Abstract
Background
Circular RNA (circRNA) has been shown to be closely associated with cancer progression, including gastric cancer (GC). However, the function of circ_0004104 in GC progression has not been clarified.
Aims
The purpose of this study was to explore the role of circ_0004104 in GC progression.
Methods
The expression levels of circ_0004104, miR-539-3p, and ring finger protein 2 (RNF2) were assessed using quantitative real-time polymerase chain reaction. Cell proliferation was measured by 3-(4,5-dimethyl-2 thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide assay, and cell migration and invasion were detected using transwell assay. The levels of glutamine, glutamate, and α-ketoglutarate were determined to evaluate the glutaminolysis of cells, and the protein levels of glutaminolysis-related markers and RNF2 were detected using western blot analysis. Furthermore, Dual-Luciferase Reporter Assay was employed to assess the interaction between miR-539-3p and circ_0004104 or RNF2. Animal experiments were carried out to evaluate the effect of circ_0004104 silencing on GC tumor growth in vivo.
Results
Circ_0004104 was upregulated in GC, and its knockdown repressed the proliferation, metastasis, and glutaminolysis of GC cells in vitro and reduced GC tumor growth in vivo. Furthermore, we discovered that circ_0004104 could sponge miR-539-3p and miR-539-3p could target RNF2. The rescue experiments suggested that miR-539-3p inhibitor could reverse the suppressive effect of circ_0004104 silencing on GC progression, and RNF2 overexpression also reversed the inhibition effect of miR-539-3p mimic on GC progression.
Conclusion
Circ_0004104 accelerated GC progression via regulating the miR-539-3p/RNF2 axis, indicating that circ_0004104 might be a potential therapeutic target for GC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) is a malignant tumor that originates from the gastric mucosa epithelium, which is the third leading cause of cancer death [1, 2]. As with traditional cancer treatment, the treatment of GC mainly is surgical resection, chemotherapy, radiotherapy, targeted therapy, and immunotherapy [3,4,5,6]. Despite considerable advances in GC treatment, metastasis and recurrence of GC are still huge problems to be faced in the treatment of GC [7, 8]. Therefore, it is necessary to identify new therapeutic targets for GC treatment.
The function of circular RNA (circRNA) in cancer has been confirmed in recent years, and it is considered to be an important biomarker for cancer [9, 10]. Differential expression of circRNA in cancer often predicts its potential role in cancer, such as participating in cancer progression as an oncogene or tumor suppressor [11, 12]. CircRNAs regulate gene expression through a variety of mechanisms, among which circRNAs have been widely studied as microRNAs (miRNAs) sponges [13, 14]. Several functional cancer-related circRNAs have been identified in GC. For example, circMTO1 had lowly expression in GC, and it could restrain the proliferation and metastasis of GC by regulating the miR-3200-5p/PEBP1 axis [15]. CircPIP5K1A served as a sponge of miR-376c-3p to enhance the progression of GC via regulating ZNF146 expression [16]. Circ-ATAD1 could act as an oncogene to promote GC tumorigenesis by targeting YY1 through sponging miR-140-3p [17]. However, there are many of circRNAs that have not been explored.
Wang et al. explored the differentially expressed (DE) circRNAs in patients with coronary heart disease (CHD) and healthy controls and found that circ_0004104 was significantly upregulated in CHD patients [18]. Using the GEO2R tool, we analyzed the DE circRNAs in GC tissues and normal samples and discovered that circ_0004104 was remarkably overexpressed in GC tissues. However, the function of circ_0004104 in GC is unclear. Therefore, our research is mainly to elucidate the role of circ_0004104 in the progression of GC and reveal its potential molecular mechanism.
Materials and Methods
Analyze the Differentially Expressed circRNA
Gene expression profiles of GC were downloaded from the GEO database (GSE141977). The GEO2R tool was used to analyze the DE circRNAs in three paired GC tissues and normal samples in GSE141977. P < 0.05 and |log2 fold change| (|log2 FC|) > 1 were set as the cutoff criterion.
Clinical Tissues Samples
50 GC patients were recruited from Chongqing Three Gorges Central Hospital. The clinicopathologic features of GC patients are shown in Table 1. After surgical resection, GC tumor tissues (Tumor) and adjacent normal tissues (Normal) were collected and immediately stored in liquid nitrogen until use. This study was approved by the Chongqing Three Gorges Central Hospital Ethics Committee and obtained written informed consent from all patients.
