko WA (2013) The population structure and recent colonization history of Oregon threespine stickleback determined using restriction-site associated DNA-sequencing. Mol Ecol 22:2864–2883. https://doi.org/10.1111/mec.12330
Collar DC, Schulte JA, O’Meara BC, Losos JB (2010) Habitat use affects morphological diversification in dragon lizards. J Evol Biol 23:1033–1049. https://doi.org/10.1111/j.1420-9101.2010.01971.x
Colombo M, Indermaur A, Meyer BS, Salzburger W (2016) Habitat use and its implications to functional morphology: niche partitioning and the evolution of locomotory morphology in Lake Tanganyikan cichlids (Perciformes: Cichlidae). Biol J Linn Soc 118:536–550
Cross FB, Mayden RL, Stewart JD (1986) Fishes in the western Mississippi basin (Missouri, Arkansas, and red rivers) the zoogeography of north American freshwater fishes:363–412
Davey JW, Blaxter ML (2010) RADSeq: next-generation population genetics. Briefings in Functional Genomics 9:416–423
Devlaming V, Grossman G, Chapman F (1982) On the use of the gonosomatic index. Comp Biochem Physiol A Physiol 73:31–39
DiBattista JD, Feldheim KA, Garant D, Gruber SH, Hendry AP (2011) Anthropogenic disturbance and evolutionary parameters: a lemon shark population experiencing habitat loss. Evol Appl 4:1–17. https://doi.org/10.1111/j.1752-4571.2010.00125.x
District LRPP, HDR Engineering I (2015) Loup Power District's Loup River Hydroelectric Project relicensing website. http://www.loup.com/relicense/index.html. Accessed 12 Dec 2016
Dynesius M, Nilsson C (1994) Fragmentation and flow regulation of river systems in the northern third of the world. Science (New York, NY) 266:753–762. https://doi.org/10.1126/science.266.5186.753
Eisenhour D (2004) Systematics, variation, and speciation of the Macrhybopsis aestivalis complex west of the Mississippi River vol:23
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Ferguson JA, Koketsu W, Ninomiya I, Rossignol PA, Jacobson KC, Kent ML (2011) Mortality of coho salmon (Oncorhynchus kisutch) associated with burdens of multiple parasite species. Int J Parasitol 41:1197–1205
Fulton C, Bellwood D, Wainwright P (2001) The relationship between swimming ability and habitat use in wrasses (Labridae). Mar Biol 139:25–33
Galat DL et al (2005) Spatiotemporal patterns and changes in Missouri River fishes. American Fisheries Society Symposium, pp 249–291
Gerrity PC, Guy CS, Gardner WM (2006) Juvenile pallid sturgeon are Piscivorous: a call for conserving native cyprinids. Trans Am Fish Soc 135:604–609. https://doi.org/10.1577/T05-122.1
Gibson-Reinemer DK, Ickes BS, Chick JH (2016) Development and assessment of a new method for combining catch per unit effort data from different fish sampling gears: multigear mean standardization (MGMS). Can J Fish Aquat Sci 74:8–14
Gilbert CR, Mayden RL, Powers SL (2017) Morphological and genetic evolution in eastern populations of the Macrhybopsis aestivalis complex (Cypriniformes: Cyprinidae), with the descriptions of four new species. Zootaxa 4247:501–555
Gimp G (2008) Image manipulation program user manual. Edge-Detect Filters, Sobel, The GIMP Documentation Team 8:8.7
Grutter A, Cribb T, McCallum H, Pickering J, McCormick M (2010) Effects of parasites on larval and juvenile stages of the coral reef fish Pomacentrus moluccensis. Coral Reefs 29:31–40
Hahn PK, Bailey RE, Ritchie A (2007) Beach seining salmonid field protocols handbook: techniques for assessing status and trends in salmon and trout populations American fisheries society. Bethesda, Maryland, pp 267–323
Harris LN, Taylor EB (2010) Pleistocene glaciations and contemporary genetic diversity in a Beringian fish, the broad whitefish, Coregonus nasus (Pallas): inferences from microsatellite DNA variation. J Evol Biol 23:72–86. https://doi.org/10.1111/j.1420-9101.2009.01858.x
Hassanin A, Kuwahara S, Tsukamoto Y, Ogawa K, Hiramatsu K, Sasaki F (2002) Gonadosomatic index and testis morphology of common carp (Cyprinus carpio) in rivers contaminated with estrogenic chemicals. J Vet Med Sci 64:921–926
He J, Zhao X, Laroche A, Lu Z-X, Liu H, Li Z (2014) Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front Plant Sci 5:484. https://doi.org/10.3389/fpls.2014.00484
Herman P, Plauck A, Utrup N, Hill T (2008) Three year summary age and growth report for Sicklefin chub (Macrohybopsis meeki). Pallid sturgeon population assessment project and associated fish community monitoring for the Missouri River. United States Fish and Wildlife Service Columbia National Fish and Wildlife Conservation Office, Columbia, MO,
