Abstract
Background
Mucin 16 (MUC16), a cell surface-associated mucin, has been implicated to be upregulated in a large repertoire of malignances. However, its function in the pathogenesis of colorectal cancer (CRC) is unknown.
Aims
Here, we explored the regulatory role of MUC16 in CRC.
Methods
First, tumor and paracancerous tissues, and serum samples from 162 CRC patients, peripheral blood samples from 48 healthy volunteers and 72 benign colorectal patients were collected. The correlation between the MUC16 expression and the clinical phenotypes of the patients was analyzed. Subsequently, HCT116 and SW480 cells with deletion of MUC16 were established to detect changes in the growth and metastatic capacities of CRC cells. The genes with the highest correlation with MUC16 were predicted by bioinformatics, and their binding relationships were detected by Co-IP and double-labeled immunofluorescence, followed by functional rescue experiments.
Results
Overexpression of MUC16 in CRC patients was positively correlated with serum biomarkers and poor prognosis of patients. It was demonstrated by in vitro and in vivo experiments that knocking-down the expression of MUC16 could significantly inhibit the growth and metastasis of CRC cells. MUC16 activated janus kinase 2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) by interacting with JAK2. Further overexpression of JAK2 in cells with poor expression of MUC16 revealed a significant increase in the proliferative and metastatic capacities of CRC cells.
Conclusions
MUC16 contributes to the development and progression of CRC by binding to JAK2, thereby promoting phosphorylation of JAK2 and further activating STAT3 phosphorylation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is still one of the driving causes of cancer-related death in a global scale, and the International Agency for Research on Cancer expected an incidence of about 1.8 million new cases of CRC and more than 860,000 CRC-related deaths worldwide for 2018 [1]. The disease is closely related to various cultural and social factors and can be prevented by living a healthy lifestyle, including avoiding the consumption of tobacco and alcohol, maintaining a healthy body weight, physical activities, and a healthy diet [2]. Chemotherapy remains the cornerstone of systemic treatment for CRC, but recent advances in molecular biology of CRC are of great importance to individualize the targeted therapies and to improve the treatment in the future [3].
Epithelia are protected from adverse conditions through a mucous barrier, which are comprised of secreted and transmembrane mucins that are effectors of carcinogenesis [4]. Mucin 16 (MUC16) represents a type I transmembrane mucin which was cloned independently by Lloyd and O'Brien in an effort toward characterizing cancer antigen 125 (CA-125), the most broadly utilized biomarker for ovarian cancer [5]. According to the sequencing data, MUC16 is a large glycoprotein with ~ 12,000 amino acids of heavily O-glycosylated N-terminal region, a tandem repeat region composing of about 60 repeats of 156 amino acids each, a transmembrane domain and a cytoplasmic tail of 32 amino acids [6]. Ectopic expression of the MUC16 was observed to enhance SKOV3 tumor cell growth, colony formation, and tumor growth and metastases in SCID mice [7]. Moreover, MUC16 has been displayed to bind strongly to mesothelin, a protein existing on peritoneal mesothelial cells, which was believed to expedite the adhesion and implantation and peritoneal spread that characterizes epithelial ovarian cancers metastasis [8]. Therefore, we postulated that MUC16 has the potency to participate in the mediation of CRC development by binding to a specific protein. Janus kinases (JAKs) encode a group of tyrosine kinases involved in a specific subset of cytokine receptor pathways, and the JAK/STAT signaling is induced in a wide variety of solid tumors, including CRC [9]. Currently, four JAK members have been identified, including JAK1, JAK2, JAK3 and TYK2 [10]. Interestingly, the correlation between MUC16 and JAK2 has been highlighted in breast cancer [11]. Therefore, we hypothesized that MUC16, together with the downstream regulators JAK2 and the JAK/STAT pathway, may be involved in regulation of CRC progression. We first detected the impact of MUC16 on proliferation, migration, invasion and epithelial-mesenchymal transition (EMT) of CRC cells, and then explored the underlying mechanism. The results would shed light on the exploration of new therapies to CRC.
Materials and Methods
Clinical Samples
The research was permitted by the Ethics Committee of the Second Hospital of Anhui Medical University, and was in accordance with the Declaration of Helsinki. All participants involved in this study signed an informed consent. Tumor tissues and corresponding paracancerous tissues, and serum samples from 162 CRC patients, peripheral blood from 48 healthy volunteers and peripheral blood from 72 benign colorectal patients were collected from the Second Hospital of Anhui Medical University. All clinical samples were preserved in liquid nitrogen until needed. No patient had undergone preoperative chemotherapy or radiotherapy, nor had other malignancies. Routine biochemical tests in fasting blood samples were performed to detect the levels of alpha fetoprotein (AFP), carcinoembryonic antigen (CEA), CA19-9, and CA72-4 in the peripheral blood of patients.