Cell Lines and Culture
Human GC cell lines (HGC-27 and AGS) and human normal gastric mucosa cell line (GES-1) were purchased from Biovector National Typical Culture Center (Beijing, China) and cultured in RPMI-1640 medium (Sigma-Aldrich, St. Louis, MI, USA) containing 10% fetal bovine serum (FBS; Sigma-Aldrich) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA) in a humidified atmosphere at 37 °C with 5% CO2.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
TRIzol reagent (Invitrogen) was employed to extract total RNA. Then, cDNA was synthesized using PrimeScript One-Step RT-PCR Kit (Takara, Dalian, China). QRT-PCR was carried out using SYBR Green Master Mix (Vazyme, Nanjing, China). GAPDH was used for normalizing the expression of circ_0004104, secreted protein acidic and rich in cysteine (SPARC), and ring finger protein 2 (RNF2), and U6 was used for normalizing the expression of miR-539-3p using the 2−ΔΔCt method. Primer sequences were summarized as follows: circ_0004104, F 5′-CCGCTTTTTCGAGACCTGTG-3′, R 5′-TTGCAAGGCCCGATGTAGTC-3′; SPARC, F 5′-GGGCTTGGATTTTCTTCCTC-3′, R 5′-GCAGGCTCATCAAACTCTCC-3′; RNF2, F 5′-TTCAGGCCTCATCCCACACT-3′, R 5′-CAGCAGCTCCGCCACTC-3′; GAPDH, F 5′-AATCCCATCACCATCTTCC-3′, R 5′-CATCACGCCACAGTTTCC-3′; miR-539-3p, F 5′-GCAGGAACGGTTCTCCACTC-3′, R 5′-GTCCCTAGGGATGTGTAAGGA-3′; U6, F 5′-CGCTTCACGAATTTGCGTGTCAT-3′, R 5′-GCTTCGGCAGCACATATACTAAAAT-3′.
Ribonuclease R (RNase R) Assay
RNase R was obtained from Lucigen (Middleton, WI, USA). In brief, extracted RNA was incubated with RNase R for 15 min. Then, qRT-PCR was carried out to assess the expression of circ_0004104 and linear SPARC. The RNA not incubated with RNase R was used as the negative control (Mock).
Cell Transfection
Circ_0004104 small interference RNA and lentiviral short hairpin RNA (si-circ_0004104 and sh-circ_0004104), miR-539-3p mimic and inhibitor (miR-539-3p and anti-miR-539-3p), RNF2 overexpression vector (RNF2), and negative controls (si-NC, sh-NC, miR-NC, anti-miR-NC, and vector) were obtained from Ribobio (Guangzhou, China). They were transfected into HGC-27 and AGS cells by Lipofectamine 3000 (Invitrogen).
3-(4,5-Dimethyl-2 thiazolyl)-2,5-diphenyl-2-H-tetrazolium Bromide (MTT) Assay
MTT Cell Proliferation and Cytotoxicity Assay Kit was purchased from Sangon Biotech (Shanghai, China). Briefly, HGC-27 and AGS cells were harvested and re-seeded into 96-well plates after transfection. After 24 h, MTT solution was added into each well and incubated for 4 h. Then, formazan solubilization solution was added into each well to dissolve the formazan. The absorbance was determined at 490 nm using an auto-microplate reader.
Transwell Assay
For migration assay, HGC-27 and AGS cells (1 × 105) were suspended with serum-free medium and seeded onto the upper chambers (BD Biosciences, San Jose, CA, USA). For invasion assay, the cells were seeded onto the upper chambers pre-coated with Matrigel (BD Biosciences). In addition, the complete medium was placed in the lower chambers. After 24 h, the migrated and invaded cells were fixed with paraformaldehyde and stained with crystal violet. The cells were photographed, and cell number was counted using a microscope (100 ×).
Detection of Glutamine, Glutamate, and α-Ketoglutarate (α-KG) Levels
Glutamine, Glutamate, and α-KG Assay Kits were obtained from Abcam (Cambridge, MA, USA). In brief, HGC-27 and AGS cells (2 × 106) were harvested and washed with cold PBS. According to the manufacturer’s instructions, the levels of glutamine, glutamate, and α-KG were determined using corresponding Assay Kits, respectively.
