Abstract
The presence of complex distributions of samples concealed in high-dimensional, massive sample-size data challenges all of the current classification methods for data mining. Samples within a class usually do not uniformly fill a certain (sub)space but are individually concentrated in certain regions of diverse feature subspaces, revealing the class dispersion. Current classifiers applied to such complex data inherently suffer from either high complexity or weak classification ability, due to the imbalance between flexibility and generalization ability of the discriminant functions used by these classifiers. To address this concern, we propose a novel representation of discriminant functions in Bayesian inference, which allows multiple Bayesian decision boundaries per class, each in its individual subspace. For this purpose, we design a learning algorithm that incorporates the naive Bayes and feature weighting approaches into structural risk minimization to learn multiple Bayesian discriminant functions for each class, thus combining the simplicity and effectiveness of naive Bayes and the benefits of feature weighting in handling high-dimensional data. The proposed learning scheme affords a recursive algorithm for exploring class density distribution for Bayesian estimation, and an automated approach for selecting powerful discriminant functions while keeping the complexity of the classifier low. Experimental results on real-world data characterized by millions of samples and features demonstrate the promising performance of our approach.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
With the constant evolution of information technologies, the large volume of data nowadays is estimated to be on the order of zettabytes, and growing at an unprecedented rate every day. For example, each day Google has more than 1 billion queries, Twitter 250 million tweets, Facebook 800 million updates and YouTube 4 billion views (Fan and Bifet 2013; Fortuny et al. 2013); every day more than one million pieces of malware are discovered by security enterprises and labs, such as McAfee, Symantec and Panda, according to their periodic threat reports.Footnote 1 At the same time, the extraordinarily high number of explanatory featuresFootnote 2 used to describe such massive data gives rise to high dimensionality. These data have also emerged from various real-life applications, such as sequence mining (Rani and Pudi 2008; Kang et al. 2006), document/text mining (Ifrim et al. 2008; Su et al. 2011) and information retrieval (Li et al. 2011; Kim et al. 2008). Here are two examples involving the classification of high-dimensional data: (1) Weinberger et al. (2009) studied a collaborative email-spam filtering task with 15 trillion unique features; (2) Dahl et al. (2013) developed large-scale neural network systems for a malware detection purpose, in which they explored patterns hidden in 2.6 million executables (i.e., samples), each typically expressed in terms of hundreds of thousands of features. Classification on such high-dimensional, massive-sample-size (HDMSS) data involving millions of features and samples is our interest here.
1.1 Contributions
In this paper, we propose a novel representation of discriminant functions in Bayesian inference for HDMSS data classification. This representation allows Multiple Bayesian decision boundaries in Subspaces per Class (MBSC for short), by incorporating the following steps:
-
Describe each class using a set of naive Bayes (NB) models, and then generate a set of Bayesian discriminant functions.
-
Select multiple optimized discriminant functions, and thereby enable the classifier to achieve a tradeoff between flexibility and generalization ability.
Along the way, three algorithms are developed to achieve these steps.
-
A robust unsupervised learning algorithm that reshapes each class into a set of high-density regions situated in different subspaces;
-
A feature-weighted Bayesian algorithm based on expectation maximization (EM) estimates an NB decision boundary by optimizing a new objective function;
-
An adapted algorithm that consecutively invokes Bayesian learning and risk measurement to preserve the advantages of NB (e.g., its simplicity and efficiency) while improving its capacity for handling complex data in the context of class-dispersion.
Running these algorithms on a target class enables a Bayesian classifier to select a class of optimal discriminant functions that are used to identify multiple piecewise-enclosed decision boundaries, each boundary in a specific subspace used to discern a part of this class from other classes. The new approach optimizes the simple, rough decision boundaries for a Bayesian classifier while preventing overfit from arising. With these boundaries, MBSC classifies an unknown sample by measuring its posterior probabilities of belonging to each of the classes. We investigate MBSC by means of a case study on a high-dimensional, low-sample-size (HDLSS) dataset and a set of comparative experimental studies on six high-dimensional, massive-sample-size (HDMSS) datasets and two low-dimensional, massive-sample-size (LDMSS) datasets. Experimental results clearly demonstrate that MBSC yields more accurate classification results in linear time.
To our best knowledge, there is as yet no documented approach addressing the class-dispersion problem in a Bayesian learning framework. In addition to addressing this problem, the approach proposed here can be advantageously applied by learning machines (e.g., model-based classifiers) to optimize decision boundaries.
1.2 Motivation
1.2.1 Needs for naive Bayes and feature weighting
The high dimensionality of data has far outpaced the processing and analytical capacities of current classifiers (Tan et al. 2014; Fan and Bifet 2013), e.g., support vector machines (SVMs) (Chang and Lin 2011), decision trees (DTs) (Zhou and Chen 2002), random forests (RFs) (Breiman 2001), neural networks (NNs) (Nakajima and Watanabe 2005) and instance-based learning (IBL) (Albert and Aha 1991). This prompts us to develop a new classification method for use on HDMSS data. On the other hand, NB (John and Langley 1995) combined with feature weighting has turned out to yield a competitive classifier of choice for high-dimensional data classification tasks. For example, many excellent results have been reported in Frank et al. (2003), Lee et al. (2011), Chen and Wang (2012a) and Chen and Wang (2012b). The simplicity and proven effectiveness of NB motivate us to use it as the base model in this study; this, however, should not be interpreted as a claim that NB is the best approach to cope with HDMSS data.
Given a high-dimensional dataset, most of the explanatory features are generally irrelevant to the class outcome. Furthermore, a number of classifiers based on measures such as the \(L_p\) distance and the cosine function tend to be invalid in high-dimensional spaces due to the curse of dimensionality (Chen et al. 2012). Examples include instance-based classifiers (Aha and Kibler 1991), model-based nearest-neighbor classifiers (Zhang et al. 2013) and centroid-based classifiers (Han and Karypis 2000; Tan 2008). To address these problems, one can resort to a feature selection technique, such as feature dependency, information gain or the gain ratio, to deal with filtering out irrelevant, redundant features and selecting the most relevant features, thus reducing the data dimensionality so that conventional classifiers are able to work in the reduced low-dimensional feature spaces. In practice, however, feature selection usually suffers from high computational complexity, because it is generally not feasible to perform an exhaustive search to find the optimal feature subsets, due to the huge number of admissible subsets which is exponential with regard to the data dimensionality (Zhang et al. 2013).
Instead, feature weighting, which assigns a continuous weighting value to each feature, has been widely employed to implement a “soft” feature selection. This can be seen as a dimensionality reduction if we select features according to the values of their weights. The rationale for using feature weighting is to shrink the correlation between features. NB thus becomes more applicable to high-dimensional data if feature weighting can be incorporated in NB’s learning procedure.
1.2.2 Need for multiple discriminant functions in Bayesian inference
The key to a reasonable learning machine for classification lies in appropriate discriminant functions to identify decision boundaries between classes. Bayesian classifiers generally encompass a small group of discriminant functions, one for formulating each decision boundary per class, e.g., the regular NB classifier (John and Langley 1995), locally weighted NB (LWNB) (Frank et al. 2003), feature-weighted NB (FWNB) (Lee et al. 2011), subspace-weighed NB (SWNB) and kernel-weighted NB (KWNB) (Chen and Wang 2012a, b). These classifiers can be reasonably applied when the samples within class are drawn from a simple density distribution. In HDMSS data, however, complex distributions are very likely for a class of samples and may lead to a class-dispersion problemFootnote 3: the samples of a class may not uniformly fill the input space or be confined to a unique subspace but may spread out into some natural high-density regions in diverse feature subspaces. The existing classifiers have not been designed to account for such a scenario and they usually suffer from an inability of tackling class dispersion, because classes characterized by complex distributions cannot be separated from each other via the single boundary per class used in these classifiers. Recalling the data-mapping approach introduced by Vilalta and Rish (2003), augmenting the number of decision boundaries according to class density distribution, e.g., the high-density regions, appears to be a straightforward way of addressing the problem.
In the language of learning theory (Vapnik 2000), the use of a single discriminant function per class can equip classifiers with a good generalization ability, but may lead to a decline in flexibility and subsequently an increase in the risk of misclassification (i.e., the probability of error), especially if a class has a complex distribution (Atashpaz-Gargari et al. 2013). The use of multiple discriminant functions per class is expected to offer the desired tradeoff between flexibility and generalization ability. In particular, since Bayesian classifiers employ simple representations, using multiple Bayesian discriminant functions is expected to allow improving the flexibility while still retaining the generalization ability that makes them suitable for wide use. Here simple classifiers are preferred to complex ones like neural networks with a large number of hidden units and nearest-neighbor classifiers with few neighbors. Such complex classifiers exhibit flexible decision boundaries but are sensitive to small variations in the data (Vilalta et al. 2003). A risk minimization that entails a balance between flexibility and generalization ability can be used, in combination with Bayes’ theorem, for learning boundaries.