Reverse Transcription-Quantitative PCR (RT-qPCR)
TRIzol reagent (Invitrogen Inc., Carlsbad, CA, USA) was utilized to separate total RNA from cells. The quality and quantity of total RNA was assessed using a NanoDrop 2000c spectrophotometer (Invitrogen). To quantify mRNA expression, total RNA was reversely transcribed to cDNA using a PrimeScript™ RT kit with gDNA eraser (TaKaRa, Dalian, China). qPCR was subsequently performed using TBGreen®Premix Ex Taq™II (TaKaRa) by using special primers (Table 1). The expression of mRNAs was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primers used in the manuscript were designed and synthesized by Shanghai Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China). Expression was quantified using the 2−ΔΔCq method.
Cell Culture
Four human CRC cell lines HCT116, SW480, HT-29, and T84 were obtained from the American Typical Culture Collection (ATCC; Manassas, VA, USA). HCT116, SW480, HT-29, and T84 cells were exposed to DMEM containing 10% FBS (Gibco, Carlsbad, CA, USA) and 1% penicillin–streptomycin (Gibco). Normal human colonic epithelial cells FHC (PCS-200–014™; ATCC) were in minimal essential medium (Gibco) plus 10% FBS and 1% penicillin–streptomycin. The culture environment was at 37 °C with 5% CO2.
Establishment of Stable Cell Lines
Short hairpin RNAs (shRNAs) designed to target MUC16 (shMUC16, shRNA-#1: CCGGACCACCAGCTCTGGATATAAACTCGAGTTTATATCCAGAGCTGGTGGTTTTTTG and shRNA-#2: CGGTCACATCTCCAATGGTTATTACTCGAGTAATAACCATTGGAGATGTGATTTTTG) and negative control (sh-NC) were from GenePharma Co., Ltd. (Shanghai, China). The JAK2 overexpression plasmid pcDNA3.1-JAK2 (oe-JAK2) and the empty pcDNA3.1 plasmid were produced by the Sangon Company. Before transfection, cells were plated into 6-well plates and incubated at 37 °C with 5% CO2. On the second day, Lipofectamine 2000 (Invitrogen) was applied for transfection. After transfection, cells were screened using puromycin for shMUC16-expressing cells and neomycin for JAK2 overexpressing cells. After screening, the culture medium was replaced with a fresh one. After 24 h, the cells were collected to detect the transfection efficiency by RT-qPCR and western blot.
Cell Counting Kit-8 (CCK-8)
Transfected cells were isolated with trypsin and collected by centrifugation 24 h after transfection. Cells were suspended in complete medium, and 100 μL suspension (2 × 103 cells) was seeded into 96-well plates. Cell viability was assessed at 0, 24, 48 and 72 h following cell seeding. At each time point, 10 µL CCK-8 (Sigma-Aldrich, St. Louis, MO, USA) was supplemented, followed by another 2-h incubation at 37 °C with 5% CO2. Finally, the optical density (OD) value at 450 nm was read using a microplate reader.
5-Ethynyl-2'-Deoxyuridine (EdU) Labeling
After transfection, the HCT116 or SW480 cells were cultured in a 24-well plate. EdU analysis was performed according to the manufacturer's protocol (Guangzhou RiboBio Co., Ltd., Guangzhou, Guangdong, China) using the Cell Light™ EdU Apollo® 488 in vitro imaging kit to analyze cell proliferation. Images of cells in four randomly selected microscopic fields were observed and counted using an Olympus DX51 fluorescence microscope (Olympus, Tokyo, Japan).
Flow Cytometry
For the determination of apoptosis levels, the cells were plated and grown overnight in six-well plates. At 48-h post-transfection, apoptosis was determined using annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) staining assay. An apoptosis detection kit (KeyGen, Nanjing, Jiangsu, China) was used according to the manufacturer's instructions. Cells were then harvested in an ethylenediaminetetraacetic acid-free trypsin solution and centrifuged at 2,000 rpm for 5 min. Cell precipitates were then resuspended in 300 µL binding buffer and incubated with 5 µL Annexin V-FITC for 15 min on ice in the dark, and 5 µL PI solution was added to each group. The rate of apoptosis in each group was determined by flow cytometry (FCM, BD Biosciences, San Jose, CA, USA).
For cell cycle determination, approximately 2 × 105 HCT116 or SW480 cells were collected and fixed in 75% ethanol at -20 °C for 24 h. Cells were then stained with DNA staining solution (LiankeBio, Hangzhou, Jiangsu, China) for 30 min, and the cycle distribution of cells in each group was determined by flow cytometry (FCM, BD).
Transwell Assay
Transwell analyses were performed using 8 µm chambers (BD) to detect cell migration and invasion abilities. First, the pre-cooled apical chamber was pretreated with Matrigel (1:20, Corning Glass Works, Corning, N.Y., USA). The upper insert was then hydrated for 1 h by Roswell Park Memorial Institute (RPMI)-1640 in the absence of FBS. Serum-free RPMI-1640 medium suspensions of HCT116 and SW480 cells were then loaded onto the apical chamber at 1 × 105 cells/well, and 10% FBS was supplemented to the culture medium in the basolateral chamber. After an incubation at 37 °C for 24 h in a 5% CO2 atmosphere, the cells in the apical chamber were carefully removed with cotton swabs, while the migrated or invaded cells were fixed in 4% paraformaldehyde for 30 min and stained with 0.5% crystal violet solution for 10 min at ambient temperature. Finally, crystal violet-positive cells in each chamber were counted in six random fields under an inverted phase contrast microscope (Olympus Optical Co., Ltd., Tokyo, Japan), and the average number of invasive or migratory cells per field was calculated.