Western Blot (WB) Analysis
HGC-27 and AGS cells were lysed with RIPA lysis buffer (Applygen, Beijing, China) to extract total proteins, and then the proteins were separated by 10% SDS-PAGE and transferred to PVDF membranes (Beyotime, Shanghai, China). The membranes were incubated with primary antibodies against ASC transporter 2 (ASCT2, Abcam, 1:500), glutaminase 1 (GLS1, Abcam, 1:2500), RNF2 (Abcam, 1:2000), or GAPDH (Abcam, 1:10,000) after blocked with 5% nonfat milk, followed by treated with Goat Anti-Rabbit IgG H&L (HRP) (Abcam, 1:50,000). The membranes were then visualized using an ECL luminescence reagent (Sangon Biotech).
Dual-Luciferase Reporter Assay
The fragments of circ_0004104 and RNF2 3′UTR containing the complementary sequences or mutant sequences of miR-539-3p were amplified and sub-cloned into the psi-CHECK reporter vector (Promega, Madison, WI, USA) to form circ_0004104 and RNF2 3′UTR wild-type (WT) or mutant-type (MUT) reporter vectors. The reporter vectors and miR-539-3p mimic or miR-NC were co-transfected into HGC-27 and AGS cells by Lipofectamine 3000. The luciferase activity of cells was utilized using Dual-Luciferase Reporter Assay System (Promega).
Tumor Transplant Models
AGS cells (5 × 106) transfected with sh-circ_0004104 or sh-NC were subcutaneously injected into the flanks of BALB/c nude mice (Cloud-clone Corp, Wuhan, China). The tumor weight and width were measured every 5 d using a caliper, and tumor volume was calculated. After 30 d, the mice were killed, and the tumor was collected and weighed. Then, qRT-PCR and WB analyses were used to measure the expression of circ_0004104, miR-539-3p, and RNF2. Animal experiments were approved by the Ethics Committee of Chongqing Three Gorges Central Hospital.
Statistical Analysis
All experiments were performed in triplicates independently. All data were presented as mean ± SD. GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA) was used for statistical analyses. Differences between two or more groups were analyzed using Student’s t test or one-way analysis of variance followed by Tukey’s post hoc analysis. Log-rank test was used for Kaplan–Meier analysis, and Pearson correlation analysis was used for analyzing the correlations among circ_0004104, miR-539-3p, and RNF2. P < 0.05 was considered statistically significant.
Results
Circ_0004104 Was Upregulated in GC Tumor Tissues and Cells
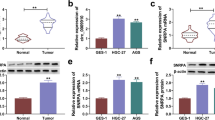
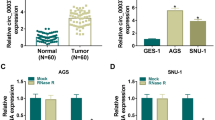
Using the GEO2R tool, we found that circ_0004104 had increased expression in GC tumor tissues (GEO accession: GSE141977) (Fig. 1a). Through genomic localization, we uncovered that circ_0004104 was derived from the Exon 2-5 of the SPARC gene (Fig. 1b). We detected the expression of circ_0004104 in 50 paired GC tumor tissues and adjacent normal tissues and found that circ_0004104 was markedly highly expressed in GC tumor tissues compared with adjacent normal tissues (Fig. 1c). Furthermore, we also examined its parent gene SPARC expression and confirmed that SPARC expression was significantly higher in GC tumor tissues than that in normal samples from the TCGA COAD dataset (Fig. 1d). According to the median of circ_0004104 expression in GC tumor tissues, all GC tumor tissues were divided into high circ_0004104 expression group (n = 25) and low circ_0004104 expression group (n = 25) (Fig. 1e), and statistical results showed that high circ_0004104 expression was closely associated with the TNM stages, lymph metastasis, and tumor size of GC patients (Table 1). Also, Kaplan–Meier analysis showed that the overall survival rate of GC patients with the high circ_0004104 expression tended to be lower (Fig. 1f). In addition, upregulation of circ_0004104 also was found in GC cell lines (HGC-27 and AGS) compared with GES-1 cells (Fig. 1g). The results of RNase R assay confirmed that circ_0004104 could resist the digestion of RNase R, indicating that circ_0004104 was indeed a circRNA (Fig. 1h–i).