The remainder of this paper is structured as follows. Section 2 is devoted to preliminaries and problem statements. Our approach is presented in Sect. 3 and the algorithms in Sect. 4. Section 5 reports our experimental results and the evaluation of MBSC. Our conclusions are given in Sect. 6.
2 Preliminaries and related work
We present the basic Bayesian inference and related work regarding the naive Bayes (NB) approaches, as well as the feature-weighting-based NB. Finally, we describe the class-dispersion problem.
2.1 Preliminaries for Bayesian inference
We begin by presenting some basic terminology and notation necessary for the understanding of subsequent sections.
2.1.1 Notation
In what follows, let \({\mathcal {A}}=(A_1,A_2,\ldots ,A_V)\) be a V-component vector-valued random variable, where each \(A_v\) represents an input attribute or an explanatory feature; the space of all possible attribute vectors is called the (attribute or feature) full space \({\mathbb {R}}^V\). Denote by \({\mathcal {D}}=\{(\mathbf{x},z)\}_{N}\) a training set of N labeled samples that pertain to K classes, or categories. Each input-output pair, \((\mathbf{x},z)\), consists of a V-dimensional vector or data point in the input space, i.e., \(\mathbf{x}=(x_{1},x_{2},\ldots ,x_{V})\in {\mathbb {R}}^V\), and its observed output attribute, i.e., class outcome \(z\in \{1,2,\ldots ,K\}\), wherein \(x_{v}\) takes the value of attribute \(A_v\), \(v\in \{1,2,\ldots ,V\}\). \(N_k\) represents the number of samples within class k which is given by \({\mathcal {C}}_k:=\{(\mathbf{x}, k)\in {\mathcal {D}}\}\), whence \(\sum \nolimits ^K_{k=1}N_k=N\).
The specific notation is outlined and explained in Table 1.
2.1.2 Bayesian decision boundary
Assigning samples in the input space to one of K classes means we divide the input space into K regions \({\mathcal {R}}_1,{\mathcal {R}}_2,\ldots ,{\mathcal {R}}_K\), such that a point falling in \({\mathcal {R}}_k\) is assigned to class \(k\in \{1,2,\ldots ,K\}\). For example, a classifier defines K functions \(f(\mathbf{x};\vartheta _1),f(\mathbf{x};\vartheta _2),\ldots ,f(\mathbf{x};\vartheta _K)\), one for each class, with a class-specific parameter \(\vartheta _k\). The classifier assigns, for a given sample, the class whose function is maximum (Domingos and Pazzani 1997), i.e., the chosen class k is the one satisfying
A decision boundary occurs at points in the input space where discriminant functions (for different classes) are equal. For example, the decision boundary between classes k and j is given by
Basically, the discriminant function may be an instantiation of the probabilistic model (e.g., Bayesian inference), and if this is indeed the case, it can be approximated by the Bayesian posterior probability, \(p(k|\mathbf{x};\vartheta _k)\), as follows:
where
-
the prior probability of class k, p(k), can be estimated in its simplest form by \(\frac{N_k}{N}\) if \(N_k>0\ \forall k\), or by a Laplace smoothing (Lee et al. 2011), e.g., \(\frac{N_k+1}{N+K}\);
-
the class-conditional probability or so-called likelihood given \(\vartheta _k\), \(p(\mathbf{x}|k;\vartheta _k)\), might be approximated through a multivariate Gaussian in V dimensions, where the parameters \(\vartheta _k\triangleq [{{\varvec{\mu }}}_k,{{\varvec{\sigma }}}_k]\) with mean \({{\varvec{\mu }}}_k=(\mu _{k1},\mu _{k2},\ldots ,\mu _{kV})\) and standard deviation \({{\varvec{\sigma }}}_k=(\sigma _{k1},\sigma _{k2},\ldots ,\sigma _{kV})\);
-
the marginal probability, \(p(\mathbf{x})\), is independent of class k.
As a result, the discriminant function of a probabilistic classifier against \(\mathbf{x}\) becomes
which can be approximated by a manipulation of the prior probability of class and the likelihood.
2.2 Related work
2.2.1 Naive Bayes (NB) and feature weighting
For the sake of simplicity, the regular NB presented in John and Langley (1995) assumes that all explanatory features are conditionally independent given the class outcome. Under this assumption, the conditional probability \(p(\mathbf{x}|k;{{\varvec{\mu }}}_k,{{\varvec{\sigma }}}_k)\) can thus be computed as a simple product of univariate Gaussians, i.e.,
Thanks to this “naive” assumption, NB is able to reduce a high-dimensional task to multiple single-dimensional tasks, thereby yielding incredible savings in the degree of complexity of the discriminant functions. The repertoire of NB’s simplicity reveals its potential capacity to adapt to classifying high-dimensional massive data, as (Zaidi et al. 2013) reported.
A variety of feature weighting approaches are commonly employed to address the fact that classes link to diverse feature subspaces, either in a local or global way. In a global feature weighting, each feature will be assigned a uniform weight over all classes, whereas a local weighting assigns each feature a set of class-dependent weights. That is, samples will be projected onto a global (sub)space by global feature weighting and onto different (sub)spaces by local feature weighting, each class of samples being in a specific (sub)space. Here are some examples of recent state-of-the-art work incorporating NB and feature weighting:
-
Chen and Wang (2012a, b) proposed subspace-weighted NB (SWNB) and kernel-weighted NB (KWNB), in which a local weighted Gaussian was thought of as the class-conditional distribution of class k, in which the predictive contributions of V features are modulated by V class-dependent weights, \({{\varvec{w}}}_k=(w_{k1},w_{k2},\ldots ,w_{kV})\), \(\forall w_{kv}\in (0,1)\).
-
Lee et al. (2011) introduced FWNB to make use of a global feature weighting to learn V continuous weights, \({{\varvec{w}}}^\text {global}=(w^{(1)},\ldots ,w^{(V)})\) for all features across the K classes, thereby yielding a global weighted Gaussian over all classes.
Table 2 summarizes various forms of boundaries used in NB-based classifiers. The characteristic worthy of note is that only one Bayesian decision boundary is sketched for each class to distinguish it from others.
2.2.2 High-dimensional data classification
The well-known classification methods, such as support vector machines (SVMs) (Chang and Lin 2011), decision trees (DTs) (Zhou and Chen 2002), random forests (RFs) (Breiman 2001), neural networks (NNs) (Nakajima and Watanabe 2005) and instance-based learning (IBL) (Albert and Aha 1991), have been shown to be successful classifiers, capable of building accurate models with practical relevance for classification. These classifiers can extend to high-dimensional data classification. For example,
-
Tan et al. (2010) proposed to learn a sparse solution with respect to input features to SVM for large-scale and very high-dimensional datasets;
-
Lin et al. (2014) explored a new representation of hash functions by training boosted decision trees for high-dimensional data;
-
Xu et al. (2012) achieved classification improvement for random forests on high dimensional data by using a feature weighting method for subspace selection;
-
Bengio and Bengio (1999) proposed to model high-dimensional data using a multi-layer neural network to represent the joint distribution of the features as the product of conditional distributions;
-
Hinneburg et al. (2000) proposed a new generalized notation for nearest neighbor search for instance-based learning in high-dimensional space.
Despites of many good performances reported in the literatures, major difficulties are encountered when these classifiers are applied to HDMSS data. These mainly fall into three broad categories:
-
inefficiency of the classification algorithms and high computational cost in learning, for SVM;
-
high complexity of models and heavy memory requirements, for DTs, RFs and NNs;
-
low validity of distance metrics and weak generalization ability, for IBL.
2.3 Examples illustrating the problem
The class dispersion in HDMSS data can be seen from the following data-mining examples:
-
In malware detection, a class of Trojans most probably consists of various malicious families. Usually, each families of Trojans can be distinguished from others by their own family-specific sequential patterns of behavior (i.e., sequence features) (Shabtai et al. 2009).
-
In healthcare prediction, electronic healthcare records (EHR) of patients from multiple parties (e.g., hospitals or health centers) are captured (Yu et al. 2008). Records for patients of distinct parties are characterized by individual party-specific values of prognostic variables (i.e., features regarding clinical symptoms), yet they may be diagnosed with the same disease.
-
In topic-based text categorization, first, documents from the same categorization most likely have various subtopics. For instance, in the widely studied hierarchical benchmark 20-Newsgroups, documents topicalized by Science are closely associated with a couple of subtopics, such as Electronics, Medicine, Cryptology and Space. The documents corresponding to particular subtopics contain a mass of their own subtopic-specific keywords (namely features in bag-of-words for text categorization) (Joachims 1996).