Western Blot
Cultured cells were lysed in radio immunoprecipitation assay buffer containing protease and phosphatase inhibitors (Beyotime, Shanghai, China). An enhanced bicinchoninic acid protein assay kit (Beyotime) was utilized to assess the total protein concentration. Equal amounts of proteins were sampled onto a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel, separated by electrophoresis and transferred to a polyvinylidene difluoride membrane (Millipore Corp, Billerica, MA, USA). The membranes were then sealed in 5% skim milk for 2 h at ambient temperature and probed with primary antibody overnight at 4 °C and with goat anti-rabbit horseradish peroxidase-coupled secondary antibody (ab205718; Abcam, Cambridge, UK). Chemiluminescence imaging analysis system (Abcam) was employed to visualize the protein bands. GAPDH was used as an internal reference control. Information on the antibodies used in the experiments is shown in Table 2.
Animal Experiments
To minimize the effects of the immune system, we used 72 nude mice (4–8 weeks old, 20 ± 2 g) purchased from the Chinese Academy of Medical Sciences (Beijing, China) for our experiments. All animal experiments were performed in accordance with the institutional guidelines and were approved by the Laboratory Animal Center of the Second Hospital of Anhui Medical University. Briefly, 5 × 106 cells were injected subcutaneously into the right axilla of nude mice (n = 6 in each group). The tumor size was calculated every five days using vernier calipers as follows: tumor volume (mm3) = (L × W2)/2, where L = long axis and W = short axis. The tumor tissue was removed after 35 days. To assess the effect of MUC16 on cell metastasis in vivo, nude mice were subjected to a tail vein injection of 5 × 106 HCT116 or SW480 cells (n = 6 each group). Mice were euthanized using an intraperitoneal injection of sodium pentobarbital (150 mg/kg). After confirming the death of the mice, tumor tissue or liver and lung tissues were extracted from the mice for subsequent experiments. Collected lung tissue and liver tissue were fixed in 4% paraformaldehyde, embedded, and sectioned, followed by hematoxylin–eosin (HE) staining to count the number of metastatic nodules.
Immunohistochemistry
Tumor tissues from each group of mice were fixed with 4% paraformaldehyde. After 24 h, the tissues were paraffin-embedded and cut into 4-μm sections. The sections were deparaffinized with xylene before being rehydrated with gradient ethanol. Then, antigen recovery was performed with 10 mM citrate buffer. The tissue sections were then incubated in 3% H2O2 for 10 min and then sealed at ambient temperature for 1 h. Afterwards, the sections were probed overnight with rabbit antibodies against Ki67 (1:100, ab243878, Abcam), E-cadherin (1:500, ab40772, Abcam) and Snai1 (1:200, #3879, CST), and with the secondary antibody for 1 h at ambient temperature. Finally, images were observed using an Olympus DX51 fluorescence microscope (Olympus), and data were analyzed using Image-Pro Plus 6.0 (Media Cybernetics, Bethesda, MD, USA).
Co-immunoprecipitation (Co-IP)
For immunoprecipitation of MUC16 and JAK2 proteins, anti-MUC16 (290-M, Sigma) and anti-JAK2 (#sc-390539, Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) agarose beads (30 µL) were used to extract MUC16 and JAK2, respectively. The beads were washed 3 times with PBS-Tween/20, and the bound proteins were denatured with 2 × SDS sample buffer. Supernatants were collected and analyzed by western blot.
Double-Labeled Immunofluorescence Staining
HCT116 or SW480 cells on coverslips were fixed with 4% paraformaldehyde and incubated with primary antibodies against MUC16 (sc-365002, Santa cruz) and JAK2 (3230, Cell Signaling Technologies, Beverly, MA, USA) at 4 °C overnight. Cells were incubated with fluorescence-coupled secondary antibody (Invitrogen) and then counter-stained with 4',6-diamidino-2-phenylindole (Life Technologies, Carlsbad, CA, USA). Images were captured using a confocal laser scanning microscope (Leica TCS SP8 STED, Leica Microsystems GmbH, Wetzlar, Germany). Fluorescence integral density was measured using ImageJ software.
Data Analysis
All values are expressed as mean ± standard deviation (SD). All experiments were performed in triplicates. A comparison of the results was performed with unpaired t test, one-way or two-way ANOVA and Tukey's multiple comparison tests. SPSS 23.0 statistical software (IBM Corp. Armonk, N.Y., USA) and GraphPad Prism 7 software (GraphPad, San Diego, CA, USA) were applied into all analysis. Statistically significant differences between groups were defined as p values less than 0.05.