Circ_0004104 was upregulated in GC. a The expression of circ_0004104 in three paired GC tumor tissues (Tumor) and adjacent normal tissues (Normal) from the GSE141977 database was shown. b The information of circ_0004104 was presented. c The expression of circ_0004104 in Tumor (n = 50) and Normal (n = 50) was detected by qRT-PCR. dThe TCGA COAD dataset showed the expression of circ_0004104 in 408 GC tissues (T) and 211 normal samples (N). e The expression distribution of circ_0004104 in 50 GC tissues was exhibited. f Kaplan–Meier analysis was used to analyze the relationship between circ_0004104 expression and the overall survival of GC patients. g QRT-PCR was performed to measure circ_0004104 expression in GC cell lines (HGC-27 and AGS) and human normal gastric mucosa cell line (GES-1). h, i RNase R assay was employed to confirm the circular characteristic of circ_0004104 in HGC-27 and AGS cells. *P < 0.05
Circ_0004104 Knockdown Repressed the Proliferation, Migration, Invasion, and Glutaminolysis of GC Cells
For investigating the role of circ_0004104 in GC, the loss-of-function experiments were performed using si-circ_0004104. After transfected with si-circ_0004104 into HGC-27 and AGS cells, we discovered that si-circ_0004104 could markedly inhibit circ_0004104 expression, confirming the transfection was successful (Fig. 2a, b). The results of MTT assay revealed that circ_0004104 knockdown significantly suppressed the proliferation of HGC-27 and AGS cells (Fig. 2c, d). Moreover, transwell assay indicated that the migration and invasion of HGC-27 and AGS cells could be repressed by circ_0004104 silencing (Fig. 2e, f). Furthermore, silenced circ_0004104 also reduced the levels of glutamine, glutamate, and α-KG in HGC-27 and AGS cells, indicating that knockdown of circ_0004104 inhibited the glutaminolysis of GC cells (Fig. 2g–l). In addition, the glutamine transporter ASCT2 and the glutaminolysis rate-limiting enzyme GLS1 also could be decreased by circ_0004104 silencing (Fig. 2m, n).
Circ_0004104 knockdown repressed the progression of GC cells. HGC-27 and AGS cells were transfected with si-NC or si-circ_0004104. a, b The expression of circ_0004104 was determined by qRT-PCR to evaluate transfection efficiency. c, d MTT assay was used to measure cell proliferation. Transwell assay was performed to detect the migration (e) and invasion (f) of cells. The levels of glutamine (g, h), glutamate (i, j), and α-KG (k, l) were examined using corresponding Assay Kits, respectively. m, n The protein levels of ASCT2 and GLS1 were tested by WB analysis. *P < 0.05
Circ_0004104 Could Serve as a Sponge of miR-539-3p
To perfect the molecular mechanism of circ_0004104 in GC, we used the StarBase v2.0 tool to predict the miRNAs that had targeted binding sites with circ_0004104. As shown in Fig. 3a, miR-539-3p was predicted to be complementary to circ_0004104. Subsequently, Dual-Luciferase Reporter Assay was performed, and the results revealed that miR-539-3p overexpression could remarkably restrain the luciferase activity of circ_0004104 WT reporter without affecting circ_0004104 MUT reporter (Fig. 3b, c). Furthermore, we detected the expression of miR-539-3p in HGC-27 and AGS cells transfected with si-circ_0004104 and discovered that its expression could be promoted by circ_0004104 silencing (Fig. 3d, e). In addition, through measuring miR-539-3p expression in GC tumor tissues and cells, we suggested that miR-539-3p was lowly expressed in GC tumor tissues and cells (Fig. 3f, g), and its expression was negatively correlated with circ_0004104 expression in GC tissues (Fig. 3h). Therefore, our results revealed that circ_0004104 could sponge miR-539-3p in GC.
