Abstract
Approximately 80% of the human genome harbors biochemical marks of active transcription that its majority transcribes to noncoding RNAs, namely long noncoding RNAs (lncRNAs). LncRNAs are heterogeneous RNA transcripts that regulate critical biological processes such as cell survival and death. They involve in the progression of different cancers by affecting transcriptional and post-transcriptional modifications as well as epigenetic control of numerous tumor suppressors and oncogenes. Recent findings show that aberrant expression of lncRNAs is associated with tumor initiation, progression, invasion, and overall survival of patients with gastrointestinal (GI) cancers. Some lncRNAs play as tumor suppressors in all GI cancers, but others play as tumor promoters. However, some other lncRNAs might function as a tumor suppressor in one GI cancer, but as a tumor promoter in another GI cancer type. This fact highlights possible context dependency of the expression patterns and roles of at least some lncRNAs in GI cancer development and progression. Here, we review the functional relation of lncRNAs involved in the development and progression of GI cancer by focusing on their roles as tumor suppressor and tumor promoter genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Failure in regulatory mechanisms involved in the maintenance of homeostasis of the human body causes cancer, the severe health problem worldwide nowadays [1, 2]. The gradual accumulation of genetic and epigenetic changes and combined interaction between two groups of critical genes named proto-oncogenes and tumor suppressor genes promote the progression of the basic properties of this serious threat of human life [3, 4]. These two types of genes usually control the main cellular processes such as growth, division, differentiation, DNA repair, survival, and programmed cell death [3, 5]. Therefore, mutations or epigenetic alterations which activate or destroy the expression of proto-oncogenes or tumor suppressors, respectively, may change the cellular destination toward the cancer initiation or progression [6].
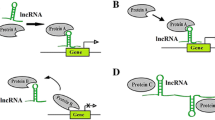
The FANTOM and ENCODE, two large‐scale genomic projects, elucidated that about 80% of the human genome harbors biochemical marks of active transcription. However, 2% of the genome codes for proteins and its majority transcribe to noncoding RNA (ncRNA) molecules [7, 8]. Long noncoding RNAs (lncRNAs), one of the major subclasses of ncRNAs, comprise a class of newly discovered effectors of gene expression [9, 10]. They are more than 200 nucleotides in length and embrace more than 80% of the cancer-associated variations [9, 11]. These regulatory molecules are usually transcribed by RNA polymerase II and undergo processing and splicing events like mRNA [9]. According to genomic origin, they are totally categorized as: (a) Intergenic (unique transcription units between two protein coding genes), (b) Sense, (c) Antisense overlapping lncRNAs (having at least one exon or part of it in common with a respective protein coding gene in the sense or antisense direction), (d) Intronic (originate from the intronic region of a gene), (e) Bidirectional (use the promoter of a neighboring gene in the opposite direction), and (f) Enhancer, 3′ untranslated region (UTR) and 5′ UTR associated lncRNAs which are transcribed from regulatory, 3′ and 5′ UTR regions of a gene, respectively [12,13,14]. These molecules implicate in many cellular processes such as growth, apoptosis, splicing, genomic imprinting, signal transduction, and disease conditions [9, 15]. They act through the direct interaction with chromatin as well as other molecules such as transcription factors, RNA-binding proteins, and micro-RNAs (miRNA) in order to regulate the structure, localization, degradation, and gene expression efficiency [9, 16]. In a consensus manner, their mechanism of action can be divided into four groups including signal which fluctuation in response to different stimuli makes it a proper marker for significant biological events, molecular decoy that acts as a molecular sponge and keep regulatory factors such as transcription factors and micro-RNAs away from their native targets in order to regulate them negatively, dynamic scaffold providing a central platform on which transient ribonucleoprotein complexes are assembled and guide which as its name suggests, directs regulatory factors to their correct location on the genome [9, 17] (Fig. 1). Due to vital regulatory roles of lncRNAs in biological processes, numerous studies have focused on their function in cancer as a disease of genome and epigenome. Moreover, it has been revealed that aberrant expression of lncRNAs is associated with tumor initiation, progression, invasion, and overall survival by mostly contributing to epigenetic control of many oncogene and tumor suppressor genes [10, 15, 18, 19].
LncRNA functional mechanism in regulation of gene expression and cellular processes. LncRNAs can affect gene expression through interaction with wide range of partners like transcription factors, chromatin modifying complexes, micro-RNAs, etc. They sponge their partners away from their true targets (decoy), make a central platform to facilitate the functional complexes assembly (scaffold) or localize a factor at proper genomic loci (guide). In this way, they control cell proliferation, apoptosis, splicing, genomic imprinting, or other cellular processes
Despite the elucidation of potential mechanistic roles, the biological functions of the vast majority of lncRNAs are not clear [20]. Due to their ability to be tissue/cell specific [3, 9, 20, 21], they might be considered as intricate molecular regulators or fine-tuners as described by Adriaens et al. [22]. In gastrointestinal (GI) cancer, a group of cancers which affect the digestive system including oral and esophageal squamous cell carcinoma (OSCC and ESCC), gastric, colorectal, hepatocellular, pancreatic, and gallbladder cancers, the implication of lncRNAs have been documented. Based on their roles, we categorized some of these lncRNAs into three groups; tumor promoter (oncogenic), tumor suppressor, and tumor promoter/suppressor which function of the last group might be considered as tissue or context dependent (Table 1). Finally, we provide further details on how the candidate lncRNA implicates in GI cancer development and progression. Some characteristics including chromosomal location, exon count, implicated GI cancer type, function, target gene(s), and implicated signaling pathway(s) of the lncRNAs have been outlined in a supplementary file 1 (sup 1). Moreover, considering that there is an approved link between inflammation and cancer susceptibility [23], supplementary file 2 (sup 2) summarizes the list of some lncRNAs involved in the principal gastrointestinal tract inflammatory diseases especially, inflammatory bowel disease (IBD).
Tumor Promoter or Oncogenic lncRNAs
Some lncRNAs were recognized as tumor promoter in all studied types of GI cancers. Table 2 summarizes their characteristics such as function, signaling pathway(s) and implicated GI cancer type and provides a brief view of the mechanism by which they exert oncogenic function. Many of the lncRNAs listed in Table 2 were reviewed elsewhere [9, 10, 14, 19, 20, 24],and hence, we discuss ones that have been reviewed less in the literature (Fig. 2).
The regulatory network of GI cancers, constructed by Cytoscape_v3.1. Most of the lncRNAs in the network benefit from the EZH2 and PTEN to control the GI cancer progression. In the network, the diamond-shaped pink nodes represent lncRNAs and the circle green, V-shaped red and rectangle blue nodes demonstrate proteins, miRNAs and molecular pathways, respectively. T-shaped arrows indicate inhibitory effect, delta-shaped arrows are promoter and square-shaped ones stabilize the target. About the last one in this figure, HuR is an exception which is sequestered by MIR22HG and cannot stabilize the B-catenin (Dash square-shaped arrow)
Plasmacytoma Variant Translocation 1 (PVT1)
PVT1 is a 1716 nucleotide length oncogenic lncRNA that maps to human chromosome 8q24 (near c.Myc). Upregulation of PVT1 shows oncogenic activity in gastric (GC) [25, 26] and colorectal cancer (CRC) [27], hepatocellular carcinoma (HCC) [28], pancreatic cancer (PC) [29], ESCC [30], and gallbladder cancer (GBC) [31] through affecting chemosensitivity, TGF-β and p53 signaling pathways, epithelial–mesenchymal transition (EMT) and also glucose metabolism. The promoter of PVT1 is inducible through the attachment of signal transducer activator phospho-STAT3 and FOXM1 (Forkhead box protein M1), two cell-cycle regulators. In a positive feedback loop, they finally lead to angiogenesis and gastric tumor invasion [32, 33]. Association of this lncRNA and Enhancer of Zeste Homolog 2 (EZH2), which epigenetically modulate the p15 and p16 expression to affect the gastric cancer development [34], is also involved in HCC progression through stabilizing of MDM2 (Mouse double minute 2 homolog) and preventing the p53 tumor suppressor signaling pathway (Fig. 2) [28]. In another mechanism, PVT1 is targeted by lots of micro-RNAs such as a cluster of miRNAs, consisting of miR‐1204, miR‐1205, miR‐1206, miR‐1207‐5p, miR‐1207‐3p, and miR‐1208 [35] in order to participate in oncogenesis. Overexpression of PVT1 in ESCC keep miR-203 away from LASP1 (LIM and SH3 domain protein 1) binding so releases LASP1 and promotes cell migration [30]. Direct interaction and negative association of PVT1 with miR-186 and miR-152 in GC cells can affect the expression of HIF-1a, CD151, and FGF2, respectively [36, 37]. Competing endogenous effect of this oncogenic lncRNA on miR-143 positively regulates the Hexokinase 2 (HK2) expression in GBC [31]. Upregulation of HK2, a key metabolic enzyme in glucose uptake, is involved in GBC malignancy via promoting glycolysis [31].