Hernandez A, Bunnell J, Sukhdeo M (2007) Composition and diversity patterns in metazoan parasite communities and anthropogenic disturbance in stream ecosystems. Parasitology 134:91–102
Hesse L (1994) The status of Nebraska fishes in the Missouri River, 5. Selected chubs and minnows (Cyprinidae): sicklefin chub (Macrhybopsis meeki), sturgeon chub (M. gelida), silver chub (M. storeriana), speckled chub (M. aestivalis), flathead chub (Platygobio gracilis), plains minnow (Hybognathus placitis), and western silvery minnow (H. argyritis). Transactions of the Nebraska Academy of Science 21:99–108
Hoffmann AA, Hercus MJ (2000) Environmental stress as an evolutionary force. Bioscience 50:217–226
Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA (2010) Population genomics of parallel adaptation in Threespine stickleback using sequenced RAD tags. PLoS Genet 6:e1000862. https://doi.org/10.1371/journal.pgen.1000862
Hooper DU, Chapin FS III, Ewel JJ, Hector A, Inchausti P, Lavorel S, Lawton JH, Lodge DM, Loreau M, Naeem S, Schmid B, Setälä H, Symstad AJ, Vandermeer J, Wardle DA (2005) Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol Monogr 75:3–35. https://doi.org/10.1890/04-0922
Hrabik R, Schainost S, Stasiak R, Peters E (2015) The Fishes of Nebraska. Mennonite Press, Inc., Newton, KS
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Irschick DJ et al (2005) A comparison of habitat use, morphology, clinging performance and escape behaviour among two divergent green anole lizard (Anolis carolinensis) populations. Biol J Linn Soc 85:223–234
Jones MD (1997) Taxonomic status and genetic structure of speckled chubs (Cyprinidae: Cf. Macrhybopsis aestivalis) in the Arkansas River Drainage
Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241. https://doi.org/10.1016/S0169-5347(02)02489-8
Kinnison MT, Hairston NG (2007) Eco-evolutionary conservation biology: contemporary evolution and the dynamics of persistence. Funct Ecol 21:444–454
Klutho MA (1983) Seasonal, daily, and spatial variation of shoreline fishes in the Mississippi River at grand tower. Southern Illinois University, Illinois
Li YF, Costello JC, Holloway AK, Hahn MW (2008) “Reverse ecology” and the power of population genomics. Evolution; Int J Organic Evol 62:2984–2994
Luikart G, England PR, Tallmon D, Jordan S, Taberlet P (2003) The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet 4:981–994
Luttrell GR, EA A, FW L (2002) Habitat correlates of the distribution of Macrhybopsis hyostoma (Teleostei: Cyprinidae) in Western reaches of the Arkansas River basin. Trans Kans Acad Sci 105:153–161
Mandrak N, Holm E (2001) COSEWIC assessment and update status report on the silver chub, Macrhybopsis storeriana. Canada, in COSEWIC assessment and update status report of the silver chub, Macrhybopsis storeriana, in Canada. Committee on the status of endangered wildlife in Canada. Ottawa,
Markle DF, Terwilliger MR, Simon DC (2014) Estimates of daily mortality from a neascus trematode in age-0 shortnose sucker (Chasmistes brevirostris) and the potential impact of avian predation. Environ Biol Fish 97:197–207
Mayden RL (1985) Biogeography of Ouachita highland fishes. Southwest Nat 30:195–211
Meirmans P (2009) GenoDive version 2.0 b14 Computer software distributed by the author Available from: http://www.bentleydrummer nl/software/software/GenoDive html. Accessed 12 Dec 2016
Meirmans PG, Van Tienderen PH (2004) GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794
Merritt R, Cummins K (1978) An introduction to the aquatic insects of North America an introduction to the aquatic insects of North America
Næsje T, Jonssons B, Skurdal J (1995) Spring flood: a primary cue for hatching of river spawning Coregoninae. Can J Fish Aquat Sci 52:2190–2196
O'Connor S (2002) The Rio Grande silvery minnow and the endangered species act University of. Colorado Law Review 73:673
Pitcher T, Parrish J (1993) Functions of shoaling behaviour in teleosts. In: Behaviour of teleost fishes. Chapman & Hall London, pp 363–439
Pracheil BM, Muzzall PM (2010) Population dynamics of larval trematodes in juvenile bluegills from three lakes II, Michigan, and the potential for overwinter parasite-induced host mortality. Trans Am Fish Soc 139:652–659
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rahel F, Thel L (2004) Sturgeon chub (Macrhybopsis gelida): a technical conservation assessment. USDA Forest Service, Rocky Mountain Region