Results
MUC16 Expresses Highly in CRC Tissues and Associated with Poor Prognosis of Patients
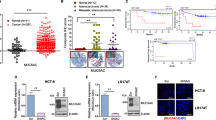
We first queried TCGA database and found significantly abnormal overexpression of MUC16 in several cancers (Fig. 1a). Subsequently, we collected tumor tissue and corresponding paracancerous tissue and serum samples from 162 patients with CRC, peripheral blood from 48 healthy volunteers and peripheral blood from 72 patients with benign colorectal lesions in a clinical setting. The expression profile of MUC16 in the serum of patients with CRC was significantly higher than those in the peripheral blood of healthy volunteers or patients with benign colorectal lesions (Fig. 1b). We further examined the levels of biomarkers AFP, CEA, CA19-9, and CA72-4 in the serum of 162 CRC patients and analyzed the correlations between the levels of tumor markers and the expression of MUC16. The expression of MUC16 was positively correlated with the levels of serum tumor markers in CRC patients (Fig. 1c). We then analyzed the relationship between the expression of MUC16 and the clinical phenotype of patients with CRC. We found that patients with high expression of MUC16 had higher tumor, node, metastases (TNM) stage, lower level of tumor differentiation, and higher risk for lymph node metastasis (Fig. 1D-F). Subsequently, we further analyzed the expression of MUC16 in FHC cells and CRC cell lines HCT116, SW480, HT-29, and T84. The expression of MUC16 was significantly higher in CRC cell lines than in FHC cells (Fig. 1G).
MUC16 overexpression predicts a poor prognosis in CRC patients. a Expression of MUC16 in a variety of tumor tissues in the TCGA database. b Expression of MUC16 in the peripheral blood of 162 CRC patients, 48 healthy volunteers and 72 benign colorectal patients by RT-qPCR. c Spearman correlation analysis of the relations between the levels of tumor markers AFP, CEA, CA19-9, CA72-4 and the MUC16 expression patterns in the serum of CRC patients. d-f Correlations between the MUC16 expression in peripheral blood of CRC patients with patients’ TNM stage (d), differentiation level (e) and lymph node metastasis (f). g Expression of MUC16 in normal colonic epithelial cells FHC and CRC cells HCT116, SW480, HT-29, and T84 detected by RT-qPCR. All values are expressed as mean ± SD. All experiments were performed in triplicates. A comparison of the results was performed with unpaired t test (f) or one-way ANOVA (b, d, e, and g) and Tukey's multiple comparison tests. **p < 0.01, ***p < 0.001
Knockdown of MUC16 Inhibits the Growth of CRC Cells
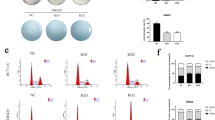
To verify the correlation between MUC16 and the growth of CRC cells, we selected HCT116 and SW480 cells with the highest expression of MUC16 to transfect shRNAs targeting MUC16 and verified the transfection efficiency by RT-qPCR. Expression of MUC16 was significantly reduced in HCT116 and SW480 cells (Fig. 2A, B). We first used CCK-8 assay to detect the growth of the cells, and we found that the growth rate of HCT116 and SW480 cells was significantly reduced in vitro after knockdown of MUC16 (Fig. 2C). Similar to this result, DNA replication activity of HCT116 and SW480 cells was also significantly decreased after knockdown of MUC16 (Fig. 2D). And then, we used flow cytometry to detect apoptosis as well as cell cycle distribution of the cells. The percentage of apoptosis in HCT116 and SW480 cells was significantly increased after knockdown of MUC16 (Fig. 2E), and the cells had significant cell cycle arrest in G2/M phase (Fig. 2F). The above results indicate that depletion of MUC16 significantly curbs the growth and metastasis of CRC cells.
Knockdown of MUC16 inhibits the growth of CRC cells in vitro. ShRNAs targeting MUC16 were transfected into HCT116 and SW480 cells. (A-B) mRNA and protein expression of MUC16 in cells by RT-qPCR (a) and western blot (b). c The growth rate of HCT116 and SW480 cells evaluated by CCK-8. d DNA replication activity in HCT116 and SW480 cells by EdU staining. e Apoptotic ratios in HCT116 and SW480 cells by flow cytometry. f Cell cycle distribution in HCT116 and SW480 by flow cytometry. All values are expressed as mean ± SD. All experiments were performed in triplicates. A comparison of the results was performed with two-way ANOVA and Tukey's multiple comparison tests. **p < 0.01
Knockdown of MUC16 Inhibits CRC Cell Invasion and Migration Abilities
We found a significant correlation between the overexpression of MUC16 and lymph node metastasis in patients with CRC in Fig. 1f. We then further examined the altered migration and invasion in HCT116 and SW480 cells, and we found that the number of cells that migrated or invaded into the basolateral chamber was significantly reduced after knocking down MUC16 (Fig. 3a, b). Then, we further analyzed the expression of factors that are closely related to EMT. The expression patterns of E-cadherin and ZO-1, which are markers of epithelial cell types, were considerably increased in HCT116 and SW480 cells with poor expression of MUC16, while the expression of mesenchymal-related factors N-cadherin, Snai1 and Slug was drastically reduced (Fig. 3c, d). The above results suggest that knockdown of MUC16 can significantly reduce the EMT of CRC cells as well as their migration and invasion in vitro.