Colon Cancer-Associated Transcript 1 (CCAT1)
Oncogenic lncRNA CCAT1 with 2628 nucleotides in length is located on “hot spot” area of chromosome 8q24.21, near to c-Myc, offering multiple genetic alternations in cancers [38]. CCAT1 upregulated in several cancers has been introduced as a potential biomarker for early detection of gastrointestinal cancers [39]. This lncRNA increases the risk of colorectal cancer by promoting IBD pathogenesis [23]. In this way, CCAT1 sequesters miR-185-3p and promotes the function of myosin light chain kinase (MLCK). Increased phosphorylation of myosin II regulatory light chain (p-MLC) following MLCK activity changes the permeability of the tight junction protein, disrupts the intestinal epithelial barrier function, and promotes the IBD pathogenesis [23]. Recently, an association between CCAT1 polymorphisms and CRC risk as well as different clinical stages of CRC in Chinese population has also been reported [40]. It was reported that aberrant expression of CCAT1 promotes cell proliferation, invasion and migration in cancers. Through E-box element in the promoter region of CCAT1, c-Myc directly binds and activates transcription of this lncRNA in gastric and colon cancer [41, 42]. CCAT1 acts as a sponge and antagonizes the effects of miR-410 on the expression of its target ITPKB in CRC [43]. Furthermore, by targeting miR-219-1 (a tumor suppressor in colon cancer), CCAT1 affects growth and metastasis in GC [44]. Likewise miRNA-218-5p and miR-490 both sponge to CCAT1, but, CCAT1/miR-490/hnRNPA1 axis appears to be more effective in promoting the migration of GC [45]. The key role of CCAT1 in GC cell proliferation and migration was highlighted by a recent study on DRE (dandelion root extract)-mediated anti-tumorigenesis effect. DRE exerted the suppression effect via targeting CCAT1 in GC cells [46]. This c-Myc-induced lncRNA also acts as a sponge of let-7 tumor suppressor and therefore rescues expression of HMGA2, c-Myc, and then itself to promote hepatocellular carcinoma progression (Fig. 2) [47, 48]. Moreover, CCAT1 plays a pivotal role in GBC malignancy via sponging of miR-218-5p and facilitating the expression of its target gene, Bmi1, a polycomb ring finger oncogene that negatively regulates cell-cycle inhibitor genes [39]. Overexpression of this lncRNA has also been detected in PC and ESCC which affects target genes especially through acting as a scaffold for polycomb repressive complex 2 [49, 50].
Gastric Adenocarcinoma Predictive Long Intergenic Noncoding RNA (GAPLINC)
This oncogenic lncRNA locates on chromosome 18p1 and encodes a 924-bp-long noncoding RNA. The strong correlation between GAPLINC expression and tumor size, patient survival, and occurrence of lymph node metastasis makes it a potential biomarker for gastric cancer and CRC diagnosis and prognosis [51, 52]. GAPLINC functions as miRNA sponge and competes with CD44 to decoy the miR-211-3p, which targets both CD44 and GAPLINC, resulting in decreased degradation of CD44 mRNA and increased gastric cancer proliferation, migration and angiogenesis [51]. GAPLINC binding to miR-378 increases MAPK1 expression that leads to advanced cell proliferation of gastric cancer cells (Fig. 2) [53]. The upregulation of GAPLINC under hypoxia revealed that HIF-1α controls transcriptional activation of GAPLINC via binding to its promoter region that leads to the acceleration of GC tumor progression and decreases its apoptosis [54]. GAPLINC also plays a key role in colorectal cancer cell invasion by binding to non-POU-domain-containing octamer binding (NONO) protein and PTB-associated splicing factor (PSF) to upregulate SNAI2 [52]. Besides, it can positively regulate the c-MET proto-oncogene through the GAPLINC/miR-34a/c-MET axis in CRC cells [18].
Small Nucleolar RNA Host Gene 20 (SNHG20)
SNHG20 is a lncRNA producing a 2183-bp transcript with chromosome 17q25 genomic location. It was originally reported as a lncRNA promoting hepatocellular carcinoma cell proliferation [55]. The upregulation of SNHG20 has been reported in multiple cancers including CRC [56]. This molecule influences cell proliferation, migration, invasion, and cell-cycle progression in CRC cells via regulating of p21 and Cyclin A1 expression which highlights its therapeutic potential [57]. A strong association between SNHG20 expression, tumor size, and lymphatic metastasis in GC patients has also been reported. Recently, it has been shown that SNHG20 acts as a competing endogenous RNA (ceRNA) for miR‐495‐3p and regulates zinc finger protein X‐linked (ZFX) expression, emphasizing the main role of SNHG20/miR‐495‐3p/ZFX in GC proliferation and invasion [58]. Furthermore, SNHG20 can bind to EZH2 and modulate the GSK-3β/β-catenin signaling pathway that leads to epigenetically silencing of the p21 and E-cadherin expression and promoting the EMT in gastric cancer (Fig. 2) [55]. On the other hand, overexpression of this lncRNA in HCC and oral squamous cell carcinoma is associated with cancer progression and stemness [56, 59, 60]. Sequestered miR-197 by an excessive level of SNHG20 leads to the release of the LIN28 stem factor which promotes cancer stem cell (CSC) properties and oncogenesis in OSCC cells [56]. Interaction between SNHG20 and EZH2- a part of the PRC2 complex- results in E-cadherin repression and EMT progression in HCC (Fig. 2) [61]. In ESCC cells, this lncRNA also participates in EMT and metastasis through enforcing the expression of ATM-JAK-PD-L1 (programmed cell death 1 ligand 1) signaling pathway effectors [62].
Focally Amplified lncRNA on Chromosome 1 (FAL1)
FAL1 or FALEC (focally amplified long noncoding RNA in epithelial cancer) encoding a 2276 bp transcript from 1q21 is remarkably upregulated in tumor tissues and serum exosome [3, 63, 64]. This lncRNA regulates the expression of many tumor proliferation and metastasis-related genes, such as Bcl-2, TGF-β, NF-κB, and PCNA [3]. Overexpression of FAL1 in ESCC is associated with reduced expression of mitochondrial fission protein DRP1 which plays an essential role in intrinsic apoptosis [65]. This lncRNA also promotes the cell proliferation of gastric cancer through regulating the phosphatase and tensin homolog (PTEN) which is the natural inhibitor of phosphoinositide 3-kinase (PI3K)/AKT pathway (Fig. 2) [66]. Moreover, FAL1 plays a critical role in CRC and HCC development and progression by acting as a ceRNA. It sponges miR-637 to rescue the expression of NUPR1 in CRC cells and miR-1236 to promote the AFP and ZEB1 effect on cell proliferation and migration of HCC [63, 64]. Downregulation of FAL1 in CRC increases the level of the epithelial marker E-cadherin and decreases the level of the mesenchymal marker Vimentin and inhibits the EMT [63]. On the other hand, It interacts with STAT3 to promote its phosphorylation, and therefore, it plays a key role in regulating the proliferation and apoptosis of colon cancer cells (Fig. 2) [3].
Gastric Carcinoma Highly Expressed Transcript 1 (GHET1)
Gastric carcinoma highly expressed transcript 1, located on the chromosome 7q36.1 with 1913 bp in length, is initially found to be overexpressed in gastric carcinoma [67]. Growing studies have reported that GHET1 is upregulated in diverse gastrointestinal carcinomas, including gastric, colorectal, hepatocellular, esophageal squamous cell and pancreatic cancer [68,69,70,71]. The knockdown of GHET1 in cancer cells inhibits cell proliferation and migration and induces cell apoptosis [68, 69]. In gastric cancer, downregulation of GHET1 is associated with reduced expression of the cell-cycle promoters such as cyclin D, CDK4, CDK6, cyclin E, and CDK2 [68]. Knockdown of this lncRNA results in induced expression of Numb protein which inhibits cell growth and stimulates apoptosis by blocking the Cyclin D1, MMP-2, and MMP-9 protein expression [72]. GHET1 mediates the physical interaction between insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) and c-Myc mRNA to increase c-MYC protein stability that leads to GC cell growth promotion in vitro and in vivo (Fig. 2) [67]. This lncRNA also affects the stability of ATF1 mRNA, a transcription factor regulating target genes related to HCC development [73], and suppresses the Krüppel-like Factor 2 (KLF2) expression by recruiting the PRC2 complex to the promoter of this gene (Fig. 2) [21].