Usually, one expects a boundary defined in a global subspace or a (local) class-dependent subspace to separate a target class from others in this (sub)space, and therefore, one resorts to a projection approach (Aggarwal et al. 2004), such as LDA (Martínez and Kak 2001) or class-dependent projection (Chen and Wang 2012a; Marchiori 2013). It is hard to identify a (sub)space in which the two classes are completely separate, however, since what is likely to happen is that some of the samples can be recognized, but others cannot. We will now give a concrete example to illustrate this problem.
Distribution of 100 samples of PSW (in \(\times \)) and 140 of Downloader (in \(\triangledown \)) in different (sub)spaces. a Distribution of 100 samples from PSW and 140 from Downloader in a 2-dimensional full space. b Distribution of PSW and Downloader in a projected subspace \(s^\text {local}\) = (0.648, 0.762). c Distribution of all 100 samples of PSW and 40 samples of Downloader in the subspace \(s_1\) = (0.653, 0.994) (left), and all 100 samples of PSW and the remaining 100 samples of Downloader in the subspace \(s_2\) = (0.918, 0.756) (right)
We selected 240 Win32 portable executables (e.g., .EXE, .COM, .DLL, .OCX, etc.). Of these executables, 100 are password-type Trojans (PSW class) and the remaining 140 are downloader-type (Downloader class). The PSW class comprises many subtypes such as FakeAIM, QQThief, YahooPass, and others; the Downloader class contains subtypes such as Agent, QQHelper, Delf, Injecter, Exchanger, etc. For ease of illustration, we disassembled these executables and then extracted the first two sequential instructions (i.e., features) \(A_1\)=“PUSH, JMP, JCC, CALL” and \(A_2\)=“RET, MOVS, CALL, POP” via principle component analysis (PCA) (Martínez and Kak 2001). In Fig. 1a, we plotted the distribution of all samples in the 2-dimensional space with regard only to \(A_1\) and \(A_2\). It can be seen from the figure that identifying boundaries to distinguish the two classes from each other becomes a troublesome task, due to the irregular sample distributions of the two classes. By means of the typical local feature weighting described in Jing et al. (2007) and Chen and Wang (2012a), we obtained an optimal subspace \(s^\text {local}=\)(0.648, 0.762) with respect to the two variants of \(A_1\) and \(A_2\), say \(A_1^\text {local}\) and \(A_2^\text {local}\), in which the two classes are mostly separate. (The subspace designated by the weights of the two features will be interpreted in detail later on.) Fig. 1b depicts the result when all samples are projected into the subspace \(s^\text {local}\). In this subspace, however, the majority of samples from Downloader and PSW occupy the same region, and therefore we draw a precise boundary between the two classes.
Things change when we take the following steps:
-
Decompose the 140 samples of Downloader into two subclasses: \(c_1\) with 40 samples and \(c_2\) with 100;
-
Project \(c_1\) and \(c_2\) separately into two subspaces: \(s_1\) designated as (0.653, 0.994) while \(s_2\) as (0.918, 0.756).
As Fig. 1c shows, the samples of \(c_1\) in subspace \(s_1\) are concentrated in the high-density region \({\mathcal {R}}_1\) and become easier to separate out; likewise, in subspace \(s_2\) this is also the case for \(c_2\) concentrated in \({\mathcal {R}}_2\). Consequently, the representation using two boundaries in the subspaces \(s_1\) and \(s_2\) for the class \({\mathcal {C}}_\text {Downloader}\) (actually for the two subclasses \(c_1\) and \(c_2\)) appears to be preferable.
This example demonstrates a universal complex class distribution of samples within high-dimensional data, where samples of a class might spread out into many regions of different subspaces. Multiple boundaries for a class are thus required, and, moreover, each boundary may be in a specific subspace. We call this representation as “multiple decision boundaries in subspaces per class”. As yet, there has been no report of learning such boundaries in the general context of Bayesian inference.
It must be emphasized that this work is not centered on class overlapping, but rather on the class dispersion concealed in HDMSS data. The primary difference between the two is that class dispersion reveals the context of within-class distribution of samples whereas class overlapping manifests the characteristic of distributions across different classes.
3 Our approach
In this section, we will first introduce a new representation of Bayesian discriminant functions used to describe decision boundaries, and then the Bayesian estimation for the parameters of discriminant functions. Finally, we present the criterion for measuring the effectiveness of discriminant functions.
3.1 Multiple Bayesian decision boundaries
Since we do not know the true class density distribution, achieving our goal inevitably depends on sketching this distribution prior to a Bayesian learning procedure. For this purpose, we propose the following definitions that can describe class density distribution:
Definition 1
(Multi-Region) For any given class k, its multi-region, \({\mathcal {R}}_k\), is a collection of \(\ell _k(1\le \ell _k\le N_k)\) high-density region(s) residing in subspace(s), i.e., \({\mathcal {R}}_k:=\{{\mathscr {R}}_{kl}\}^{\ell _k}_{l=1}\). The high-density region, given by \({\mathscr {R}}_{kl}\triangleq [c_{kl},s_{kl}], \forall l=1,2,\ldots ,\ell _k\), is represented by subclass \(c_{kl}\) and subspace \(s_{kl}\) where the samples of \(c_{kl}\) are concentrated.
-
The number, \(\ell _k\), of high-density region(s) for class k indicates how many discriminant functions used to describe decision boundaries for class k. An appropriate \(\ell _k\) should lead to a balance between model’s flexibility and generalization ability. Thus, we will determine \(\ell _k\) via structural risk minimization (SRM) in Bayesian model learning. More details can be seen in the following section.
-
The total \(\ell _k\) subclasses are subject to the following constraints:
-
Every subclass belongs to a class: \(\bigcup ^{\ell _k}_{l=1}c_{kl}={\mathcal {C}}_k\).
-
All subclasses are pairwise disjoint: \(c_{kl}\cap c_{kl'}=\varnothing \ \forall l, l', l\ne l'\).
-
There are no empty subclasses: \(c_{kl}\ne \varnothing \ \forall l\).
-
-
Each subspace s is designated by a V-dimensional vector with respect to V nonuniform squared feature weights, i.e., \((\sqrt{w_{kl1}},\sqrt{w_{kl2}},\ldots ,\sqrt{w_{klV}})\). For readability and simplicity, we drop the subscript kl and denote such subspace by
$$\begin{aligned} (\sqrt{w_1},\sqrt{w_2},\ldots ,\sqrt{w_V})\triangleq \sqrt{\mathbf{w}}. \end{aligned}$$(2)Following up on the regularization step introduced by Kooij et al. (2007), we regularize the feature weights by imposing the constraint that
$$\begin{aligned} \Vert \mathbf{w}\Vert _1=\mathbf{1}_V^\top \mathbf{w}:=\sqrt{V},\ \mathbf{w}\in (0,1]^V. \end{aligned}$$(3)
Return to the example shown in Fig. 1, the multi-region for the class Downloader (140 samples) indicates the subclass \(c_1\) (40 samples) in subspace \(s_1\) while \(c_2\) (the remaining 100 samples) is in \(s_2\). The subspace \(s_1\), given by (0.653, 0.994), means that the squared weight for feature \(A_1\) is 0.653 while for feature \(A_2\) it is 0.994. In contrast, for the subspace \(s_2\), given by (0.918, 0.756), we have \(\sqrt{w_{A_1}}=0.918\) and \(\sqrt{w_{A_2}}=0.756\).
Unlike the existing feature-weighting methods (Domeniconi et al. 2007; Jing et al. 2007), which require a sum-to-one constraint, our regulator stretches up to a V-dependent value in case numerical underflow occurs in the context of high dimensionality. Note that, \(w_v\) tells us the relevance of the feature v to c. The higher the relevance of a feature, the more likely that the feature is related to the region.
A close inspection of \({\mathscr {R}}=[c,s]\) shows that we are able to modulate \({\mathscr {R}}\) through a subspace NB parameterized as
Looking more closely at Eq. 4, one might notice that our approach discards a vector of standard deviations, e.g., \({{\varvec{\sigma }}}=(\sigma _1,\sigma _2,\ldots ,\sigma _V)\), with respect to the V features. Instead, we consider a uniform global standard deviation, as Eq. 5 shows. Unlike the use of an empirical constant or the simple Laplace estimation (Seeger 2006), the features whose variances equate to 0 will gain their non-zero projected variance, according to the feature’s proper contribution to classification.
Definition 2
(Multiple Bayesian Discriminant Functions per Class) The union of \(\ell _k\) Bayesian discriminant functions for class k is given by
Details regarding the NB prior probability, \(p(\Theta )\), and the likelihood, \(p(\mathbf{x};\Theta )\), will be presented later.