MUC16 knockdown inhibits CRC cell aggressiveness. a The migration of HCT116 and SW480 cells examined by Transwell assay. b The invasion of HCT116 and SW480 cells examined by Transwell assay. c mRNA expression of E-cadherin, ZO-1, N-cadherin, Snai1, and Slug in HCT116 and SW480 cells by RT-qPCR. d Protein expression of E-cadherin, ZO-1, N-cadherin, Snai1, and Slug in HCT116 and SW480 cells. All values are expressed as mean ± SD. All experiments were performed in triplicates. A comparison of the results was performed with two-way ANOVA and Tukey's multiple comparison tests. **p < 0.01
Knockdown of MUC16 Inhibits CRC Development and Metastasis In Vivo
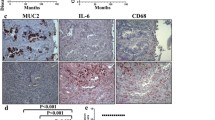
To further clarify the effect of MUC16 on the development and metastasis of CRC cells in vivo, we inoculated HCT116 and SW480 cells with stable low expression of MUC16 into nude mice by subcutaneous injection or tail vein injection. After subcutaneous inoculation of cells, tumor volumes were measured every 5 days for assessing the growth rate of cells in vivo. The growth rate of tumors formed by HCT116 and SW480 cells was significantly reduced in the presence of shMUC16 (Fig. 4a, b). We subsequently extracted tumor tissues for immunohistochemical detection of positive staining rates for KI67, Snai1, and E-cadherin. It was found that the positive rates for KI67 and Snai1 were significantly lower, and the positive rate for E-cadherin was significantly higher in tumors formed by HCT116 and SW480 cells with poor expression of MUC16 (Fig. 4c-e). Then, we extracted liver and lung tissues from nude mice 45 days after tail vein injection of the cells and detected the number of metastatic nodules in liver and lung tissues by HE staining. The metastasis of HCT116 and SW480 cells with knockdown of MUC16 was significantly reduced in lung and liver tissues (Fig. 4f, g).
Knockdown of MUC16 inhibits CRC cell growth and metastasis in vivo. a Growth rate curves of tumors formed by HCT116 and SW480 cells. b Weight of tumors formed by HCT116 and SW480 cells. c–e Positive staining rate of KI67 (c), Snai1 (d), and E-cadherin (e) in tumor tissues by immunohistochemistry. f–g) Lung (F) and liver (G) metastatic nodules of nude mice observed by HE staining. Each group contained 6 mice. All values are expressed as mean ± SD. A comparison of the results was performed with two-way ANOVA and Tukey's multiple comparison tests. **p < 0.01
MUC16 Activates the JAK/STAT Signaling Pathway by Binding to JAK2
To clarify the role of MUC16 in influencing the growth and metastasis of CRC, we further analyzed the genes with correlation coefficient > 0.5 with MUC16 in the TCGA-colorectal adenocarcinoma (COAD) database. The JAK/STAT signaling pathway was significantly enriched (Fig. 5A). Moreover, we further used the Co-IP assay and found that the complexes pulled-down by MUC16 antibodies had significant JAK2 expression, while the complexes pulled-down by JAK2 antibodies also showed MUC16 expression (Fig. 5B). To further verify the sublocalization of MUC16 and JAK2 in CRC cells, double-labeled immunofluorescence was carried out. Both red fluorescence (MUC16) and green fluorescence (JAK2) were significantly expressed in the cytoplasm, and yellow fluorescence appeared after merged (Fig. 5C). The above results indicated that MUC16 could bind to JAK2.
MUC16 activates the JAK/STAT signaling pathway by binding to JAK2. a KEGG enrichment analysis of genes with correlation coefficients greater than 0.5 with MUC16 in the TCGA-COAD database. b Co-IP analysis of the binding of MUC16 to JAK2 protein in HCT116 and SW480 cells. c Subcellular localization of JAK2 and MUC16 by double-labeled immunofluorescence detection. d The extents of JAK2 and STAT3 phosphorylation in HCT116 and SW480 cells assessed by western blot. e–f mRNA and protein expression of Bcl-2, p21, and SOCS1 in HCT116 and SW480 cells by RT-qPCR (e) and western blot (f). All values are expressed as mean ± SD. All experiments were performed in triplicates. A comparison of the results was performed with two-way ANOVA and Tukey's multiple comparison tests. **p < 0.01
Subsequently, we further examined the activation of the JAK/STAT signaling in CRC cells using western blot. The extents of JAK2 and STAT3 phosphorylation were significantly reduced after knockdown of MUC16 (Fig. 5d). We further examined the expression of genes downstream of the JAK/STAT pathway, Bcl-2, p21, and SOCS1, which also showed significantly reduced expression after MUC16 knockdown (Fig. 5e, f). The above results suggest that MUC16 can promote the growth and metastasis of CRC cells by binding to JAK2, thereby promoting the activation of the JAK/STAT signaling pathway.