Double Homeobox A Pseudogene 10 (DUXAP10)
DUXAP10 is a pseudogene which derived from long noncoding RNA located on 14q11.2 and has 2398 nt length [74]. Several studies have shown that the downregulation of DUXAP10 leads to decreased cell proliferation and metastasis, as well as increased apoptosis in cancer cells [75,76,77]. Mechanistically, DUXAP10 can interact directly with polycomb repressive complex 2 members like EZH2 and also LSD1 to epigenetically block the expression of tumor suppressor genes like p21, LATS1, KLF2, SMAD4, and PTEN in ESCC, gastric, pancreatic cancers, and CRC (Fig. 2) [75,76,77,78].
Cancer Susceptibility Candidate 9 (CASC9)
The chromosome region contributed to the CASC9 lncRNA (8q21.11) is highly conserved among primates, which indicates the evolutionary importance of it [79, 80]. Multiple studies have found that CASC9 is overexpressed in cancer cells and tumors. It has been revealed that the higher expression level of this lncRNA is associated with deep invasion, poor differentiation, and lymph node metastases [79, 81, 82] through multiple mechanisms. CASC9 mediates the EZH2 enrichment on the promoter region of PDCD4, leading to the downregulation of this tumor suppressor gene and inducing the cell proliferation of ESCC (Fig. 2) [80]. CASC9 interacts with the transcriptional coactivator, CREB-binding protein (CBP), concentrating on the LAMC2 promoter, which leads to the modification of histone acetylation in its promoter and finally upregulation of the LAMC2 expression [83]. Upon LAMC2–integrin ligation, focal adhesion kinase (FAK) is phosphorylated and activated PI3K/AKT signaling pathway in HCC (Fig. 2) [84]. It also physically interacts with HNRNPL. This protein which participates in RNA processing regulates the expression of target genes associated with PI3K/AKT signaling pathway in cancers including HCC [81, 85]. Moreover, CASC9 lncRNA can activate the PI3K/AKT/mTOR signaling pathway in OSCC [86].
FOXD2 Adjacent Opposite Strand RNA 1 (FOXD2-AS1)
FOXD2-AS1 lncRNA that located on the human chromosome 1p33 has only a transcript length of 2527 nucleotides [87]. Downregulation of FOXD2-AS1 suppresses cell proliferation, invasion, and migration in vitro and in vivo [88,89,90,91]. FOXD2-AS1 boosts CRC development via EMT and Notch signaling pathways, because its knockdown influences the expression levels of EMT-related proteins: E-cadherin and N-cadherin, as well as notch signaling-related proteins: Hes-1 and NICD [88]. It attracts EZH2 and LSD1 to the EphB3 promoter and induces histone modification and finally inhibits EphB3 transcription in order to gastric carcinoma promotion (Fig. 2) [91]. Acting as a ceRNA, in CRC, FOXD2-AS1 sequesters miR-185-5p to regulate the expression of CDC42 (cell division control protein 42) which is predicted to be a miRNA-185-5p direct target [92], and sponges miR-206 and so increases the level of ANXA2 through decoying of this tumor suppressor micro-RNA in HCC [89].
Tumor Suppressor lncRNAs
Expression of lncRNAs might be downregulated in cancer, acting as a tumor suppressor. Some lncRNAs that function as a tumor suppressor in all studied GI cancers are listed in Table 3. Here we provide some details on how they affect GI cancer development and progression (Fig. 2).
HAND2 Antisense RNA 1 (HAND2-AS1)
HAND2-AS1 lncRNA that also known as UPH, DEIN, and NBLA00301 is located on chromosome 4q34.1, and its role as a tumor suppressor has been demonstrated in several cancers such as osteosarcoma and endometrioid endometrial carcinoma [93]. Studies on gastrointestinal cancers such as esophageal squamous cell carcinoma, colorectal cancer, and hepatocellular carcinoma have revealed that downregulation of HAND2-AS1 in cancerous tissues is negatively correlated with the tumors invasion and advanced tumor stages [93,94,95]. Functionally, this lncRNA attenuates tumor progression and invasion through sponging an oncogenic micro-RNA, miR-21, in ESCC [93], facilitating the expression of KLF14, as a member of Kruppel like factor family (KLF) of transcription factors in CRC [95]. Both KLF14 and HAND2-AS1 are the target of miR-1275, so overexpression of HAND2-AS1 suppresses the effect of miR-1275 on KLF14 and its downstream targets [93] to detract CRC tumor progression.
HOXA Transcript Antisense RNA, Myeloid-Specific 1 (HOTAIRM1)
The short arm of the chromosome 7 (7p15.2) between HOXA1 and HOXA2 genes, codes for a tumor suppressor lncRNA named HOTAIRM1 or HOXA1-AS1 in colorectal tissue [96]. Decreased expression of this lncRNA is associated with tumor progression especially through affecting the PI3K/AKT pathway in gastric cancer. HOTAIRM1 acts as a competing endogenous RNA for miR-17-5p which is related to the PI3K/AKT pathway by suppressing phosphatase and tensin homolog (PTEN)—the negative regulator of the cell-cycle expression (Fig. 2) [97]. Although Zhou et al. [98] reported that upregulation of HOTAIRM1 was correlated with the pancreatic ductal adenocarcinoma progression, and nonetheless, its expression decreases in HCC, CRC, and GC [99]. It was documented that HOTAIRM1 downregulation in HCC is associated with the declined expression level of apoptosis driver factor Bax and increased expression of Bcl2 and Bid through affecting the Wnt pathway [99].
MIR22 Host Gene (MIR22HG)
MIR22HG lncRNA codes from chromosome 17p13.3 and was first discovered as a tumor suppressor in hepatocellular carcinoma [100], and afterward, its downregulation effect was reported in more detail in HCC [101, 102], endometrial [103] and lung [104] cancers. MIR22HG which is also known as C17orf91 inhibits HCC tumor progression by two mechanisms. The first mechanism is mediated by HMGB1 (High mobility group box 1) where MIR22HG promotes the expression of miR-22-3p to target the 3ˋ UTR of the HMGB1 mRNA and inhibit the HMGB1-mediated gene expression of matrix metallopeptidase 9 (MMP9) and ERK [105] which are involved in epithelial to mesenchymal transition and cell proliferation, respectively (Fig. 2). In the second mechanism, MIR22HG competes with oncogenes such as Bcl2 and B-catenin to bind HuR protein and reduces the mRNA stability of these oncogenes (Fig. 2) [105]. ELAV-like protein 1 or HuR is an RNA-binding protein that plays key roles in the cellular proliferation, growth, and survival pathways by stabilizing the bound RNA [106]. It has also been reported that sponging of miR-10a-5p is able to suppress the Wnt pathway by inducing the NCOR2 expression as a transcriptional co-repressor in HCC [107]. Low expression of this lncRNA in gastric cancer decreases the 5-year overall survival rate and indicates poor prognosis of the patients. Moreover, MIR22HG silencing promotes gastric cancer cell proliferation and invasion through NOTCH2 signaling pathway via enhancing the nuclear localization of NOTCH2 and inducing its downstream targets especially HEY1 (Hes-Related Family BHLH Transcription Factor With YRPW Motif 1) transcriptional repressor expression [108].
Fer-1-Like Family Member 4, Pseudogene (FER1L4)
FER1L4 is a 6.7-Kb lncRNA on chromosome 20q11.22 which was first detected as the most downregulated lncRNA in gastric cancer [109, 110]. Loss of its expression and tumor suppressor role has been reported in gastric, colon, liver, and esophageal cancers [111]. This lncRNA acts as a ceRNA and inhibits the proliferation and invasion of the above-mentioned tumors mainly through the sponging of miR-106a-5p [110,111,112,113]. MiR-106a-5p as an oncogenic micro-RNA suppresses the expression of tumor suppressor genes such as PTEN and RB, hence promoting cell growth by inducing the G0/G1 to S phase transition [111,112,113,114]. Overall, the best-established mechanism of action of FER1L4 is the sequestration of miR-106a-5p and making the PTEN free to prevent the activation of PI3K/AKT pathway and cell proliferation in gastrointestinal cancers (Fig. 2) [112, 115].