Circ_0004104 could sponge miR-539-3p. a The binding sites of circ_0004104 on miR-539-3p were presented. b, c The interaction between circ_0004104 and miR-539-3p was verified using Dual-Luciferase Reporter Assay. d, e The expression of miR-539-3p in HGC-27 and AGS cells transfected with si-NC or si-circ_0004104 was measured by qRT-PCR. f The expression of miR-539-3p in Tumor (n = 50) and Normal (n = 50) was detected by qRT-PCR. g QRT-PCR was used to determine the expression of miR-539-3p in GC cell lines (HGC-27 and AGS) and GES-1 cells. h Pearson correlation analysis was used to analyze the correlation between circ_0004104 and miR-539-3p in GC tissues. *P < 0.05
MiR-539-3p Inhibitor Partially Reversed the Inhibition Effect of Circ_0004104 Silencing on the Proliferation, Migration, Invasion, and Glutaminolysis of GC Cells
To confirm that circ_0004104 regulated GC progression via targeting miR-539-3p, we co-transfected with si-circ_0004104 and anti-miR-539-3p into HGC-27 and AGS cells to carry out the rescue experiments. Compared with the control group, the significant decrease of miR-539-3p expression in the si-circ_0004104 + anti-miR-539-3p group confirmed the success of anti-miR-539-3p transfection (Fig. 4a, b). The results of MTT assay and transwell assay indicated that miR-539-3p inhibitor could partially reverse the suppressive effect of circ_0004104 silencing on the proliferation, migration, and invasion of HGC-27 and AGS cells (Fig. 4c–h). Similarly, the inhibitory effect of circ_0004104 knockdown on the levels of glutamine, glutamate, and α-KG also could be partially reversed by miR-539-3p inhibitor in HGC-27 and AGS cells (Fig. 4i–n). At the same time, we also discovered that the protein levels of ASCT2 and GLS1 were improved in the si-circ_0004104 + anti-miR-539-3p group compared with the control group (Fig. 4o, p). Hence, all data indicated that miR-539-3p was involved in the regulation of circ_0004104 on GC progression.
Effects of circ_0004104 silencing and miR-539-3p inhibitor on the progression of GC cells. HGC-27 and AGS cells were transfected with si-NC, si-circ_0004104, si-circ_0004104 + anti-miR-NC, or si-circ_0004104 + anti-miR-539-3p. a, b The expression of miR-539-3p was determined by qRT-PCR to evaluate transfection efficiency. c, d Cell proliferation was examined using MTT assay. Transwell assay was used to measure the migration e, f and invasion g, h of cells. Corresponding Assay Kits were performed to assess the levels of glutamine (i, j), glutamate (k, l), and α-KG (m, n), respectively. o, p WB analysis was employed to detect the protein levels of ASCT2 and GLS1. *P < 0.05
RNF2 Was a Target of miR-539-3p
To search for the targets of miR-539-3p, the Targetscan.org tool was used and RNF2 was found to have binding sites with miR-539-3p (Fig. 5a). Then, we performed the Dual-Luciferase Reporter Assay and discovered that miR-539-3p overexpression only could inhibit the luciferase activity of RNF2 3′UTR WT reporter, while no effect on the luciferase activity of RNF2 3′UTR MUT reporter (Fig. 5b, c). Besides, in HGC-27 and AGS cells transfected with miR-539-3p mimic, we found that miR-539-3p overexpression could markedly suppress RNF2 expression, both in mRNA and protein levels (Fig. 5d–f). Meanwhile, we also uncovered that circ_0004104 silencing could significantly decrease the mRNA and protein levels of RNF2 in HGC-27 and AGS cells, while the addition of miR-539-3p inhibitor could partially reverse this effect (Fig. 5g–i). In GC tumor tissues, we found that the mRNA expression of RNF2 was higher than adjacent normal tissues (Fig. 5j), which was consistent with the results of the TCGA COAD dataset (Fig. 5k). Also, the detection results of RNF2 protein level were also similar to the results of mRNA level (Fig. 5l). Furthermore, we discovered a significant increase of RNF2 expression in GC cell lines compared with GES-1 cells, as demonstrated by qRT-PCR and WB analysis (Fig. 5m, n). In addition, correlation analysis revealed that RNF2 expression was negatively correlated with miR-539-3p expression and positively correlated with circ_0004104 expression in GC tissues (Fig. 5o, p).