Overexpression of JAK2 Restores the Malignant Biological Behaviors of CRC Cells
In order to examine the binding of MUC16 to JAK2 in CRC cells, we further overexpressed JAK2 in cells with stable low expression of MUC16 and verified the transfection efficiency by western blot analysis (Fig. 6A). Moreover, we found that the phosphorylation levels of JAK2 and STAT3 had a relative augment after overexpression of JAK2 (Fig. 6A). The expression of Bcl-2, p21, and SOCS1 was also significantly increased after overexpression of JAK2 (Fig. 6B). We further analyzed the activity and DNA proliferation capacity of HCT116 and SW480 cells and observed that overexpression of JAK2 significantly attenuated the inhibitory effect of shMUC16 on cell activity (Fig. 6C, D). Moreover, the results of flow cytometry showed that the percentage of apoptosis in HCT116 and SW480 cells was significantly reduced after overexpression of JAK2, and that overexpression of JAK2 significantly alleviated the cell cycle arrest of G2/M (Fig. 6E, F). We further analyzed the migration and invasion abilities of HCT116 and SW480 cells using Transwell assay. Overexpression of JAK2 significantly increased the number of cells migrated or invaded into the basolateral chamber of Transwell assay in HCT116 and SW480 cells (Fig. 6G, H). In summary, MUC16 contributes to the development and progression of CRC by binding to JAK2, thereby promoting the phosphorylation of JAK2 and further activating the phosphorylation of STAT3.
Overexpression of JAK2 rescues the inhibitory effect of shMUC16 on CRC aggressiveness in vitro. HCT116 and SW480 cells were co-transfected with shMUC16 + oe-NC or oe-JAK2. a The extents of JAK2 and STAT3 phosphorylation in HCT116 and SW480 cells assessed by western blot. b mRNA expression of Bcl-2, p21, and SOCS1 in HCT116 and SW480 cells by RT-qPCR. c The growth rate of HCT116 and SW480 cells evaluated by CCK-8. d DNA replication activity in HCT116 and SW480 cells by EdU staining. e Apoptotic ratios in HCT116 and SW480 cells by flow cytometry. f Cell cycle distribution in HCT116 and SW480 by flow cytometry. g The migration ability of HCT116 and SW480 cells in vitro examined by Transwell assay. h The invasion ability of HCT116 and SW480 cells in vitro examined by Transwell assay. All values are expressed as mean ± SD. All experiments were performed in triplicates. A comparison of the results was performed with two-way ANOVA and Tukey's multiple comparison tests. **p < 0.01
Discussion
CRC, a leading cause of cancer-related death in the world range, results from the accumulation of a series of genetic alterations in the normal colonic epithelium, contributing to the progression of colorectal adenomas and invasive adenocarcinomas [12]. MUC16, a cell surface glycoprotein, is strongly expressed on ovarian tumors where it is shed from the tumor surface in a proteinase-dependent manner [13]. We observed that MUC16, overexpressed in CRC tissues and cell systems, was positively linked to tumor serum biomarkers, including AFP, CEA, CA19-9, and CA72-4 content, advanced TNM stage and higher potentials for lymph node metastasis, which suggests that MUC16 may exert a critical role in the CRC pathogenesis.
The linkage between mucin glycoproteins and CRC has been widely documented [14,15,16,17,18,19]. Nevertheless, its role in CRC remains poorly characterized. Streppel et al. revealed that MUC16 was detected in 64.1% of the COAD samples, and focal MUC16 in COAD was significantly correlated with the outcomes of patients [20]. Consistently, the MUC16 expression was a prognostic factor in pancreatic cancer, independent of other factors, by analysis of TCGA [21]. Here, we observed that cells with MUC16 knockdown showed less growth and tumorigenic properties than control cells transfected with sh-NC. In addition, cells with MUC16 knockdown demonstrated significantly reduced migration and invasion relative to scramble cells, which suggests that MUC16 may be involved in CRC cancer metastasis. Upregulation of MUC16 cytoplasmic tail domain expedited the proliferation and migration capacities of SKOV3 cells, while the loss of MUC16 reversed this effect [22]. Mechanistically, both Akita et al. and Comamala et al. reported that interactions between MUC16 and E-cadherin and β-catenin in human epithelial cancer cells and ovarian carcinoma cells, which might explain the EMT and migration events [23, 24]. Further, EMT markers were significantly altered after MUC16 depletion in our study. The epithelial markers E-cadherin and ZO-1 were enhanced, and the mesenchymal markers N-cadherin, Snai1 and Slug were reduced in CRC cells after MUC16 knockdown, thereby suggesting that MUC16 may be involved in the EMT during CRC cell metastasis. The anti-metastatic role of shMUC16 was then substantiated in vivo, as evidenced by lower KI67- and Snai1-positive rates and higher E-cadherin-positive rates, along with reduced number of metastatic nodules in liver and lung tissues of nude mice.