Low Expression in Tumor (LncRNA LET)
NPTN intronic transcript 1 or NPTN-IT1 mostly known as lncRNA LET is located on chromosome 15q24.1. It has been initially reported as a hypoxia inhibited tumor suppressor in HCC [116, 117] following by several other cancers such as ESCC [116, 118], gastric cancer [119], and gallbladder cancer [120]. The significant effect of this lncRNA on tumor growth prevention makes it the main target of oncogenic genes like miR-548 k [119] and lncRNA DANCR [121]. The depletion of lncRNA LET expression as a result of overexpressed miR-548 k is related to poor overall survival in ESCC [119]. In this axis, lncRNA LET acts as the regulator of p53 and NF90. Downregulation of this lncRNA decreases p53, one of the main regulators of gene expression which inhibits cell cycle and promotes cell death, and increases NF90 expression hence, promoting the proliferation and invasion of ESCC cells [116, 119]. NF90 which is ubiquitylated and degraded after binding to lncRNA LET is a double-stranded RNA-binding protein that involves in stabilization, transport, and translation of some metastasis mediator genes in HCC. Decreased expression of LET in HCC stabilizes NF90 and leads to hypoxia-induced tumor invasion and metastasis [117, 120]. Downregulation of LET in gastric cancer [119, 122] resulted from overexpression of lncRNA DANCR stimulates the migration of GC cells [121]. Mechanistically, DANCR acts as a scaffold to target the EZH2 and Histone Deacetylase 4 (HDAC4) to the promoter of lncRNA LET to epigenetically silence its transcription and influence the proliferation and apoptosis of GC cells [119, 121] (Fig. 2). HDAC4 also involves in hypoxia-induced HCC progression via preventing the lncRNA LET expression [123]. Hypoxia-induced lncRNA LET downregulation in GBC is also associated with tumor progression and poor prognosis. Furthermore, it regulates the cell cycle and apoptosis by affecting the expression of p21, Bcl2, Bax, and caspase 3 [120].
Functionally Context-Dependent LncRNAs
Although some lncRNAs play an obvious tumor promotion or tumor suppression role in GI cancers, there are lncRNAs that their function might be considered as tumor or context dependent. The expression patterns of these lncRNAs depend on the types of GI cancers; upregulating in one or some types while downregulating in the other types of GI cancers. Table 4 summarizes the characteristics of some context-dependent lncRNAs in GI cancers.
Terminal Differentiation-Induced ncRNA (TINCR)
TINCR which primarily involves in human epidermal differentiation has been reported as an upregulated LncRNA in gastric cancer [124], ESCC [125], and HCC [126]. This 3700-bp lncRNA is expressed from chromosome 19p13.3. Induced expression of TINCR mediated by nuclear transcription factor SP1 and E2F1 involves proliferation, invasion, migration, and apoptosis of cancer cells [124, 127]. Interaction of TINCR with KLF2 recruits double-stranded RNA (dsRNA) detecting protein, Staufen 1 (STAU1), leading to the silencing of KLF2 in gastric cancer cells (Fig. 2) [124]. Therefore TINCR affects the oncogenic potential of GC cells through inhibiting the KLF2 regulated gene expression such as CDKN1A/p21 and CDKN2B/p15 cell-cycle regulators [124]. This regulatory lncRNA suppresses the expression of CDKN2B through the E2F1/TINCR/STAU1/CDKN2B signaling axis too. In this pathway, E2F1-promoted expression of TINCR influences the CDKN2B mRNA stability by recruiting STAU1 [127]. Moreover, TINCR can act as a ceRNA and be targeted by miR-375. Sponging of miR-375 by TINCR leads to the upregulation of PDK1 in gastric cancer cells, which highlights the oncogenic role of TINCR in gastric cancer pathogenesis [128]. Furthermore, Tian et al. [126] showed that the expression of TINCR can be repressed by miR-133a and miR-137 in hepatocellular carcinoma, and contributed a tumor-promoting role for TINCR in HCC differentiation, invasion, and metastasis. On the other hand, TINCR plays a tumor suppressor role in CRC hence, its expression downregulates in that cancer [129]. In CRC, TINCR acts as a Wnt signaling inhibitor and its transcription is controlled by c-Myc-regulated SP1 (Fig. 2). The c-Myc-mediated repression of SP1 results in TINCR downregulation, leading to the declined EpCAM protein stability, followed by accumulation of EpICD which colocalizes with B-catenin and induces Wnt downstream gene expression in colorectal cancer [129]. Moreover, the tumor-suppressing function of TINCR in CRC was also demonstrated by overexpression of TINCR which attenuated CRC tumor growth through miR-107 sponging [130].
TP73 Antisense RNA1 (TP73-AS1)
LncRNA TP73-AS1 which is transcribed from chromosome 1p36 deregulates in cancer [16]. It was first reported to be downregulated in oligodendroglial tumors [131]. Moreover, in colorectal cancer, it functions as a tumor suppressor by sponging of miR-103, which results in PTEN escape and halting of the PI3/AKT signaling pathway (Fig. 2) [132]. In contrast, some studies have indicated a tumor-promoting role for this lncRNA in gastric [16], colorectal [133], and hepatocellular cancers [134]. It promotes cell proliferation by sponging the miRNAs as well as the regulation of tumor suppressor genes. For example, TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE/NF-κB regulation (Fig. 2) [134]. Furthermore, it was reported that suppression of TP73-AS1 expression improves the chemotherapeutic response of gastric cancer cells to Cisplatin [135].
Long Intergenic Non-protein Coding RNA 52 (LINC00052)
LINC00052 or TMEM83 codes for a 1786-bp-long noncoding RNA from chromosome 15q25.3. It plays as a tumor suppressor in HCC and pancreatic cancer cell proliferation and migration [136,137,138,139]. LINC00052 sequesters a set of miRNAs such as miR-330-3p and miR-452-5p to prevent their tumorigenesis activity in HCC and pancreatic cancer [136, 138]. It triggers apoptosis and prevents G1 phase progression via sponging of miR-330-3p in pancreatic cancer [136]. Besides, reciprocal repression between miR-452-5p and lncRNA LINC00052 in HCC forces the depletion of EPB4K3 and tumor progression [138]. Moreover, overexpression of LINC00052 might block the MAPK pathway. It sponges miR-128 and miR-485 to the liberation of NTRK3, which follows by negative control on ERK1/2 and attenuation of the cell invasion and migration of HCC (Fig. 2) [137]. Furthermore, this lncRNA plays as a tumor suppressor by inducing miR-101-3p activity which diminishes the oncogenic expression of SOX9 [139]. On the other hand, LINC00052 plays as a tumor promoter in gastric cancer. Its high expression levels in gastric cancer cells as well as in tumor tissues associate with carcinogenesis and poor survival of the patients [140]. LINC00052 promotes gastric cancer cell proliferation and metastasis via activating the Wnt/β-catenin signaling pathway. It interacts with SMYD2 methyltransferase and improves β-catenin stability by methylation, leading to the activation of oncogenic Wnt signaling [140].
LncRNAs and Drug Resistance in GI Cancer
Dysfunction of lncRNAs is also associated with GI tumor relapse and progression through contributing to drug resistance. Therefore, it is crucial to explore the molecular mechanisms of this event in order to improve the treatment and patient survival rate. Here we further discussed the molecular pathways related to some of the above-mentioned lncRNAs involvement in chemoresistance (Fig. 3).
Some lncRNAs interact with anticancer drugs. The influence of lncRNAs on drug resistance affects the response of patients to treatment. According to the network, some lncRNAs such as PVT1, CASC9, and GHET1 are involved in multidrug resistance by activating of MDR1 and MRP1 expression. It was also shown that colon cancers with high level of CCAT1 are more sensitive to JQ1. Triangle orange nodes represent the chemotherapy drugs. The diamond-shaped pink, circle green and V-shaped red nodes demonstrate lncRNAs, proteins, and miRNAs, respectively. T-shaped arrows indicate inhibitory effect and delta-shaped arrows are promoters. Octagon light green shapes are cancer types. GI cancer: gastrointestinal cancer, GC gastric cancer, ESCC esophageal squamous cell carcinoma, PC: pancreatic cancer
PVT1 One of the mainly used drugs in gastric cancer chemotherapy is cisplatin (DDP) [26] which triggers apoptosis via cross-linking of the DNA and causing strand breaks during cell-cycle progression [141]. According to the research [26], PVT1 can affect the gastric cancer response to this agent. Increased level of PVT1 in cisplatin-resistant patients and cells is associated with apoptosis inhibition through activating several multidrug resistance (MDR)-related proteins mainly multidrug resistance-1 (MDR1) and multidrug resistance-associated protein 1 (MRP1) [26]. MDR1, as an ATP-dependent efflux pump, forces DDP out of the cell and prevents its access to DNA and apoptosis promoting [26] (Fig. 3). This lncRNA is also involved in gemcitabine resistance of pancreatic cancer by blocking apoptosis [142]. Gemcitabine which acts through the interfering with nucleic acid synthesis is the first-line therapeutic agent in PC. Although the molecular mechanism of PVT1 involvement in PC cell resistance against this drug needs further investigation, You and his colleagues demonstrated the PVT1 regulatory role on Gemcitabine sensitivity in PC cells (Fig. 3) [142].