RNF2 was a target of miR-539-3p. a The binding sites of RNF2 3′UTR on miR-539-3p were shown. b, c Dual-Luciferase Reporter Assay was used to verify the interaction between RNF2 and miR-539-3p. The mRNA and protein levels of RNF2 were detected by qRT-PCR (d) and WB analysis (e, f) in HGC-27 and AGS cells transfected with miR-NC or miR-539-3p mimic. QRT-PCR (g) and WB analysis (h, i) were used to determine the mRNA and protein levels of RNF2 in HGC-27 and AGS cells transfected with si-NC, si-circ_0004104, si-circ_0004104 + anti-miR-NC, or si-circ_0004104 + anti-miR-539-3p. j The mRNA expression of RNF2 in tumor (n = 50) and normal (n = 50) was determined by qRT-PCR. k The TCGA COAD dataset indicted the expression of RNF2 in 408 GC tissues (t) and 211 normal samples (N). l WB analysis was used to measure the protein expression of RNF2 in tumor (n = 50) and normal (n = 50). The mRNA and protein levels of RNF2 in GC cell lines (HGC-27 and AGS) and GES-1 cells were assessed using qRT-PCR (m) and WB analysis (n). o, p The correlation between RNF2 and miR-539-3p or circ_0004104 was analyzed using Pearson correlation analysis. *P < 0.05
Suppressive Effect of miR-539-3p Overexpression on GC Progression Could Be Partially Reversed by RNF2 Overexpression
We co-transfected with miR-539-3p mimic and RNF2 overexpression plasmid into HGC-27 and AGS cells to explore whether miR-539-3p regulated GC progression by targeting RNF2. The detection results of RNF2 protein level revealed that RNF2 overexpression plasmid could invert the inhibition effect of miR-539-3p overexpression on RNF2 expression, confirming the transfection efficiency of both was good (Fig. 6a, b). Through MTT assay and transwell assay, we discovered that miR-539-3p overexpression markedly inhibited the proliferation, migration and invasion of HGC-27 and AGS cells, while this effect could be reversed by the addition of RNF2 overexpression plasmid (Fig. 6c–h). Also, overexpressed RNF2 reversed the suppression effect of miR-539-3p overexpression on the levels of glutamine, glutamate, and α-KG (Fig. 6i–n), as well as the protein levels of ASCT2 and GLS1 in HGC-27 and AGS cells (Fig. 6o, p). These data showed that miR-539-3p restrained GC progression by targeting RNF2.
Effects of miR-539-3p overexpression and RNF2 overexpression on the progression of GC cells. HGC-27 and AGS cells were transfected with miR-NC, miR-539-3p, miR-539-3p + vector, or miR-539-3p + RNF2. a, b The protein level of RNF2 was measured by WB analysis to evaluate transfection efficiency. c, d MTT assay was performed to test cell proliferation. Transwell assay was employed to detect the migration (e, f) and invasion (g, h) of cells. Corresponding Assay Kits were performed to determine the levels of glutamine (i, j), glutamate (k, l), and α-KG (m, n), respectively. o, p The protein levels of ASCT2 and GLS1 were assessed using WB analysis. *P < 0.05
Silencing of Circ_0004104 Inhibited the Growth of GC Tumor In Vivo
To further confirm the role of circ_0004104 in GC, we constructed GC tumor transplant models. After 30 days of measurement, we found that the tumor volume of the sh-circ_0004104 group was obviously lower than the sh-NC group (Fig. 7a). Meanwhile, the tumor weight in the sh-circ_0004104 group also was reduced compared with the sh-NC group (Fig. 7b). To confirm the success of sh-circ_0004104 transfection, we detected circ_0004104 expression in the tumors and discovered that the expression of circ_0004104 in the sh-circ_0004104 group was remarkably decreased compared with the sh-NC group (Fig. 7c). In addition, we also determined the expression of miR-539-3p and RNF2 in the tumors. As presented in Fig. 7d–f, we uncovered that miR-539-3p expression was markedly promoted, while RNF2 mRNA and protein levels were significantly suppressed in the sh-circ_0004104 group. Therefore, our data indicated that circ_0004104 knockdown reduced GC tumor growth through inhibiting RNF2 by promoting miR-539-3p.