Since the oncogenic function of MUC16 on CRC has been uncovered, the underlying mechanism was then elucidated. Integrated TCGA-COAD dataset enquiry and experimental validation disclosed that MUC16 interacts with JAK2 to mediate the JAK/STAT pathway. The involvement of JAK2 and the JAK/STAT pathway has been indicated in the progression of CRC via different mechanisms of action [25,26,27,28]. Our western blot analysis presented that MUC16 knockdown reduced the extent of JAK2 and STAT3 phosphorylation as well as the downstream genes of the JAK/STAT pathway, including Bcl-2, p21, and SOCS1. Likewise, IL-6 treatment of CRC cells increased the expression of MUC13 by activating the JAK2/STAT5 signaling pathway [29]. As for MUC16, it facilitated cervical cancer progression through JAK2/STAT3 phosphorylation-modulated cyclooxygenase-2 expression [30]. Moreover, MUC16-induced activation of STAT3 via JAK2 is responsible for increased cell proliferation in cancers [31]. Interestingly, overexpression of MUC16-carboxyl terminal in MUC16 knockdown cells showed activation of JAK2 (Y1007/1008) and STAT3 (Y705) in lung cancer cells [32], indicating the indispensable role of the carboxyl terminal in the function of MUC16. While the underlying mechanism awaits further validation. To corroborate that it is JAK2 that connects MUC16 and the JAK/STAT pathway in CRC, we performed rescue experiments. Overexpression of JAK2 was found to reverse the carcinostatic role of shMUC16 to enhance cell viability and prevent apoptosis. Also, JAK2 was indicated to express highly in liver cancer cell lines relative to the normal liver cells, and JAK2 knockout led to promoted apoptosis and autophagy through inducing Caspase-3, Cyclin-D1 and mTOR regulated by STAT3/5 signaling pathway [33]. However, further experiments are needed to confirm whether this is the main mechanism by which JAK2 participates in CRC progression.
Conclusion
In conclusion, the findings of the present study establish that increased expression of MUC16 may be associated with a poor prognosis in CRC patients. Furthermore, the MUC16 mediates the JAK/STAT pathway via upregulation of JAK2 expression in CRC cells. These results may provide new insights into mechanisms related to CRC development and metastasis. This knowledge may also be beneficial for developing new therapeutic targets for CRC.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Hull R, Francies FZ, Oyomno M, Dlamini Z. Colorectal cancer genetics, incidence and risk factors: in search for targeted therapies. Cancer Manag Res 2020;12:9869–9882.
Binefa G, Rodriguez-Moranta F, Teule A, Medina-Hayas M. Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol. 2014;20:6786–6808.
Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–885.
O’Brien TJ, Beard JB, Underwood LJ, Shigemasa K. The CA 125 gene: a newly discovered extension of the glycosylated N-terminal domain doubles the size of this extracellular superstructure. Tumour Biol. 2002;23:154–169.
Das S, Batra SK. Understanding the unique attributes of MUC16 (CA125): potential implications in targeted therapy. Cancer Res. 2015;75:4669–4674.
Theriault C, Pinard M, Comamala M et al. MUC16 (CA125) regulates epithelial ovarian cancer cell growth, tumorigenesis and metastasis. Gynecol Oncol. 2011;121:434–443.
Rump A, Morikawa Y, Tanaka M et al. Binding of ovarian cancer antigen CA125/MUC16 to mesothelin mediates cell adhesion. J Biol Chem. 2004;279:9190–9198.
Perner F, Perner C, Ernst T, Heidel FH. Roles of JAK2 in aging, inflammation, hematopoiesis and malignant transformation. Cells. 2019;8.
He HL, Lee YE, Liang PI et al. Overexpression of JAK2: a predictor of unfavorable prognosis for nasopharyngeal carcinoma. Future Oncol. 2016;12:1887–1896.
Lakshmanan I, Ponnusamy MP, Das S et al. MUC16 induced rapid G2/M transition via interactions with JAK2 for increased proliferation and anti-apoptosis in breast cancer cells. Oncogene. 2012;31:805–817.
Jung G, Hernandez-Illan E, Moreira L, Balaguer F, Goel A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020;17:111–130.
Bruney L, Conley KC, Moss NM, Liu Y, Stack MS. Membrane-type I matrix metalloproteinase-dependent ectodomain shedding of mucin16/ CA-125 on ovarian cancer cells modulates adhesion and invasion of peritoneal mesothelium. Biol Chem. 2014;395:1221–1231.
Byrd JC, Bresalier RS. Mucins and mucin binding proteins in colorectal cancer. Cancer Metastasis Rev. 2004;23:77–99.
Chen Q, Li D, Ren J, Li C, Xiao ZX. MUC1 activates JNK1 and inhibits apoptosis under genotoxic stress. Biochem Biophys Res Commun. 2013;440:179–183.
Das S, Rachagani S, Sheinin Y et al. Mice deficient in Muc4 are resistant to experimental colitis and colitis-associated colorectal cancer. Oncogene. 2016;35:2645–2654.
Reynolds IS, Fichtner M, McNamara DA, Kay EW, Prehn JHM, Burke JP. Mucin glycoproteins block apoptosis; promote invasion, proliferation, and migration; and cause chemoresistance through diverse pathways in epithelial cancers. Cancer Metastasis Rev. 2019;38:237–257.
Sheng YH, He Y, Hasnain SZ et al. MUC13 protects colorectal cancer cells from death by activating the NF-kappaB pathway and is a potential therapeutic target. Oncogene. 2017;36:700–713.