CCAT1 Increased expression of CCAT1 in ESCC cells is closely related to Cisplatin drug resistance. CCAT1 knockdowned cells display better response to DDP treatment through upregulating of apoptosis factors like Bax and downregulating of cell-cycle-related ones [143]. In these cells, CCAT1 sequesters a tumor suppressor miR-143 to promote the Polo Like Kinase 1 (PLK1)/BUBR1 signaling. Given that PLK1 plays a significant role in provoking cell-cycle progression specially M-phase and inhibiting apoptosis, this signaling axis makes ESCC cells resistant to DDP (Fig. 3) [143]. These results suggest that CCAT1 may be a suitable target to defeat chemoresistance in esophageal cancer [143]. This lncRNA also affects the chemosensitivity of colon cancer. JQ1 blocks cell growth in CIMP (CpG island methylator phenotype) positive colon cancers by interfering with the Bromodomain-containing protein 4 (BRD4) effect on the c-Myc transcription. C-Myc expression in CIMP+ cells is dependent on the CCAT1 expression, a superenhancer of c-Myc transcription [144]. JQ1 inhibits CCAT1 expression which leads to the inhibition of the c-Myc oncogenic effects on the CIMP+ colon cancer cells. Therefore, CCAT1 may serve as a predictive biomarker to determine the tumors which are more sensitive to JQ1 than others (Fig. 3) [144].
SNHG20 Overexpression of SNHG20 in GC cells causes Fluorouracil (5-FU) resistance through targeting miR-140-5p [145]. 5-FU which is analog of uracil exerts its cytotoxicity in multiple ways such as misincorporation into DNA or RNA structure and also blocking of thymidine synthesis. Its anticancer effect is the result of the interruption in DNA replication and repair [146]. In gastric cancer, SNHG20 induces the expression of the NDRG3, one of the main oncogenic targets of miR-140-5p [145]. The N-myc downstream-regulated gene 3 (NDRG3) is a member of the NDRG family which are involved in lots of cellular processes such as cell proliferation and differentiation [147, 148]. Enhanced expression of NDRG3 following the sequestration of the miR-140-5p by SNHG20 stimulates 5-FU resistance in GC through regulating cell-cycle progression in favor of carcinogenesis (Fig. 3) [145].
GHET1 cisplatin resistance is a common event and major obstacle in gastric cancer treatment [149] and multidrug resistance factors including MDR1 and MRP1 play a significant role in this event [150]. Aberrantly expression of GHET1 in gastric carcinoma benefits these factors in order to affect the sensitivity of GC cells to DDP [150]. Upregulation of this lncRNA, which is associated with GC cell proliferation, bigger tumor size, and worse survival [67], is able to promote the development of multidrug resistance [150]. It also exerts its effect by affecting the Bax and Bcl2 as well as MDR1 and MRP1 expression (Fig. 3) [150].
CASC9 Paclitaxel (PTX) as a microtubule-stabilizing drug and adriamycin (ADR) or doxorubicin, an anthracycline, make their anticancer effect through chromosome missegregation on multipolar spindles and mitotic arrest [151] and intercalation into DNA and topoisomerase II-related poisoning [152], respectively. CASC9 knockdown elevates the chemosensitivity of GC-resistant cells to paclitaxel and adriamycin through decreased expression of MDR1 protein (Fig. 3) [79]. Although the direct regulatory role of CASC9 on MDR1 needs further investigation, eightfold high expression of this lncRNA in GC cells makes exactly a relation between MDR1 expression and GC chemoresistance [79].
Conclusion
LncRNAs implicated in GI cancer development and progression can be classified functionally as a tumor promoter, tumor suppressor, or tumor/context dependent. While the first and second classes played as tumor promoters and tumor suppressors, respectively, at all reported GI cancer research, the third class showed a tumor-type-specific function in GI cancer. This highlights further complications in lncRNA function and implication in GI cancer development and progression which needs to be considered in diagnosis, prognosis, and therapy of GI cancers.
References
Xie H, Ma B, Gao Q, Zhan H, et al. Long non-coding RNA CRNDE in cancer prognosis: review and meta-analysis. Clinica Chimica Acta. 2018;485:262–271.
Aguadé-Gorgorió G, Solé R. Adaptive dynamics of unstable cancer populations: the canonical equation. Evol Appl. 2018;11:1283–1292.
Wu K, Zhang N, Ma J, Huang J, et al. Long noncoding RNA FAL1 promotes proliferation and inhibits apoptosis of human colon cancer cells. IUBMB Life. 2018;70:1093–1100.
Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12.
Lee EY, Muller WJ. Oncogenes and tumor suppressor genes. Cold Spring Harbor Perspect Biol. 2010;2:a003236.
Baeissa HM, Benstead-Hume G, Richardson CJ, Pearl FM. Mutational patterns in oncogenes and tumour suppressors. Biochem Soc Trans. 2016;44:925–931.
Djebali S, Davis CA, Merkel A, Dobin A, et al. Landscape of transcription in human cells. Nature. 2012;489:101.
Consortium F. A promoter-level mammalian expression atlas. Nature. 2014;507:462.
Balas MM, Johnson AM. Exploring the mechanisms behind long noncoding RNAs and cancer. Non-Coding RNA Res. 2018;3:108–117.
Sun W, Yang Y, Xu C, Xie Y, Guo J. Roles of long noncoding RNAs in gastric cancer and their clinical applications. J Cancer Res Clin Oncol. 2016;142:2231–2237.
Lu S, Su Z, Fu W, Cui Z, et al. Altered expression of long non-coding RNA GAS5 in digestive tumors. Biosci Rep. 2019;39:BSR20180789.
Zeng S, Xiao Y-F, Tang B, Hu C-J, et al. Long noncoding RNA in digestive tract cancers: function, mechanism, and potential biomarker. The Oncologist. 2015;20:898–906.
Parsons C, Tayoun AM, Benando B, Ragusa G, et al. The role of long noncoding RNAs in cancer metastasis. J Cancer Metastasis Treat. 2018;4:1–23.
Fanelli GN, Gasparini P, Coati I, Cui R, et al. LONG-NONCODING RNAs in gastroesophageal cancers. Non-coding RNA Res. 2018;3:195–212.
Zheng Y, Yang C, Tong S, Ding Y, et al. Genetic variation of long non-coding RNA TINCR contribute to the susceptibility and progression of colorectal cancer. Oncotarget. 2017;8:33536.
Ding Z, Lan H, Xu R, Zhou X, Pan Y. LncRNA TP73-AS1 accelerates tumor progression in gastric cancer through regulating miR-194-5p/SDAD1 axis. Pathol-Res Pract. 2018;214:1993–1999.
Zhan L, Li J, Wei B. Long non-coding RNAs in ovarian cancer. J Exp Clin Cancer Res. 2018;37:120.
Luo Y, Ouyang J, Zhou D, Zhong S, et al. Long noncoding RNA GAPLINC promotes cells migration and invasion in colorectal cancer cell by regulating miR-34a/c-MET signal pathway. Digest Dis Sci. 2018;63:890–899.
Chen S, Zhu J, Wang F, Guan Z, et al. LncRNAs and their role in cancer stem cells. Oncotarget. 2017;8:110685.
Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–2100.
Jin L, He Y, Tang S, Huang S. LncRNA GHET1 predicts poor prognosis in hepatocellular carcinoma and promotes cell proliferation by silencing KLF2. J Cell Physiol. 2018;233:4726–4734.
Adriaens C, Marine J-C. NEAT1-containing Paraspeckles: central hubs in stress response and tumor formation. Cell Cycle. 2017;16:137.
Ma D, Cao Y, Wang Z, He J, et al. CCAT1 lncRNA promotes inflammatory bowel disease malignancy by destroying intestinal barrier via downregulating miR-185-3p. Inflam Bowel Dis. 2019;25:862–874.
Huang T, Wang M, Huang B, Chang A, et al. Long noncoding RNAs in the mTOR signaling network: biomarkers and therapeutic targets. Apoptosis. 2018;23:255–264.
Ding J, Li D, Gong M, Wang J, et al. Expression and clinical significance of the long non-coding RNA PVT1 in human gastric cancer. OncoTargets Ther. 2014;7:1625.