Silencing of circ_0004104 inhibited the growth of GC tumor in vivo. AGS cells transfected with sh-NC or sh-circ_0004104 were injected into the flank of nude mice. Tumor volume (a) and tumor weight (b) were determined after 30 d. c–e QRT-PCR was used to measure the expression of circ_0004104, miR-539-3p, and RNF2 in the tumors. f WB analysis was performed to detect the protein level of RNF2 in the tumors. *P < 0.05
Discussion
As a common malignant tumor of the digestive system, the occurrence of GC seriously harms human health [19]. More and more studies show that non-coding RNAs (ncRNAs) have an essential regulatory function in various processes, including cancer progression [20]. At present, molecular targeted therapy of ncRNA has been proven to have great potential in cancer treatment and can serve as a direction for future research [21, 22]. CircRNA is one of the most widely studied ncRNA in recent years. In our study, we selected an upregulated circRNA, circ_0004104, in GC, and explored its role in GC. Our results discovered that circ_0004104 was highly expressed in GC, which was similar with the expression trend in CHD [18]. In addition, we found that high circ_0004104 level was related to the poor prognosis of GC patients. Functional experiments confirmed that circ_0004104 silencing could hinder the proliferation, metastasis, and glutaminolysis of GC cells in vitro and reduce GC tumor growth in vivo, suggesting that circ_0004104 might play a pro-cancer role in GC.
The hypothesis of the circRNA-miRNA-mRNA axis has been regarded as an important way to elucidate the molecular mechanism of circRNAs [13, 14]. For revealing the molecular mechanism of circ_0004104, we performed bioinformatics prediction and experimental verification and uncovered that circ_0004104 could serve as a sponge of miR-539-3p, and miR-539-3p could target RNF2. Previous studies suggested that miR-539 was differentially expressed in H. pylori-induced gastritis and MALT lymphoma, suggesting that it might be related to the malignant transformation of gastritis [23]. Studies had indicated that miR-539-3p could suppress GC proliferation and metastasis to restrain the progression of GC by targeting CTBP1 and SOX5 [24, 25]. And the results of Jin et al. revealed that low miR-539 expression could accelerate GC progression [26]. Therefore, miR-539-3p might function as a tumor suppressor in GC. Our research revealed that circ_0004104 negatively regulated miR-539-3p expression in vitro and in vivo and confirmed that circ_0004104 regulated GC progression by sponging miR-539-3p. Importantly, the anticancer effect of miR-539-3p could help us better understand the cancer-promoting mechanism of circ_0004104 in GC.
RNF2, a catalytic subunit of polycomb repressive complex 1 with E3 ligase activity, has been shown to be able to participate in a variety of biological functions, such as cell proliferation, apoptosis, and metastasis [27, 28]. Increasing evidence indicates that RNF2 is significantly overexpressed in various types of cancer and is closely related to cancer progression [29]. In GC, Qi et al. suggested that long ncRNA LINC00665 could promote RNF2 expression to enhance the tumorigenesis of GC via sponging miR-149-3p [30], and Zhang et al. showed that RNF2 was upregulated in GC and its silencing could inhibit cell viability and induce cell cycle arrest in GC cells [31]. These revealed that RNF2 was an oncogene in GC. Consistent with the previous study, we found that RNF2 had elevated expression in GC. In addition, our study confirmed that circ_0004104 could positively regulate RNF2 expression to promote GC progression by sponging miR-539-3p.
In conclusion, our study is the first to clarify the role of circ_0004104 in GC. Our results confirmed that circ_0004104 promoted the proliferation, metastasis, glutaminolysis of GC by regulating the miR-539-3p/RNF2 axis. Our findings enrich the research gaps in circ_0004104 and provide a new potential therapeutic target for GC treatment.
References
Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664.
Zhang XY, Zhang PY. Gastric cancer: somatic genetics as a guide to therapy. J Med Genet. 2017;54:305–312.
Kinami S, Funaki H, Fujita H, Nakano Y, Ueda N, Kosaka T. Local resection of the stomach for gastric cancer. Surg Today. 2017;47:651–659.
Ratti M, Lampis A, Hahne JC, Passalacqua R, Valeri N. Microsatellite instability in gastric cancer: molecular bases, clinical perspectives, and new treatment approaches. Cell Mol Life Sci CMLS. 2018;75:4151–4162.
Coutzac C, Pernot S, Chaput N, Zaanan A. Immunotherapy in advanced gastric cancer, is it the future? Crit Rev Oncol/Hematol. 2019;133:25–32.
Patel TH, Cecchini M. Targeted therapies in advanced gastric cancer. Curr Treat Opt Oncol. 2020;21:70.