Definition 3
(MBSC) Multiple Bayesian decision boundaries in their individual subspaces for class k consist of \(\ell _k\) piecewise-enclosed boundaries in different subspaces, defined as
It is essential to note that here our approach recognizes a target class via multiple boundaries rather than a single one.
Definition 4
(MBSC Classification) With multiple discriminant functions for each class, Bayes’ theorem predicts the class outcome z of a data point \(\mathbf{x}\) as the class k, in the following way:
3.2 Bayesian estimation
A key issue in learning a Bayesian decision boundary in subspace is what the “best” values are for \(\mathbf{w}\), \({{\varvec{\mu }}}\) and \(\sigma \) of \(\Theta \). In Bayesian learning, one can either maximize the likelihood (ML) or maximize a posterior probability (MAP) to achieve optimal model parameters. In general, MAP estimation allows us to incorporate the prior knowledge into the classification or prediction and therefore might produce better results than ML estimation (Seeger 2006). Hence, we choose the MAP estimation to optimize the model parameter \(\widehat{\Theta }\), and then obtain
with the prior probability \(p(\Theta )\) and the likelihood \(p(c|\Theta )\). Eq. 7 takes a logarithm, since decision boundaries will not be changed by monotonic transformations of the discriminant functions.
We shall assume without loss of generality that samples of c are drawn from a Gaussian distribution. As previously argued, c corresponds to a certain subspace s; thus, such a distribution can be named as a subspace Gaussian. We abbreviate this as
where the parameter, \(\mathbf{w}\), quantifies the subspace s. Next, we will figure out the probability density function of this subspace Gaussian through the transformations below:
-
1.
Suppose \(\mathbf{y}\) is the projection of \(\mathbf{x}\) into the subspace s and can be obtained by imposing \(\sqrt{\mathbf{w}}\) on all features; that is,
$$\begin{aligned} \mathbf{x}\xrightarrow {s}\mathbf{y}:\ \mathbf{y}=\sqrt{\mathbf{w}}\circ \mathbf{x}. \end{aligned}$$Assume that V variables of \(\mathbf{y}\) are drawn from a V-dimensional Gaussian distribution whose probability density is a function of the form:
$$\begin{aligned} G(\mathbf{y};\bar{\mathbf{y}},\sigma )&=\frac{\;1\;}{\;\sqrt{2\pi }\sigma \;}\exp \left( -\frac{1}{2\sigma ^2}(\mathbf{y}-\bar{\mathbf{y}})(\mathbf{y}-\bar{\mathbf{y}})^\top \right) \\&=\frac{\;1\;}{\;\sqrt{2\pi }\sigma \;}\exp \left( -\frac{\;1\;}{\;2\sigma ^2\;}\mathbf{w}\circ (\mathbf{x}-{{\varvec{\mu }}})(\mathbf{x}-{{\varvec{\mu }}})^\top \right) \\&=\frac{\;1\;}{\;\sqrt{2\pi }\sigma \;}\exp \left( -\frac{\;1\;}{\;2\sigma ^2\;}(\mathbf{x}-{{\varvec{\mu }}})\mathsf {diag}(\mathbf{w})(\mathbf{x}-{{\varvec{\mu }}})^\top \right) . \end{aligned}$$where \(\bar{\mathbf{y}}\) and \(\sigma \) are the associated mean and standard deviation, respectively.
-
2.
Referring to Straub (2009), we know the multivariate Gaussian integrals
$$\begin{aligned} \int G(\mathbf{y};\bar{\mathbf{y}},\sigma )\mathsf{d}\mathbf{y}=\sqrt{2\pi }\sigma \bigl |\mathsf {diag}(\mathbf{w})\bigr |^{-1/2}=\sqrt{2\pi }\sigma \bigl |\mathsf {diag}(\sqrt{\mathbf{w}})\bigr |^{-1}. \end{aligned}$$By transferring \(\mathbf{y}\) back to \(\mathbf{x}\), the subspace Gaussian distribution of \(\mathbf{x}\in c\) can be written as
$$\begin{aligned} G(\mathbf{x};\mathbf{w},{{\varvec{\mu }}},\sigma )&=\frac{\;\bigl |\mathsf {diag}(\sqrt{\mathbf{w}})\bigr |\;}{\;\sqrt{2\pi }\sigma \;}G(\mathbf{y};\bar{\mathbf{y}},\sigma )\nonumber \\&=\frac{\;\bigl |\mathsf {diag}(\sqrt{\mathbf{w}})\bigr |\;}{\;\sqrt{2\pi }\sigma \;}\exp \left( -\frac{\;1\;}{\;2\sigma ^2\;}(\mathbf{x}-{{\varvec{\mu }}})\mathsf {diag}(\mathbf{w})(\mathbf{x}-{{\varvec{\mu }}})^\top \right) . \end{aligned}$$(8)
Besides the above assumption, in order to allow a fully Bayesian approach, the prior distribution for \(\Theta \) must also be specified. Taking into account that within \(\Theta \) the \(\sigma \) is a constant, and \({{\varvec{\mu }}}\) depends only on the above-specified distribution of \(\mathbf{x}\in c\), we switch to the prior distribution of the V feature weights. In this study, we considered the Dirichlet prior (Rao and Wu 2010) for use. Later, for ease of use, our approach was simplified such that the user-defined hyperparameters, \(\alpha _v(v=1,2,\ldots ,V)\), concentrate upon a uniform value \(\alpha \), i.e., \(\alpha _v:=\alpha \) for all v, thereby meeting a criterion of the symmetric Dirichlet prior (Teh et al. 2006; Hsu et al. 2003), that is,
Here, \(Beta(\alpha )\) denotes the Beta function with the concentration factor \(\alpha \). The choice of a symmetric Dirichlet distribution is not self-evident but may be suggested by understanding of data-fit. The factor \(\alpha \) technically determines the subspace in which the c can be fitted well. It acts upon the goodness of local data fitting, and contributes to our learning algorithm. We will discuss this merit in more detail in Sect. 4.
With the definite likelihood and prior provided by Eqs. 8 and 9, the best realization for \(\mathbf{w}\), \({{\varvec{\mu }}}\) and \(\sigma \) can then be found as a solution of
Solving this problem requires an algorithm which simultaneously maximizes the prior and the likelihood; we will discuss this in Sect. 4.
3.3 Measuring the effective discriminant functions
The paradigm for depicting the decision boundaries for class k entails learning from samples a class of Bayesian discriminant functions \({\mathcal {F}}_k\). This learning comprises three elements:
-
an input sample \(\mathbf{x}\in {\mathcal {C}}_k\), drawn independently from an unknown distribution \(P(\mathbf{x},k)\) (note that \(P(\mathbf{x},k)\) is different from the distribution of samples \(\mathbf{x}\in {\mathcal {D}}\), i.e., \(p(\mathbf{x})\) of Eq. 1);
-
an observer that returns a class outcome z to every input \(\mathbf{x}\);
-
a learning machine capable of implementing \({\mathcal {F}}^{(\ell _k)}_k(\mathbf{x})=\{f(\mathbf{x};\Theta )\}_{\ell _k}\).