Zhang X-W, Bu P, Liu L, Zhang X-Z, Li J. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Biophys Res Commun. 2015;462:227–232.
Takahashi Y, Sawada G, Kurashige J, Uchi R, et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110:164.
Guo J, Hao C, Wang C, Li L. Long noncoding RNA PVT1 modulates hepatocellular carcinoma cell proliferation and apoptosis by recruiting EZH2. Cancer Cell Int. 2018;18:98.
Wu B-Q, Jiang Y, Zhu F, Sun D-L, He X-Z. Long noncoding RNA PVT1 promotes EMT and cell proliferation and migration through downregulating p21 in pancreatic cancer cells. Technol Cancer Res Treatment. 2017;16:819–827.
Li P-D, Hu J-L, Ma C, Ma H, et al. Upregulation of the long non-coding RNA PVT1 promotes esophageal squamous cell carcinoma progression by acting as a molecular sponge of miR-203 and LASP1. Oncotarget. 2017;8:34164.
Chen J, Yu Y, Li H, Hu Q, et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Molecular Cancer. 2019;18:33.
Zhao J, Du P, Cui P, Qin Y, et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37:4094.
Xu M-D, Wang Y, Weng W, Wei P, et al. A positive feedback loop of lncRNA-PVT1 and FOXM1 facilitates gastric cancer growth and invasion. Clin Cancer Res. 2017;23:2071–2080.
Kong R, Zhang E-B, Yin D-D, You L-H, et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer. 2015;14:82.
Huppi K, Volfovsky N, Runfola T, Jones TL, et al. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol Cancer Res. 2008;6:212–221.
Li T, Meng X-L, Yang W-Q. Long noncoding RNA PVT1 acts as a “Sponge” to inhibit microRNA-152 in gastric cancer cells. Digest Dis Sci. 2017;62:3021–3028.
Huang T, Liu HW, Chen JQ, Wang SH, et al. The long noncoding RNA PVT1 functions as a competing endogenous RNA by sponging miR-186 in gastric cancer. Biomed Pharmacother. 2017;88:302–308.
Xin Y, Li Z, Shen J, Chan MT, Wu WKK. CCAT 1: a pivotal oncogenic long non-coding RNA in human cancers. Cell Prolifer. 2016;49:255–260.
Mizrahi I, Mazeh H, Grinbaum R, Beglaibter N, et al. Colon cancer associated transcript-1 (CCAT1) expression in adenocarcinoma of the stomach. J Cancer. 2015;6:105.
Li Y, Jing F, Ding Y, He Q, et al. Long noncoding RNA CCAT1 polymorphisms are associated with the risk of colorectal cancer. Cancer Genet. 2018;222:13–19.
Yang F, Xue X, Bi J, Zheng L, et al. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–445.
He X, Tan X, Wang X, Jin H, et al. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumor Biol. 2014;35:12181–12188.
Li B, Shi C, Zhao J, Li B. Long noncoding RNA CCAT1 functions as a ceRNA to antagonize the effect of miR-410 on the down-regulation of ITPKB in human HCT-116 and HCT-8 cells. Oncotarget. 2017;8:92855.
Li Y, Zhu G, Ma Y, Qu H. LncRNA CCAT1 contributes to the growth and invasion of gastric cancer via targeting miR-219-1. J Cell Biochem. 2017.
Zhou B, Wang Y, Jiang J, Jiang H, et al. The long noncoding RNA colon cancer-associated transcript-1/miR-490 axis regulates gastric cancer cell migration by targeting hnRNPA1. IUBMB Life. 2016;68:201–210.
Zhu H, Zhao H, Zhang L, Xu J, et al. Dandelion root extract suppressed gastric cancer cells proliferation and migration through targeting lncRNA-CCAT1. Biomed Pharmacother. 2017;93:1010–1017.
Zhu H-Q, Zhou X, Chang H, Li H-G, et al. Aberrant expression of CCAT1 regulated by c-Myc predicts the prognosis of hepatocellular carcinoma. Asian Pac J Cancer Prev. 2015;16:5181–5185.
Deng L, Yang S-B, Xu F-F, Zhang J-H. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18.
Yu Q, Zhou X, Xia Q, Shen J, et al. Long non-coding RNA CCAT1 that can be activated by c-Myc promotes pancreatic cancer cell proliferation and migration. Am J Transl Res. 2016;8:5444.
Zhang E, Han L, Yin D, He X, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucl Acids Res. 2016;45:3086–3101.
Hu Y, Wang J, Qian J, Kong X, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890–6902.
Yang P, Chen T, Xu Z, Zhu H, et al. Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO. Oncotarget. 2016;7:42183.
Diao L, Wang S, Sun Z. Long noncoding RNA GAPLINC promotes gastric cancer cell proliferation by acting as a molecular sponge of miR-378 to modulate MAPK1 expression. OncoTargets Ther. 2018;11:2797.
Liu L, Zhao X, Zou H, Bai R, et al. Hypoxia promotes gastric cancer malignancy partly through the HIF-1α dependent transcriptional activation of the long non-coding RNA GAPLINC. Front Physiol. 2016;7:420.
Liu J, Liu L, Wan J-X, Song Y. Long noncoding RNA SNHG20 promotes gastric cancer progression by inhibiting p21 expression and regulating the GSK-3β/β-catenin signaling pathway. Oncotarget. 2017;8:80700.
Wu J, Zhao W, Wang Z, Xiang X, et al. Long non-coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR-197/LIN28 axis. J Cell Mol Med. 2019;23:680–688.
Li C, Zhou L, He J, Fang X-Q, et al. Increased long noncoding RNA SNHG20 predicts poor prognosis in colorectal cancer. BMC Cancer. 2016;16:655.
Cui N, Liu J, Xia H, Xu D. LncRNA SNHG20 contributes to cell proliferation and invasion by upregulating ZFX expression sponging miR-495-3p in gastric cancer. J Cell Biochem. 2019;120:3114–3123.
Gao P, Fan R, Ge T. SNHG20 serves as a predictor for prognosis and promotes cell growth in oral squamous cell carcinoma. Oncol Lett. 2019;17:951–957.
Zhang D, Cao C, Liu L, Wu D. Up-regulation of LncRNA SNHG20 predicts poor prognosis in hepatocellular carcinoma. J Cancer. 2016;7:608.
Liu J, Lu C, Xiao M, Jiang F, et al. Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and promotes cell invasion by regulating the epithelial-to-mesenchymal transition. Biomed Pharmacother. 2017;89:857–863.
Zhang C, Jiang F, Su C, Xie P, Xu L. Upregulation of long noncoding RNA SNHG20 promotes cell growth and metastasis in esophageal squamous cell carcinoma via modulating ATM-JAK-PD-L1 pathway. J Cell Biochem. 2019;120:11642–11650.
Wang L, Jiang F, Xia X, Zhang B. LncRNA FAL1 promotes carcinogenesis by regulation of miR-637/NUPR1 pathway in colorectal cancer. Int J Biochem Cell Biol. 2019;106:46–56.
Li B, Mao R, Liu C, Zhang W, et al. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018;197:122–129.
Liu T, Wang Z, Zhou R, Liang W. Focally amplified lncRNA on chromosome 1 regulates apoptosis of esophageal cancer cells via DRP1 and mitochondrial dynamics. IUBMB Life. 2019;71:254–260.
Zhu C, Xiao D, Dai L, Xu H, et al. Highly expressed lncRNA FAL1 promotes the progression of gastric cancer by inhibiting PTEN. Eur Rev Med Pharmacol Sci. 2018;22:8257–8264.
Yang F, Xue X, Zheng L, Bi J, et al. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802–813.
Xia Y, Yan Z, Wan Y, Wei S, et al. Knockdown of long noncoding RNA GHET1 inhibits cell-cycle progression and invasion of gastric cancer cells. Mol Med Rep. 2018;18:3375–3381.
Liu H, Zhen Q, Fan Y. LncRNA GHET1 promotes esophageal squamous cell carcinoma cells proliferation and invasion via induction of EMT. Int J Biol Mark. 2017;32:403–408.
Zhou J, Li X, Wu M, Lin C, et al. Knockdown of long noncoding RNA GHET1 inhibits cell proliferation and invasion of colorectal cancer. Oncol Res Featur Preclin Clin Cancer Therap. 2016;23:303–309.
Li J, Jiang X, Li Z, Huang L, et al. Long noncoding RNA GHET1 in human cancer. Clinica Chimica Acta. 2018.