Zhu X, Long X, Luo X, Song Z, Li S, Wang H. Abrogation of MUC5AC expression contributes to the apoptosis and cell cycle arrest of colon cancer cells. Cancer Biother Radiopharm. 2016;31:261–267.
Streppel MM, Vincent A, Mukherjee R et al. Mucin 16 (cancer antigen 125) expression in human tissues and cell lines and correlation with clinical outcome in adenocarcinomas of the pancreas, esophagus, stomach, and colon. Hum Pathol. 2012;43:1755–1763.
Liang C, Qin Y, Zhang B et al. Oncogenic KRAS targets MUC16/CA125 in pancreatic ductal adenocarcinoma. Mol Cancer Res. 2017;15:201–212.
Chen X, Li X, Wang X, Zhu Q, Wu X, Wang X. MUC16 impacts tumor proliferation and migration through cytoplasmic translocation of P120-catenin in epithelial ovarian cancer cells: an original research. BMC Cancer. 2019;19:171.
Akita K, Tanaka M, Tanida S, Mori Y, Toda M, Nakada H. CA125/MUC16 interacts with Src family kinases, and over-expression of its C-terminal fragment in human epithelial cancer cells reduces cell-cell adhesion. Eur J Cell Biol. 2013;92:257–263.
Comamala M, Pinard M, Theriault C et al. Downregulation of cell surface CA125/MUC16 induces epithelial-to-mesenchymal transition and restores EGFR signalling in NIH:OVCAR3 ovarian carcinoma cells. Br J Cancer. 2011;104:989–999.
Lai F, Deng W, Fu C, Wu P, Cao M, Tan S. Long non-coding RNA SNHG6 increases JAK2 expression by targeting the miR-181 family to promote colorectal cancer cell proliferation. J Gene Med. 2020;22:e3262.
Li S, Xu Z, Guo J, Zheng J, Sun X, Yu J. Farnesoid X receptor activation induces antitumour activity in colorectal cancer by suppressing JAK2/STAT3 signalling via transactivation of SOCS3 gene. J Cell Mol Med. 2020.
Zhang ZH, Li MY, Wang Z et al. Convallatoxin promotes apoptosis and inhibits proliferation and angiogenesis through crosstalk between JAK2/STAT3 (T705) and mTOR/STAT3 (S727) signaling pathways in colorectal cancer. Phytomedicine. 2020;68:153172.
Zhao T, Li Y, Zhang J, Zhang B. PD-L1 expression increased by IFN-gamma via JAK2-STAT1 signaling and predicts a poor survival in colorectal cancer. Oncol Lett. 2020;20:1127–1134.
Gupta BK, Maher DM, Ebeling MC et al. Functions and regulation of MUC13 mucin in colon cancer cells. J Gastroenterol. 2014;49:1378–1391.
Shen H, Guo M, Wang L, Cui X. MUC16 facilitates cervical cancer progression via JAK2/STAT3 phosphorylation-mediated cyclooxygenase-2 expression. Genes Genomics. 2020;42:127–133.
Felder M, Kapur A, Gonzalez-Bosquet J et al. MUC16 (CA125): tumor biomarker to cancer therapy, a work in progress. Mol Cancer. 2014;13:129.
Lakshmanan I, Salfity S, Seshacharyulu P et al. MUC16 regulates TSPYL5 for lung cancer cell growth and chemoresistance by suppressing p53. Clin Cancer Res. 2017;23:3906–3917.
Xu Y, Lv SX. The effect of JAK2 knockout on inhibition of liver tumor growth by inducing apoptosis, autophagy and anti-proliferation via STATs and PI3K/AKT signaling pathways. Biomed Pharmacother. 2016;84:1202–1212.
Acknowledgments
The authors would like to thank the National Clinical Key Specialty Construction Project (2013-544), the Anhui Natural Science Foundation Project (1408085MKL70) and the grants of the Scientific Research of BSKY (XJ201935).
Funding
This work was supported by the National Clinical Key Specialty Construction Project (2013–544), the Anhui Natural Science Foundation Project (1408085MKL70) and the grants of the Scientific Research of BSKY (XJ201935).
Author information
Authors and Affiliations
Contributions
ZNL is the guarantor of integrity of the entire study and contributed to the concepts; YMG and XHL contributed to the design and definition of intellectual content of this study; LBZ, XHC, HJ, YH and YFZ contributed to the experimental studies, data acquisition and statistical analysis; TTX, WSY and QH contributed to the source, validation and methodology. All authors contributed to the manuscript preparation, read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no potential conflicts of interest.
Ethics approval
The research was permitted by the Ethics Committee of the Second Hospital of Anhui Medical University, and was in accordance with the Declaration of Helsinki. All participants involved in this study signed an informed consent. All animal experiments were performed in accordance with the institutional guidelines and were approved by the Laboratory Animal Center of the Second Hospital of Anhui Medical University.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Z., Gu, Y., Li, X. et al. Mucin 16 Promotes Colorectal Cancer Development and Progression Through Activation of Janus Kinase 2. Dig Dis Sci 67, 2195–2208 (2022). https://doi.org/10.1007/s10620-021-07004-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-021-07004-3