The goal of learning is to choose one of \({\mathcal {F}}^{(1)}_k,{\mathcal {F}}^{(2)}_k,\ldots ,{\mathcal {F}}^{(\ell _k)}_k,\ldots \) which minimizes the local probability of error, i.e., samples of class k being classified incorrectly; for example, assigning \(\mathbf{x}\) to \({\mathcal {C}}_{\forall j\ne k}\) when it actually belongs to \({\mathcal {C}}_k\) (\(\mathbf{x}\) is in the region \({\mathcal {R}}_{\forall j\ne k}\) when it belongs to class k). Suppose that we have functions \({\mathcal {F}}^{(\ell _k)}_k\) and then consider the expected value of the loss or discrepancy \(L(z,{\mathcal {F}}^{(\ell _k)}_k(\mathbf{x}))\), given by
The smallest possible value for \({\mathcal {E}}({\mathcal {F}}^{(\ell _k)}_k)\) (Atashpaz-Gargari et al. 2013; Boyd and Vandenberghe 2004) is apparently required for learning. In practice, however, one has no access to \({\mathcal {E}}({\mathcal {F}}^{(\ell _k)}_k)\), since \(P(\mathbf{x},k)\) is unknown and the only available information is contained in the training data. In Vapnik (1999) the structural risk turns out to be an upper bound on this expected risk, with probability of at least \(1-\eta \) (in our case, \(\eta =0.05\)), i.e.,

which sums the following two quantities:
-
\({\mathcal {E}}_{emp}({\mathcal {F}}^{(\ell _k)}_k)\): the frequency of error over class k for \({\mathcal {F}}^{(\ell _k)}_k\), as measured the probability of disagreements between the class outcome z (it is actually k for \(\mathbf{x}\in {\mathcal {C}}_k\)) of the observer and the classification output \(z|{\mathcal {F}}^{(\ell _k)}_k\) provided by the learning machine. It in general simplifies to the fraction of incorrectly predicated samples to all samples of \({\mathcal {C}}_k\) such that
$$\begin{aligned} {\mathcal {E}}_{emp}({\mathcal {F}}^{(\ell _k)}_k)&=\frac{\;1\;}{\;N_k\;}\sum _{\mathbf{x}\in {\mathcal {C}}_k}\;{\mathbb {I}}(z|{\mathcal {F}}^{(\ell _k)}_k\ne k).\nonumber \end{aligned}$$The independence of the learning procedure on each class allows us to evaluate the frequency of error over \({\mathcal {C}}_k\), i.e., the local empirical risk, rather than over the training data \({\mathcal {D}}\). Consequently, learning from the training data can be reduced to an easy problem of one-class classification (Manevitz and Yousef 2002), in which \({\mathcal {F}}^{(\ell _k)}_k\) is utilized to recognize only the samples \(\mathbf{x}\in {\mathcal {C}}_k\) through the rule
$$\begin{aligned} z|{\mathcal {F}}^{(\ell _k)}_k&\triangleq {\left\{ \begin{array}{ll} k, &{} \quad \text {if}\ \max \;{\mathcal {F}}^{(\ell _k)}_k(\mathbf{x})>\max \;{\mathcal {F}}^{(1)}_j(\mathbf{x})\ \forall j\ne k\\ \text {NULL}, &{} \quad \text {otherwise} \end{array}\right. } \end{aligned}$$(12) -
\({ Conf}(N_k,d,{\mathcal {F}}^{(\ell _k)}_k,\eta )\): the Vapnik–Chervonenkis (VC) confidence interval on the difference between the training error and the real error. It is measured by the VC-dimension of the class of functions, d, and the size of the target data, \(N_k\), determined as
$$\begin{aligned} { Conf}(N_k,d,{\mathcal {F}}^{(\ell _k)}_k,\eta )\simeq {\left\{ \begin{array}{ll} \sqrt{\frac{\;d(\ln (2N_k/d)+1)-\ln \eta \;}{\;N_k\;}},&{}\text {if}\ N_k/d\ \text {is large (near 0.5)}\\ \frac{\;d(\ln (2N_k/d)+1)-\ln \eta \;}{\;N_k\;}, &{}\text {otherwise} \end{array}\right. } \end{aligned}$$
The minimization of the bound given by Eq. 11 yields the so-called principle of structural risk minimization (SRM). The optimal discriminant functions should be found by striking a tradeoff between the flexibility and the generalization ability of learning (Vapnik 1992). The basic argument is that the flexibility (determined by the complexity) and generalization ability are incompatible properties. The functions should not be expected to generalize very well if they possess high enough flexibility (complexity) to fit every possible dataset. On the contrary, if they are characterized by low complexity (particularly in the VC-dimension, regardless of the dimensionality of the feature space) but able to explain some particular datasets, they would also work well on the unseen data.
The VC-dimension is a more “sophisticated” measure of the complexity of a learning machine (or its discriminant functions) than dimensionality or number of parameters. Theoretical estimates of the VC-dimension have been obtained for only handful of simple classes of functions, most notably the class of linear discriminant functions. For our case, we use a simulation methodology proposed by Vapnik et al. (1994) which is well suited for estimating the VC-dimension of any learning machine; the approximation steps are outlined below.
-
1.
Generate 2m random independent samples of vectors (the \(\mathbf{x}\)s) and their class outcomes (the zs, each \(z\in \{k,\text {NULL}\}\)):
$$\begin{aligned} {\mathcal {Z}}^t_{2m}=\left\{ (\mathbf{x}^t_1,z^t_1),\ldots ,(\mathbf{x}^t_m,z^t_m), (\mathbf{x}^t_{m+1},z^t_{m+1}),\ldots ,(\mathbf{x}^t_{2m},z^t_{2m})\right\} . \end{aligned}$$Both \(\mathbf{x}\) and z are generated randomly: the \(\mathbf{x}\)s as a Gaussian noise, and the zs by Bernoulli trials with a probability of success value of 0.5.
-
2.
Study the maximum deviation of error rates between the two independent half-samples over a given class of discriminant functions \({\mathcal {F}}^{(\ell _k)}_k\):
$$\begin{aligned} \delta ({\mathcal {Z}}^t_{2m})&=\max _{{\mathcal {F}}^{(\ell _k)}_k}\left\{ \xi ^m_1\left( {\mathcal {Z}}^t_{2m};{\mathcal {F}}^{(\ell _k)}_k\right) -\xi ^m_2\left( {\mathcal {Z}}^t_{2m};{\mathcal {F}}^{(\ell _k)}_k\right) \right\} \\ \xi ^m_1\left( {\mathcal {Z}}^t_{2m};{\mathcal {F}}^{(\ell _k)}_k\right)&=\frac{\;1\;}{\;m\;}\sum ^m_{i=1}{\mathbb {I}}\left( z_i|{\mathcal {F}}^{(\ell _k)}_k=\text {NULL}\right) \\ \xi ^m_2\left( {\mathcal {Z}}^t_{2m};{\mathcal {F}}^{(\ell _k)}_k\right)&=\frac{\;1\;}{\;m\;}\sum ^{2m}_{i=m+1}{\mathbb {I}}\left( z_i|{\mathcal {F}}^{(\ell _k)}_k=\text {NULL}\right) \end{aligned}$$where \(\xi ^m_1\) and \(\xi ^m_2\) denote the frequencies of erroneous classifications on the two half-samples of \({\mathcal {Z}}^t_{2m}\).
-
3.
Approximate the expectation by an average over T independently generated sets of size 2m via
$$\begin{aligned} \mathbf{E}[\delta (m)]=\frac{\;1\;}{\;T\;}\sum ^T_{t=1}\delta ({\mathcal {Z}}^t_{2m}). \end{aligned}$$According to the theoretical findings of Vapnik et al. (1994), \(\mathbf{E}[\delta (m)]\) is bounded as follows:
$$\begin{aligned} \mathbf{E}[\delta (m)]&\le \Phi \left( \frac{\;m\;}{\;d\;}\right) \\ \Phi \left( \frac{\;m\;}{\;d\;}\right)&= {\left\{ \begin{array}{ll} 1, &{}\text { if }\frac{\;m\;}{\;d\;}<0.5\\ \epsilon _1\frac{\;\ln (2m/d)+1\;}{\;m/d-\epsilon _3\;}\left( \sqrt{1+\frac{\;\epsilon _2(m/d-\epsilon _3)\;}{\;\ln (2m/d)+1\;}}+1\right) , &{}\text {otherwise} \end{array}\right. }\nonumber \end{aligned}$$(13)The parameters can be set as \(\epsilon _1=0.16\), \(\epsilon _2=1.2\) and \(\epsilon _3\) chosen based on the condition of continuity at point \(m/d=0.5\), where \(\Phi (0.5)=1\), according to the results provided by the authors.
Since the bound Eq. 13 is tight, we can safely assume \(\mathbf{E}[\delta (m)]\simeq \Phi \left( \frac{\;m\;}{\;d\;}\right) \). Hence, we can find the effective VC-dimension that provides the best fit between \(\Phi (m/d)\) and \(\mathbf{E}[\delta (m)]\) by repeating the above procedure for various values of m (i.e., \(m_1, m_2, \ldots , m_h\)), i.e.,
4 Learning algorithm
For now, the only unaddressed problem for our work is how to learn the discriminant functions that achieve the minimum structural risk of misclassification. A new learning framework addressing this problem is outlined in Fig. 2. Within the framework, Algorithm 1 successively performs the following two routines independently on each of K classes:
-
(re)model multi-region via the unsupervised learning shown in Algorithm 3;
-
learn subspace NB via the EM estimation shown in Algorithm 2.
4.1 Selection of discriminant functions (Algorithm 1)
We recommend the following iteration to choose, from a set of discriminant functions \({\mathcal {F}}^{(1)}_k,{\mathcal {F}}^{(2)}_k,\ldots ,{\mathcal {F}}^{(\ell _k)}_k\), the one which best approximates the outcome (i.e., lowest risk).
-
1.
It starts by learning \({\mathcal {F}}^{(\ell _k=1)}_k\) composed of only one discriminant function \(f(\mathbf{x};\Theta )\).
-
2.