Figueiredo C, Camargo MC, Leite M, Fuentes-Panana EM, Rabkin CS, Machado JC. Pathogenesis of gastric cancer: genetics and molecular classification. Curr Top Microbiol Immunol. 2017;400:277–304.
Hamashima C. Current issues and future perspectives of gastric cancer screening. World J Gastroenterol. 2014;20:13767–13774.
Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14:514–521.
Chen B, Huang S. Circular RNA: an emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41–50.
Jin C, Zhao W, Zhang Z, Liu W. CircLMTK2 acts as a tumor suppressor in prostate cancer via regulating the expression of microRNA-183. Life Sci. 2020;241:117097.
Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20.
Qiu Y, Pu C, Li Y, Qi B. Construction of a circRNA-miRNA-mRNA network based on competitive endogenous RNA reveals the function of circRNAs in osteosarcoma. Cancer Cell Int. 2020;20:48.
Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79.
Hu K, Qin X, Shao Y, Zhou Y, Ye G, Xu S. Circular RNA MTO1 suppresses tumorigenesis of gastric carcinoma by sponging miR-3200-5p and targeting PEBP1. Mol Cell Probes 2020:101562.
Ma Y, Cong X, Zhang Y, Yin X, Zhu Z, Xue Y. CircPIP5K1A facilitates gastric cancer progression via miR-376c-3p/ZNF146 axis. Cancer Cell Int. 2020;20:81.
Zhang L, Chang X, Zhai T, Yu J, Wang W, Du A, et al. A novel circular RNA, circ-ATAD1, contributes to gastric cancer cell progression by targeting miR-140-3p/YY1/PCIF1 signaling axis. Biochem Biophys Res Commun 2020.
Wang L, Shen C, Wang Y, et al. Identification of circular RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis. 2019;286:88–96.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: A Cancer J Clin 2018;68:7–30.
Ling H, Girnita L, Buda O, Calin GA. Non-coding RNAs: the cancer genome dark matter that matters! Clin Chem Lab Med. 2017;55:705–714.
Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474:4219–4251.
Chen L, Dzakah EE, Shan G. Targetable long non-coding RNAs in cancer treatments. Cancer Lett. 2018;418:119–124.
Thorns C, Kuba J, Bernard V, et al. Deregulation of a distinct set of microRNAs is associated with transformation of gastritis into MALT lymphoma. Virchows Archiv Int J Pathol. 2012;460:371–377.
Zhou J, Su M, Zhang H, Wang J, Chen Y. miR-539-3P inhibits proliferation and invasion of gastric cancer cells by targeting CTBP1. Int J Clin Exp Pathol. 2019;12:1618–1625.
Ding S, Zhang Y. MicroRNA539 inhibits the proliferation and migration of gastric cancer cells by targeting SRYbox 5 gene. Mol Med Rep. 2019;20:2533–2540.
Jin W, Han H, Liu D. Downregulation miR-539 is associated with poor prognosis of gastric cancer patients and aggressive progression of gastric cancer cells. Cancer Biomark Sect A Dis Mark. 2019;26:183–191.
Vidal M. Role of polycomb proteins Ring1A and Ring1B in the epigenetic regulation of gene expression. Int J Dev Biol. 2009;53:355–370.
Wen W, Peng C, Kim MO, et al. Knockdown of RNF2 induces apoptosis by regulating MDM2 and p53 stability. Oncogene. 2014;33:421–428.
Su WJ, Fang JS, Cheng F, Liu C, Zhou F, Zhang J. RNF2/Ring1b negatively regulates p53 expression in selective cancer cell types to promote tumor development. Proc Natl Acad Sci USA. 2013;110:1720–1725.
Qi H, Xiao Z, Wang Y. Long non-coding RNA LINC00665 gastric cancer tumorigenesis by regulation miR-149-3p/RNF2 axis. OncoTargets Therapy. 2019;12:6981–6990.
Zhang J, Sun Z, Han Y, et al. Rnf2 knockdown reduces cell viability and promotes cell cycle arrest in gastric cancer cells. Oncol Lett. 2017;13:3817–3822.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yue, F., Peng, K., Zhang, L. et al. Circ_0004104 Accelerates the Progression of Gastric Cancer by Regulating the miR-539-3p/RNF2 Axis. Dig Dis Sci 66, 4290–4301 (2021). https://doi.org/10.1007/s10620-020-06802-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-020-06802-5