Rice WR (1989) Analyzing tables of statistical tests. Evolution; Int J Org Evol 43:223–225
Schneider JC (2000) Manual of fisheries survey methods II: with periodic updates. vol 25. Michigan Department of Natural Resources, Fisheries Division,
Service USFaW (2001) Updated status review of Sicklefin and sturgeon chub in the United States. United States Department of the Interior U.S. Fish and Wildlife Service,
Southwood TR (1977) Habitat, the templet for ecological strategies? J Anim Ecol 46:337–365
Starrett W (1950) Food relationships of the minnows and darters of the Des Moines River, Iowa. Ecology 31:216–233
Steffensen KD, Shuman DA, Stukel S (2014) The status of fishes in the Missouri River, Nebraska: shoal chub (Macrhybopsis hyostoma), sturgeon chub (M. gelida), Sicklefin chub (M. meeki), silver chub (M. storeriana), Flathead chub (Platygobio gracilis), plains minnow (Hybognathus placitus), Western silvery minnow (H. argyritis), and brassy minnow (H. hankinsoni)
Team RC (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria 2013. ISBN 3-900051-07-0,
Therneau TM, Atkinson B, Ripley B (2010) rpart: Recursive Partitioning. R package version 3.1-46 Computer software program retrieved from http://CRAN R-project org/package= rpart. Accessed 12 Dec 2016
Thomas P (1988) Evidence for baseline flow spikes as spawning cues for Colorado squawfish in the Yampa River, Colorado. In: American Fisheries Society Symposium, 1988. pp 68–79
Townsend CR, Hildrew AG (1994) Species traits in relation to a habitat templet for river systems. Freshw Biol 31:265–275
Underwood DM, Echelle AA, Eisenhour DJ, Jones MD, Echelle AF, Fisher WL (2003) Genetic variation in western members of the Macrhybopsis aestivalis complex (Teleostei: Cyprinidae), with emphasis on those of the red and Arkansas river basins. Copeia 2003:493–501
Weitzell R, Khoury M, Gagnon P, Schreurs B, Grossman D, Higgins J (2003) Conservation priorities for freshwater biodiversity in the upper Mississippi River basin. Nature serve and the nature conservancy. 2003 NatureServe is a non-profit organization dedicated to providing the scientific knowledge that forms the basis for effective conservation action. NatureServe 1101:22203–21606
Welker T, Drobish M (2010) Missouri River standard operating procedures for fish sampling and data collection US Army Corps of Engineers, Omaha District, Yankton, South Dakota
Welker T, Drobish M (2011) Pallid sturgeon population assessment project US Army Corps of Engineers, Yankton, SD
Wentworth CK (1922) A scale of grade and class terms for clastic sediments. J Geology 30:377–392
Wickham H, Chang W, Wickham MH (2013) Package ‘ggplot2’
Wiggins GB (1977) Larvae of the north American caddisfly genera (Trichoptera). J Fisheries Board Canada 35
Wiley E, Mayden RL (1985) Species and speciation in phylogenetic systematics, with examples from the north American fish fauna. Ann Mo Bot Gard 72:596–635
Wood S, Wood MS (2015) Package ‘mgcv’ R package version:1.7–29
Acknowledgements
We are grateful to George Cunningham, Jim Lamer at the Kibbe Research Field Station, John West and Dave Herzog with the Missouri Department of Conservation for assistance with specimen collection. Specimen collection and experimentation was approved by the University of Nebraska Omaha’s Institutional Animal Care and Use Committee under protocol number 14-005-02-EP. This study was supported through funding from the University of Nebraska Omaha Graduate Research and Creative Activity, University of Nebraska Omaha University Committee on Research and Creative Activity, Office of Graduate Studies Rhoden Fellowship and the Department of Biology. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government. The Nebraska Cooperative Fish and Wildlife Research Unit is jointly supported by a cooperative agreement between the U.S. Geological Survey, the Nebraska Game and Parks Commission, the University of Nebraska−Lincoln, the U.S. Fish and Wildlife Service and the Wildlife Management Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure statement
The authors report no conflict of interest. The authors are solely responsible for the content and writing of this manuscript.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gaughan, S., Steffensen, K. & Lu, G. Habitat use and population structure of the shoal chub (Macrhybopsis hyostoma) in the upper Mississippi River basin. Environ Biol Fish 102, 901–914 (2019). https://doi.org/10.1007/s10641-019-00878-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10641-019-00878-3