It then learns the succeeding \({\mathcal {F}}^{(\ell _k+1)}_k\), which includes one more function than the current \({\mathcal {F}}^{(\ell _k)}_k\). The iteration goes on if \({\mathcal {F}}^{(\ell _k+1)}_k\) is superior to \({\mathcal {F}}^{(\ell _k)}_k\), and ends up with \({\mathcal {F}}^{(\ell _k)}_k\) otherwise.
Algorithm 1, written in pseudo-code, implements a greedy approach to selecting preferable boundaries per class. This is done by comparing the risks of every pair of discriminant functions, e.g., \({\mathcal {F}}^{(\ell _k+1)}_k\) and \({\mathcal {F}}^{(\ell _k)}_k\), generated by two consecutive iterations. Finally, the one who firstly reaches the lowest structural risk is retained. To compare the predictive abilities of \({\mathcal {F}}^{(\ell _k+1)}_k\) and \({\mathcal {F}}^{(\ell _k)}_k\), we let the discriminant functions for class \(\forall j\ne k\) (cf. the \({\mathcal {F}}_j\) in Eq. 12) be taken over by \({\mathcal {F}}^{(1)}_j\) throughout the learning. This allows the selection procedure to be implemented in a parallel computing infrastructure when \(\ell _k>1\), because it evolves into a one-class learning, where all other classes are to be discarded, or wasted. This trick is reserved for extremely massive data classification in actual practice, as we note that it might be impractical to learn the discriminant functions for all classes, or even store them in memory, on a single machine.

4.2 Subspace NB learning (Algorithm 2)
Let us return to the problem, foreshadowed at Eq. 10, of estimating Bayesian parameters in \(\Theta \). We replace \(p(\Theta )\) with the Dirichlet probability function given by Eq. 9, and \(p(c|\Theta )\) with the probability density function in Eq. 8, arriving at a new objective function
where
-
the column vector, \(\mathbf{X}=(\mathbf{x}_1,\mathbf{x}_2,\ldots ,\mathbf{x}_{n})^\top \), relates to the n samples of c;
-
\({\varvec{\Sigma }}\) represents the standard deviation of these n samples on their features, given by
$$\begin{aligned} {\varvec{\Sigma }}=(\mathbf{X}\ominus {{\varvec{\mu }}})^\top \triangleq (\mathbf{x}_1-{{\varvec{\mu }}},\mathbf{x}_2-{{\varvec{\mu }}},\ldots ,\mathbf{x}_{n}-{{\varvec{\mu }}})^\top ; \end{aligned}$$ -
\(\ln \mathbf{w}\triangleq (\ln {w_1},\ln {w_2},\ldots ,\ln {w_V})\).
By plugging a Lagrange multiplier \(\lambda \) into \(J(\Theta )\) to impose the constraints illustrated by Eq. 3, one can obtain
A closer look at \((2\alpha -1)\mathbf{1}_V^\top \ln \mathbf{w}\) part of \(J(\Theta )\) reveals that this algebraic term essentially trades off bias and variance. Three effects on \(\mathbf{w}\) should be noted:
-
The weights are mainly concentrated in the values near the mean, e.g., \(\frac{1}{\sqrt{V}}\) in our case, when \(\alpha >1\) holds; that is, all weights closely approach to each other.
-
The weights spread across the interval (0,1) when \(\alpha <1\). In fact, only a few take large values, whereas the rest will be assigned a group of quite small weights close to zero.
-
The weights regress to a uniform distribution only if \(\alpha =1\), and consequently the MAP will reduce to an ML estimation.
Take the case where \(\alpha <1\) as an example. The learning machine may reduce bias, but should also increase variance on feature weights and, consequently, the chances of overfitting the regions. Details about this view can be derived from the statements indicated in Kohavi et al. (1997) and the ridge regularization approach used in regression analysis (Verweij and Houwelingen 1994). In this study we will selection a value of \(\alpha \) via cross-validation.
Set the derivatives of \({\mathcal {J}}\) with respect to \({{\varvec{\mu }}}\), \(\sigma \), \(\mathbf{w}\) and \(\lambda \) to zero. Some algebraic manipulations then yield the update equations required in our algorithm for Bayesian learning; these are summarized below.
Here, \((\bullet )^{-\mathbbm {1}}\) means that taking reciprocal for each entry of vector/matrix; for example, \((2,3,4)^{-\mathbbm {1}}=(\frac{1}{2},\frac{1}{3},\frac{1}{4})\). With \(\frac{\;\partial {\mathcal {J}}\;}{\;\partial \lambda \;}=0\), we further conclude from Eqs. 3 and 16 that the optimized value of \(\lambda \) corresponds to the root of
To solve the convex, differentiable nonlinear function, \(\varphi (\lambda )\), one can resort to a common numerical algorithm, e.g., the well-known Newton–Raphson or quasi-Newton method (Navon et al. 1988).
Given the derived update equations, it is possible to determine a desired estimate via Algorithm 2 which recurrently invokes each of these equations. The algorithm is an instantiation of the EM algorithm, which alternates between computing expectations for the unobserved values using the current parameters and computing the maximum MAP parameters of \({\mathcal {J}}\) using the current expectations (Dempster et al. 1977). For example, the weights are re-estimated by Eq. 16 and subsequently used for computation in the next iteration. This iterative process runs until a user-defined halting criterion that determines the convergence of Algorithm 2 when the change of weights between the two successive iterations is smaller than \(10^{-5}\). Since the numbers of iterations differ widely over the regions, we have imposed an additional criterion for convergence by limiting the maximum number of iterations to 100.

4.3 Remodeling multi-region (Algorithm 3)
For each iteration of the repeat-until loop in Algorithms 1 and 3 seeks a new set of \(\ell _k+1\) regions based on the \(\ell _k\) known pairs of means, \({\mathcal {U}}^{(\ell _k)}\triangleq \{{{\varvec{\mu }}}\}_{\ell _k}\), and weights, \({\mathcal {W}}^{(\ell _k)}\triangleq \{\mathbf{w}\}_{\ell _k}\), generated by Algorithm 2. For this purpose, Algorithm 3 includes the following two steps:
-
Update \({\mathcal {U}}^{(\ell _k+1)}\) and \({\mathcal {W}}^{(\ell _k+1)}\triangleq \{\mathbf{w}\}_{\ell _k}\), where the mean relevant to the \(\Theta \) with the worst F1-measure on class k is replaced by the two points of c; likewise, the associated weight is switched to the two initialized weights. The F1-measure on \({\mathcal {C}}_k\) is redefined as
$$\begin{aligned} \text {F1}({\mathcal {C}}_k;\Theta )&=\frac{\;2\times { recall}\times { precision}\;}{\;{ recall}+{ precision}\;}\\ { recall}&=\frac{\;1\;}{\;n\;}\sum _{\mathbf{x}\in c}\,{\mathbb {I}}(p(\mathbf{x}|\Theta )\text { is maximal})\\ { precision}&=\frac{\;\sum _{\mathbf{x}\in c}{\mathbb {I}}(p(\mathbf{x}|\Theta )\text { is maximal})\;}{\;\sum _{\mathbf{x}\in {\mathcal {C}}_k}{\mathbb {I}}(p(\mathbf{x}|\Theta )\text { is maximal})\;}. \end{aligned}$$ -
With the updated \({\mathcal {U}}^{(\ell _k+1)}\) and \({\mathcal {W}}^{(\ell _k+1)}\), the repeat-until loop remodels \(\ell _k+1\) regions for \({\mathcal {C}}_k\) by means of clustering.
Algorithm 3 resembles well-known paradigms such as top-down clustering (Parsons et al. 2004) and class-splitting (Gopal et al. 2012), but differs from them in two respects:
-
In comparison to top-down clustering, each iteration of remodeling starts from scratch, because all previously generated regions are no longer of use.
-
The traditional way of splitting a class would be too restrictive, as it updates only the worst model and stores the remaining ones. In contrast, our algorithm does not allow this because the class regions are highly constrained.
Strictly speaking, this algorithm can be thought of as a heuristic recursive learning that yields a quick convergence of Algorithm 1 and requires fewer runs to learn the final boundaries.

4.4 Algorithmic time complexity
A classifier customized for processing HDMSS should be fast, because we expect a simple, time-efficient classification. Notwithstanding the search for efficient algorithms in data mining, in practice many exhibit an exponential runtime. Here we show that our algorithm is linear with respect to the numbers of features and samples.
Two major computational iterations are considered in learning \(\ell _k\) decision boundaries per class: 1) the procedure of remodeling \(\ell _k\) regions takes \({\mathcal {O}}(\ell _kN_kV)\); 2) learning \(\ell _k\) Bayesian discriminant functions would be done in \({\mathcal {O}}(\ell _kVn)\), calling Algorithm 2 \(\ell _k\)-rounds. Now, for a K-class problem, we need to run Algorithm 1 K-rounds, and hence the total runtime requirements should be given precisely by \({\mathcal {O}}(K\ell _k^2VN_k)+{\mathcal {O}}(K\ell _k^2Vn)\), which is linear with respect to the number of training samples and the number of features, where K and \(\ell _k\) are in general much smaller than n, \(N_k\) and V.