Huang H, Liao W, Zhu X, Liu H, Cai L. Knockdown of long noncoding RNA GHET1 inhibits cell activation of gastric cancer. Biomed Pharmacother. 2017;92:562–568.
Ding G, Li W, Liu J, Zeng Y, et al. LncRNA GHET1 activated by H3K27 acetylation promotes cell tumorigenesis through regulating ATF1 in hepatocellular carcinoma. Biomed Pharmacother. 2017;94:326–331.
Wei C-C, Nie F-Q, Jiang L-L, Chen Q-N, et al. The pseudogene DUXAP10 promotes an aggressive phenotype through binding with LSD1 and repressing LATS2 and RRAD in non small cell lung cancer. Oncotarget. 2017;8:5233.
Wang Z, Ren B, Huang J, Yin R, et al. LncRNA DUXAP10 modulates cell proliferation in esophageal squamous cell carcinoma through epigenetically silencing p21. Cancer Biol Ther. 2018;19:998–1005.
Lian Y, Xiao C, Yan C, Chen D, et al. Knockdown of pseudogene derived from lncRNA DUXAP10 inhibits cell proliferation, migration, invasion, and promotes apoptosis in pancreatic cancer. J Cell Biochem. 2018;119:3671–3682.
Xu Y, Yu X, Wei C, Nie F, et al. Over-expression of oncigenic pesudogene DUXAP10 promotes cell proliferation and invasion by regulating LATS1 and β-catenin in gastric cancer. J Exp Clin Cancer Res. 2018;37:13.
Lian Y, Xu Y, Xiao C, Xia R, et al. The pseudogene derived from long non-coding RNA DUXAP10 promotes colorectal cancer cell growth through epigenetically silencing of p21 and PTEN. Sci Rep. 2017;7:7312.
Shang C, Sun L, Zhang J, Zhao B, et al. Silence of cancer susceptibility candidate 9 inhibits gastric cancer and reverses chemoresistance. Oncotarget. 2017;8:15393.
Wu Y, Hu L, Liang Y, Li J, et al. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer. 2017;16:150.
Klingenberg M, Groß M, Goyal A, Polycarpou-Schwarz M, et al. The lncRNA CASC9 and RNA binding protein HNRNPL form a complex and co-regulate genes linked to AKT signaling. Hepatology. 2018;68:1817–1832.
Pan Z, Mao W, Bao Y, Zhang M, et al. The long noncoding RNA CASC9 regulates migration and invasion in esophageal cancer. Cancer Med. 2016;5:2442–2447.
Liang Y, Chen X, Wu Y, Li J, et al. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018;25:1980.
Leng C, Zhang Z-G, Chen W-X, Luo H-P, et al. An integrin beta4-EGFR unit promotes hepatocellular carcinoma lung metastases by enhancing anchorage independence through activation of FAK–AKT pathway. Cancer Lett. 2016;376:188–196.
Fei T, Chen Y, Xiao T, Li W, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Pro Natl Acad Sci. 2017;114:E5207–E5215.
Yang Y, Chen D, Liu H, Yang K. Increased expression of lncRNA CASC9 promotes tumor progression by suppressing autophagy-mediated cell apoptosis via the AKT/mTOR pathway in oral squamous cell carcinoma. Cell Death Dis. 2019;10:41.
Chen G, Sun W, Hua X, Zeng W, Yang L. Long non-coding RNA FOXD2-AS1 aggravates nasopharyngeal carcinoma carcinogenesis by modulating miR-363-5p/S100A1 pathway. Gene. 2018;645:76–84.
Yang X, Duan B, Zhou X. Long non-coding RNA FOXD2-AS1 functions as a tumor promoter in colorectal cancer by regulating EMT and Notch signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:3586–3591.
Chang Y, Zhang J, Zhou C, Qiu G, et al. Long non-coding RNA FOXD2-AS1 plays an oncogenic role in hepatocellular carcinoma by targeting miR-206. Oncol Rep. 2018;40:3625–3634.
Bao J, Zhou C, Zhang J, Mo J, et al. Upregulation of the long noncoding RNA FOXD2-AS1 predicts poor prognosis in esophageal squamous cell carcinoma. Cancer Biom. 2018;21:527–533.
Xu T-P, Wang W-Y, Ma P, Shuai Y, et al. Upregulation of the long noncoding RNA FOXD2-AS1 promotes carcinogenesis by epigenetically silencing EphB3 through EZH2 and LSD1, and predicts poor prognosis in gastric cancer. Oncogene. 2018;37:5020–5036.
Zhu Y, Qiao L, Zhou Y, Ma N, et al. Long non-coding RNA FOXD 2-AS 1 contributes to colorectal cancer proliferation through its interaction with micro RNA-185-5p. Cancer Sci. 2018;109:2235–2242.
Yan Y, Li S, Wang S, Rubegni P, et al. Long noncoding RNA HAND2-AS1 inhibits cancer cell proliferation, migration, and invasion in esophagus squamous cell carcinoma by regulating microRNA-21. J Cell Biochem. 2019;120:9564–9571.
Shi B, Zhang X, Chao L, Zheng Y, et al. Comprehensive analysis of key genes, microRNAs and long non-coding RNAs in hepatocellular carcinoma. FEBS Open Biol. 2018;8:1424–1436.
Zhou J, Lin J, Zhang H, Zhu F, Xie R. LncRNA HAND2-AS1 sponging miR-1275 suppresses colorectal cancer progression by upregulating KLF14. Biochem Biophys Res Commun. 2018;503:1848–1853.
Wan L, Kong J, Tang J, Wu Y, et al. HOTAIRM 1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J Cell Mol Med. 2016;20:2036–2044.
Lu R, Zhao G, Yang Y, Jiang Z, et al. Long noncoding RNA HOTAIRM1 inhibits cell progression by regulating miR-17-5p/PTEN axis in gastric cancer. J Cell Biochem. 2019;120:4952–4965.
Zhou Y, Gong B, Jiang Z-L, Zhong S, et al. Microarray expression profile analysis of long non-coding RNAs in pancreatic ductal adenocarcinoma. Int J Oncol. 2016;48:670–680.
Zhang Y, Mi L, Xuan Y, Gao C, et al. LncRNA HOTAIRM1 inhibits the progression of hepatocellular carcinoma by inhibiting the Wnt signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:4861–4868.
Liu L, Zhang D, Wu D. 748 long non-coding RNA miR22HG represses hepatocellular carcinoma cell invasion by deriving miR-22 and targeting HMGB1. Gastroenterology. 2016;150:S1045–S1046.
Dong Y, Yan W, Zhang S-L, Zhang M-Z-H, et al. Prognostic values of long non-coding RNA MIR22HG for patients with hepatocellular carcinoma after hepatectomy. Oncotarget. 2017;8:114041.
Wu Y, Wang PS, Wang BG, Xu L, et al. Genomewide identification of a novel six-LncRNA signature to improve prognosis prediction in resectable hepatocellular carcinoma. Cancer Med. 2018;7:6219–6233.
Cui Z, An X, Li J, Liu Q, Liu W. LncRNA MIR22HG negatively regulates miR-141-3p to enhance DAPK1 expression and inhibits endometrial carcinoma cells proliferation. Biomed Pharmacother. 2018;104:223–228.
Su W, Feng S, Chen X, Yang X, et al. Silencing of long noncoding RNA MIR22HG triggers cell survival/death signaling via oncogenes YBX1, MET, and p21 in lung cancer. Cancer Res. 2018;78:3207–3219.
Zhang D-Y, Zou X-J, Cao C-H, Zhang T, et al. Identification and functional characterization of long non-coding RNA MIR22HG as a tumor suppressor for hepatocellular carcinoma. Theranostics. 2018;8:3751.
Dai W, Zhang G, Makeyev EV. RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucl Acids Res. 2011;40:787–800.
Wu Y, Zhou Y, Huan L, Xu L, et al. LncRNA MIR22HG inhibits growth, migration and invasion through regulating the miR-10a-5p/NCOR2 axis in hepatocellular carcinoma cells. Cancer Sci. 2019;110:973.
Li H, Wang Y. Long noncoding RNA (lncRNA) MIR22HG suppresses gastric cancer progression through attenuating NOTCH2 signaling. Med Sci Monitor Int Med J Exp Clin Res. 2019;25:656.
Song H, Sun W, Ye G, Ding X, et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225.
Yue B, Sun B, Liu C, Zhao S, et al. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer Sci. 2015;106:1323–1332.
Ma W, Zhang C, Li H, Gu J, et al. LncRNA FER1L4 suppressed cancer cell growth and invasion in esophageal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:2638–2645.
Xia T, Chen S, Jiang Z, Shao Y, et al. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci Rep. 2015;5:13445.