5 Experiments
In this section, we evaluate our method in two sets of experiments. The first was designed to provide deeper insight into the functionality of MBSC through a case study, and the second, to evaluate how well MBSC performs in practical HDMSS data classification through a group of comparative experiments on a variety of HDMSS data.
5.1 Datasets and preprocessing
In the case study, we used a typical high-dimensional, low-sample-size (HDLSS) dataset. Such datasets can sometimes be more intractable to cope with than HDMSS data, due to the phenomenon of “empty space” (aka the curse of dimensionality) (Indyk and Motwani 1998). Comparative experiments were conducted on six HDMSS datasets, four real-world high-dimensional benchmark datasets and two semi-synthetic high-dimensional datasets, each dataset involving several million features. The two low-dimensional, massive-sample-size (LDMSS) datasets were also used to investigate how well MBSC performs on general low-dimensional data in general. The statistical characteristics of the datasets are summarized in Table 3.
-
One real-world HDLSS dataset for case study: The well-known Ovarian Cancer datasetFootnote 4 was used. It comprises proteomic spectra generated by mass spectroscopy for 162 ovarian cancers (Cancer) and 91 controls (Normal). The raw spectral data for each observation contains the relative amplitude of the intensity at each molecular mass/charge (M/Z) identity. There are a total of 15,154 M/Z identities (i.e., features). The aim of applying a classification method to this data is to recognize proteomic patterns that are relevant to a particular disease.
-
Four real-world HDMSS datasets commonly used to test the classification power of classifiers:
-
The 20 Newsgroups datasetFootnote 5 is a collection of approximately 20,000 newsgroup documents, which has become a popular data set for experiments in text applications of machine learning techniques, such as text classification and text clustering. We used the News20 dataset which was thoroughly well-processed by Chang and Lin (2011) and represented by the bag-of-words model.
-
The Yahoo-Korea dataset includes documents in a hierarchy of classes (Lin et al. 2007). We considered the largest branch from the root node (i.e., the branch including the largest number of classes) as positive, and all others as negative. By this treatment, the samples involved in this dataset belong to two classes.
-
The Bank dataset provides eleven months of payment transactions from over 31 million consumers to merchants or other persons, to predict the buying of a pension fund product or a long-term deposit (Martens and Provost 2011). The payment receivers were treated as features.
-
The URL Reputation dataset was collected by a time-order system over 120 days, and is used for classifying URLs as malicious (spam, phishing, exploits and so forth) and non-malicious (benign), where malicious URLs were obtained from a large web mail provider,and benign URLs from Yahoo’s directory listing. Here, we used only 60 subsets, namely URL-D60, of data collected on days 1 through 60. The set contains 64 real variables and the rest are in binary (i.e., the feature can take only two values: either one or zero). Since a large number of binary features have values of zero over all 60 subsets, we removed these non-active features according to the Bernoulli method established by Fortuny et al. (2013).
-
-
Two semi-synthetic HDMSS datasets extracted from Win32 portable executables for classifying (or detecting) malware (we collected the raw malware data from VX Heavens,Footnote 6 and then disassembled them and extracted frequent sequences, i.e., features) at the byte level (Masud et al. 2008):
-
The Trojan-\(*\) dataset comprise 8 types (classes) of Trojans: Banker, Clicker, Downloader, Dropper, PSW, GameThief, Proxy and Spy. Each feature/sequence is represented by a 4-g assembly instruction (cf. “PUSH, JMP, JCC, CALL” shown in Fig. 1c).
-
The Malware dataset contains 5 common types of malicious codes: Backdoor, Worm, Trojan, Virus and Rootkit. We reorganized each class by incorporating malware families composed of less than 20 executables into large similar families. For example, the family GetQQPass with only two PEs was merged into QQPass which contains 1308 PEs. Each feature of this dataset is a 6-g opcode (e.g., “4BE2F082DA74”).
-
-
Two LDMSS datasets available from UCI Machine Learning Repository:
-
The Covertype dataset contains 581,012 samples of 7 cover types (i.e., 7 classes) described by 54 explanatory features. The types Spruce-Fir and Lodgepole Pine, comprising 211,840 and 283,301 samples respectively, are much larger than the other 5 types. That is, Covertype is an extremely imbalanced dataset and poses challenges to classifiers. To reduce the difficulties of analyzing such extremely imbalanced data in comparative experiments, we extracted these two largest classes from the original data to yield the Covertype-Binary dataset, which consists of 495,141 samples belonging to two different classes.
-
The original Poker-Hand dataset includes 1,025,010 records, each being a sample of 5 playing cards drawn from a standard deck of 52. Each card is described using two features (suit and rank) and thus each sample is represented by a total of 10 explanatory features. All of these samples belong to one of 10 classes. Similar to the Covertype-Binary dataset, we extracted the two largest classes Nothing-in-hand” (513,702 samples) and One-pair (433,097 samples), yielding the Poker-Binary dataset with total 946,799 samples for classification analysis.
-
A few missing data in the datasets would lead to inaccurate discriminant functions. Data imputation can be used to remedy these missing data. We filled in missing entries via the linear regression introduced by Yuan (2010). As a post-processing step to impute discrete-valued features, we rounded the imputed values to the nearest discrete value. In addition, to obtain a meaningful set of weights, normalization was performed over all samples of each dataset for all variables via min-max-scaling. After normalization, each feature value falls within the range 0 to 1.
5.2 Setup
Each dataset was classified by algorithms for ten executions using tenfold cross-validation, and the average results were reported in the form mean (standard deviation). Three evaluation measures used are respectively 0–1 loss (Friedman 1997), F1-measure (Zhang et al. 2013) and AUC (Fawcett 2006). We report the classification accuracies, in terms of 0–1 loss, on the two balanced LDMSS datasets L1–L2, since it is an appropriate measure when classes are balanced. We also report the F1-measure on the imbalanced datasets Ovarian Cancer (binary-class) and S1–S2 (multi-class), and the AUC on the imbalanced datasets R1–R4 (binary-class).
The regular NB approach and its variations incorporated into feature weighting, subspace projection, kernel estimation, and multi-model schemes were selected as competing classifiers. SVM and the logistic regression model were also used for comparative evaluation.
-
NB: the regular NB, which uses a Gaussian distribution modeling the continuous features. We set \(\sigma _{kv}=10^{-5}\) in case it yields values of 0 on some features.
-
SWNB: an NB-based classifier using class-dependent projection (Chen and Wang 2012a), which automatically optimizes local feature subspaces for high-dimensional data. The standard deviation of the weights and the bandwidth factor, corresponding to a constraint on the weights, were set to 0.5 and 7, respectively.
-
FWNB: an NB-based method designed for categorical data classification using heuristic feature weighting, where the weights are calculated by the Kullback-Leibler measure (Lee et al. 2011). Because it is designed for categorical features, we discretized all continuous features except the binary ones, in accordance with the authors’ suggestion.
-
KWNB: a semi-naive Bayesian classifier using an embedded feature weighting. The weights are estimated by the kernel density function (Chen and Wang 2012b). We set the factor controlling weight distribution to \(\beta =2\) in the light of our previous experiences.
-
LWL+MNB: a basic locally weighted learning (LWL) which uses Weka’s Multinomial NB as the base classifier.
-
MapNB: an NB-based classifier which maps the data in hand into a new dataset by a class decomposition that decomposes each class into many clusters (Vilalta and Rish 2003). In this experiment, the number of clusters is estimated using cross-validation.
-
SVMLIN: the linear classifier (Lin et al. 2007), which performs as well as kernelized ones on large-scale data. We discarded the non-linear SVM in view of its time-consuming learning process.
-
DLR+SGD: The logistic regression (LR) model using stochastic gradient descent (SGD) learning. SGD has been successfully applied to large-scale machine learning problems. For large dataset, we used SGDClassifier with “log-loss” and filtered the features via discretization (Fayyad and Irani 1993) to allow easy use of this model.
All experiments ran on a Linux workstation with an Intel Core i7-4770S quad-core processor operating at 3.1GHz base clock, 16GB of main memory and a Samsung SSD with up to 550MB/s sequential read speed. The source codes for MBSC and the datasets for case study, including the original Ovarian Cancer dataset and its two reduced datasets extracted by our model, have been made publicly available.Footnote 7
5.3 A case study
Surveys were designed to study the following issues of interest, each linked to behaviors of MBSC:
-
Survey 1 MBSC is intended to address the class-dispersion issue. Does this issue actually arise in high-dimensional massive data, and what does it look like?