Wu J, Huang J, Wang W, Xu J, et al. Long non-coding RNA Fer-1-like protein 4 acts as a tumor suppressor via miR-106a-5p and predicts good prognosis in hepatocellular carcinoma. Cancer Biom. 2017;20:55–65.
Xia T, Liao Q, Jiang X, Shao Y, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088.
Sun X, Zheng G, Li C, Liu C. Long non-coding RNA Fer-1-like family member 4 suppresses hepatocellular carcinoma cell proliferation by regulating PTEN in vitro and in vivo. Mol Med Rep. 2019;19:685–692.
Wang PL, Liu B, Xia Y, Pan CF, et al. Long non-coding RNA-Low Expression in Tumor inhibits the invasion and metastasis of esophageal squamous cell carcinoma by regulating p53 expression. Mol Med Rep. 2016;13:3074–3082.
Weidle UH, Birzele F, Kollmorgen G, Rueger R. Long non-coding RNAs and their role in metastasis. Cancer Genom-Proteom. 2017;14:143–160.
Chen Z, Lin J, Wu S, Xu C, et al. Up-regulated miR-548 k promotes esophageal squamous cell carcinoma progression via targeting long noncoding RNA-LET. Exp Cell Res. 2018;362:90–101.
Tian J, Hu X, Gao W, Zhang J, et al. Identification of the long non-coding RNA LET as a novel tumor suppressor in gastric cancer. Mol Med Rep. 2017;15:2229–2234.
Ma MZ, Kong X, Weng MZ, Zhang MD, et al. Long non-coding RNA-LET is a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer. Mol Carcinogen. 2015;54:1397–1406.
Mao Z, Li H, Du B, Cui K, et al. LncRNA DANCR promotes migration and invasion through suppression of lncRNA-LET in gastric cancer cells. Biosci Rep. 2017;37:BSR20171070.
Zhou B, Jing X-Y, Wu J-Q, Xi H-F, Lu G-J. Down-regulation of long non-coding RNA LET is associated with poor prognosis in gastric cancer. Int J Clin Exp Pathol. 2014;7:8893.
He Y, Meng X-M, Huang C, Wu B-M, et al. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27.
Xu T, Liu X, Xia R, Yin L, et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648.
Xu Y, Qiu M, Chen Y, Wang J, et al. Long noncoding RNA, tissue differentiation-inducing nonprotein coding RNA is upregulated and promotes development of esophageal squamous cell carcinoma. Dis Esophagus. 2016;29:950–958.
Tian F, Xu J, Xue F, Guan E, Xu X. TINCR expression is associated with unfavorable prognosis in patients with hepatocellular carcinoma. Biosci Rep. 2017;37:301.
Xu T-P, Wang Y-F, Xiong W-L, Ma P, et al. E2F1 induces TINCR transcriptional activity and accelerates gastric cancer progression via activation of TINCR/STAU1/CDKN2B signaling axis. Cell Death Dis. 2017;8:e2837.
Chen Z, Liu H, Yang H, Gao Y, et al. The long noncoding RNA, TINCR, functions as a competing endogenous RNA to regulate PDK1 expression by sponging miR-375 in gastric cancer. OncoTargets Ther. 2017;10:3353.
Zhang Z-y, Lu Y-x, Zhang Z-y, Chang Y-y, et al. Loss of TINCR expression promotes proliferation, metastasis through activating EpCAM cleavage in colorectal cancer. Oncotarget. 2016;7:22639.
Zhang X, Yao J, Shi H, Gao B, Zhang L. LncRNA TINCR/microRNA-107/CD36 regulates cell proliferation and apoptosis in colorectal cancer via PPAR signaling pathway based on bioinformatics analysis. Biol Chem. 2019;400:663–675.
Pang JCS, Li KKW, Lau KM, Ng YL, et al. KIAA0495/PDAM is frequently downregulated in oligodendroglial tumors and its knockdown by siRNA induces cisplatin resistance in glioma cells. Brain Pathol. 2010;20:1021–1032.
Jia Z, Peng J, Yang Z, Chen J, et al. Long non-coding RNA TP73-AS1 promotes colorectal cancer proliferation by acting as a ceRNA for miR-103 to regulate PTEN expression. Gene. 2019;685:222–229.
Cai Y, Yan P, Zhang G, Yang W, et al. Long non-coding RNA TP73-AS1 sponges miR-194 to promote colorectal cancer cell proliferation, migration and invasion via up-regulating TGFα. Cancer Biom. 2018(Preprint):1–12.
Li S, Huang Y, Huang Y, Fu Y, et al. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. 2017;36:51.
Peng J. si-TP73-AS1 suppressed proliferation and increased the chemotherapeutic response of GC cells to cisplatin. Oncol Lett. 2018;16:3706–3714.
Xiong X, Shi Q, Yang X, Wang W, Tao J. LINC00052 functions as a tumor suppressor through negatively modulating miR-330-3p in pancreatic cancer. J Cell Physiol. 2019;234:15619–15626.
Xiong D, Sheng Y, Ding S, Chen J, et al. LINC00052 regulates the expression of NTRK3 by miR-128 and miR-485-3p to strengthen HCC cells invasion and migration. Oncotarget. 2016;7:47593.
Zhu L, Yang N, Chen J, Zeng T, et al. LINC00052 upregulates EPB41L3 to inhibit migration and invasion of hepatocellular carcinoma by binding miR-452-5p. Oncotarget. 2017;8:63724.
Yan S, Shan X, Chen K, Liu Y, et al. LINC00052/miR-101-3p axis inhibits cell proliferation and metastasis by targeting SOX9 in hepatocellular carcinoma. Gene. 2018;679:138–149.
Shan Y, Ying R, Jia Z, Kong W, et al. LINC00052 promotes gastric cancer cell proliferation and metastasis via activating the Wnt/β-catenin signaling pathway. Oncol Res Featur Preclin Clin Cancer Therap. 2017;25:1589–1599.
Gold JM, Raja A. Cisplatin (Cisplatinum). StatPearls [Internet]: StatPearls Publishing; 2019.
You L, Chang D, Du H-Z, Zhao Y-P. Genome-wide screen identifies PVT1 as a regulator of Gemcitabine sensitivity in human pancreatic cancer cells. Biochem Biophys Res Commun. 2011;407:1–6.
Hu M, Zhang Q, Tian XH, Wang JL, et al. lncRNA CCAT1 is a biomarker for the proliferation and drug resistance of esophageal cancer via the miR-143/PLK1/BUBR1 axis. Mol Carcinogen. 2019;58:2207–2217.
McCleland ML, Mesh K, Lorenzana E, Chopra VS, et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J Clin Investig. 2016;126:639–652.
Yu J, Shen J, Qiao X, Cao L, et al. SNHG20/miR-140-5p/NDRG3 axis contributes to 5-fluorouracil resistance in gastric cancer. Oncol Lett. 2019;18:1337–1343.
Zhang N, Yin Y, Xu S-J, Chen W-S. 5-Fluorouracil: mechanisms of resistance and reversal strategies. Molecules. 2008;13:1551–1569.
Wang W, Li Y, Li Y, Hong A, et al. NDRG3 is an androgen regulated and prostate enriched gene that promotes in vitro and in vivo prostate cancer cell growth. Int J Cancer. 2009;124:521–530.
Jing J-S, Li H, Wang S-C, Ma J-M, et al. NDRG3 overexpression is associated with a poor prognosis in patients with hepatocellular carcinoma. Biosci Rep. 2018;38:1–9.
Verstraelen J, Reichl S. Multidrug resistance-associated protein (MRP1, 2, 4 and 5) expression in human corneal cell culture models and animal corneal tissue. Mol Pharm. 2014;11:2160–2171.
Zhang X, Bo P, Liu L, Zhang X, Li J. Overexpression of long non-coding RNA GHET1 promotes the development of multidrug resistance in gastric cancer cells. Biomed Pharmacother. 2017;92:580–585.
Weaver BA. How Taxol/paclitaxel kills cancer cells. Mol Biol Cell. 2014;25:2677–2681.
Thorn CF, Oshiro C, Marsh S, Hernandez-Boussard T, et al. Doxorubicin pathways: pharmacodynamics and adverse effects. Pharmacogenet Genom. 2011;21:440.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Khajehdehi, M., Khalaj-Kondori, M., Ghasemi, T. et al. Long Noncoding RNAs in Gastrointestinal Cancer: Tumor Suppression Versus Tumor Promotion. Dig Dis Sci 66, 381–397 (2021). https://doi.org/10.1007/s10620-020-06200-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-020-06200-x