-
Survey 2 The classifier selects for class k a set of \(\ell _k\) discriminant functions, \({\mathcal {F}}^{(\ell _k)}_k\), by a specific risk criterion. With varying values of \(\ell _k\), what are the changes in the risk of misclassification by using \({\mathcal {F}}^{(\ell _k)}_k\)? Do these low-risk discriminant functions achieve high classification accuracy and low complexity?
-
Survey 3 A local feature weighting is embedded with Bayesian learning. What are the advantages of feature weighting in comparison to other methods, e.g., feature selection? The unique Bayesian prior parameter of the learning machine, \(\alpha \), has been claimed to work with this feature weighting. What are the changes of MBSC’s performance with this parameter?
-
Survey 4 Feature weighting quantifies the significance of features. What prominent enhancement can classifiers achieve if they are applied to the reduced dataset with only those features which are significant?
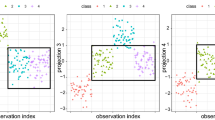
For Survey 1, Fig. 3 shows the five high-density regions and their individual subspaces discovered from the data. We vectorized a subspace explicitly as a group of non-uniform feature weights and each subspace simplifies to the five most relevant features that are most heavily weighted, where these weights are within the range 0 to 0.07. It can be clearly seen from the figure that samples of each class naturally yield multiple regions and, more importantly, they link to different sets of features. For example, the Cancer class groups into two regions \([c_1,s_1]\) and \([c_2,s_2]\), where the subclass \(c_1\) fills the subspace \(s_1\) relevant to the five features colored in cyan while \(c_2\) fills the subspace \(s_2\) relevant to the five features colored in brown. The weights assigned to the five features for \(s_1\) are approximately 0.06 and for \(s_2\) 0.045. Three subclasses discovered from the Normal class are residing in three different subspaces whose feature weights are much larger. This reveals a typical class dispersion in the sense we have described. This is the reason why our approach neither forces all classes into the full space nor arbitrarily constrains a class to a whole. Rather, it proposes to explore the class density distribution.
For Survey 2, Fig. 4 depicts the changes in the structural risk and classification accuracy on the two classes with varying \(\ell _k\). From the figure it can be clearly seen that the classification accuracy in terms of the F1-measure depends heavily upon \({\mathcal {E}}_k\); in this regard, it is reasonable for us to set up risk-oriented learning. The two F1-measure curves increase slightly for smaller \(\ell _k\), although not much, and subsequently go down for the larger one, because a large \(\ell _k\) will lead to a scarcity of known samples for learning each subclass and its corresponding subspace. Specifically, the F1-measure on the Cancer class increases from 0.93 when \(\ell _\text {Cancer}=1\) to the maximal 0.96 in the context of \(\ell _\text {Cancer}=3\). Meanwhile, on the Normal class, it increases from 0.82 to the maximal 0.94. The change in structural risk is actually a consequence of the interaction between empirical risk and VC confidence. Figure 5 shows that the structural risk can be approximated by the empirical risk in the context of a small \(\ell _k\), whereas with increasing \(\ell _k\) the VC-dimension of the learning machine grows such that the VC confidence plays an increasing role in the structural risk, despite of a lower empirical risk. We can see from the figure a sharp growth of structural risk because the VC confidence of the learning machine increases rapidly. This is a typical scenario of data overfitting. For an intuitive explanation, we compared the F1-measures of MBSC conducted on the training and test datasets and plotted the results in Fig. 6. In can be seen from the figure that the learning machine achieves over 90 % F1-measure accuracies on both the training and test sets in the context of three discriminant functions used to identify decision boundaries. However, the F1-measure will drop down to around 60 % when quite a few decision boundaries are built; this reveals that the learning machine ceases to be effective if its complexity exceeds a certain value, in contrast to the superior performance on the training data.
For Survey 3, Fig. 7 shows the distribution of weight values for all features, given varying \(\alpha \). In order to prevent MAP reducing to ML estimation, we required \(\alpha \ne 1\). Moreover, because the Newton method probably yields many weights below 0 when \(\alpha \le 0.5\), we specifically assert that \(\alpha >0.5\) in the experiments. The figure clearly shows that most of the weights take values that approximate the mean \(\sqrt{V}/V\approx 0.008\) in the case of a larger \(\alpha \); i.e., the weights are likely to be evenly distributed. At the opposite extreme, when \(\alpha \) takes a quite small value, the vast majority of the weights are concentrated in small values, generally less than 0.008, while a few take large values. By this property we might be able to easily filter out most of the irrelevant features that contribute weakly to classification. In Fig. 8 we can see, with varying \(\alpha \), the changes of classification accuracy in terms of the F1-measure. MBSC appears to no longer perform well when \(\alpha \) becomes small, probably because a small \(\alpha \) leads to large variance of feature weights and consequently to overly fitted regions. The best performances occur in the case where \(\alpha \) is at a value near 0.85, corresponding to a balance between bias and variance. The sudden change occurs when \(\alpha =1\) because MAP reduces to ML estimation. With increasing \(\alpha \), the variance of weights tends towards zero and the F1-measure towards stability.
For Survey 4, key to our approach is whether or not the training algorithm produces an optimal subspace for each region; that is, whether we can or cannot acquire a set of appropriate weights for features for each region. We therefore observed the behavior of the classifiers on the full space and on the two reduced subspaces. Taking advantage of the properties indicated in Survey 3, we selected the 41 most relevant features assigned large weights in the phase of learning Bayesian functions \({\mathcal {F}}^{(1)}_\text {Cancer}\) and \({\mathcal {F}}^{(1)}_\text {Normal}\), and the 115 features involved in learning the final discriminant functions \({\mathcal {F}}^{(2)}_\text {Cancer}\) and \({\mathcal {F}}^{(3)}_\text {Normal}\). Table 4 illustrates the classification accuracies of MBSC, in comparison to the other approaches. It appears that all of the classifiers are strengthened when they are performing in reduced subspaces. In general, all of the classifiers achieve higher F1-measures on the data with 41 and 115 selected features. In particular, these selected features allow SVMLIN and DLR+SGD to maintain an F1-measure of 100 %. This table demonstrates that learning multiple decision boundaries is able to detect the different significance of features for different classes to improve classification accuracy.
5.4 Comparative evaluation
5.4.1 Accuracy
Figure 9 reports the AUC of each classifier on the four binary high-dimensional massive data classification problems. We observed that MBSC achieves 94.7, 88.2, 88.7 and 93.1 % AUC on R1, R2, R3 and R4, respectively. It significantly outperforms all competing NB-based classifiers and DLR+SGD, as well as SVMLIN on all datasets but R4. The single maximum-margin hyperplane used by SVMLIN and the use of linear regression by DLR+SGD lead to high misclassification rates because the two classes are non-linearly separable in the full space (Joachims 1998). Looking more deeply, MBSC shows greater general stability with lower variances, because the classifier performs an SRM in learning decision boundaries. FWNB weights the features based on the gain ratio of features over classes, and hence acquires a model with many cases the model does not classify correctly (Chen and Wang 2012a). In contrast, the local weighting technique helps LWL+MNB, SWNB and KWNB achieve better results than FWNB. MBSC yields an approximate 12 % average improvement in comparison to NB, 5 % to SWNB, 10.5 % to FWNB, 4 % to KWNB, 7.5 % LWL+MNB, 5 % to MapNB, revealing the effectiveness of using multiple discriminant functions.
Table 5 demonstrates that MBSC achieves 94 % Micro-F1 and 92.5 % Macro-F1 on the S1 dataset and 96.4 % Micro-F1 and 93.3 % Macro-F1 on S2; it scores a clear win over the others on the two semi-synthetic datasets. In these two datasets, most classes cannot be distinguished from each other, mainly due to many dispersed subclasses in each class and similarity in the behavior of samples between subclasses. In order to recognize different classes of samples, the mapping procedure used in MapNB generally decomposes classes into an excessive number of shattered clusters, and thus, the classifier seems to have learnt quite a number of discriminant functions (Vilalta and Rish 2003). That is why we see the performance instability on the test sets caused by the poor generalization. However, MBSC is immune to this scenario because learning multiple decision boundaries for each class in different subspaces allows MBSC to find out the underlying malicious families of executables. The class dispersion makes SVMLIN less effective, as compared to the preceding binary classification datasets. The results demonstrate that the proposed method significantly improves classification performance on datasets with a substantial number of features and complex class distributions of samples.
Table 6 shows the comparison of classification accuracy in terms of 0–1 loss on the two LDMSS datasets. On the L1 dataset, 16.8 % samples are classified incorrectly by MBSC and 16.6 % by KWNB, which perform better than others. On the L2 dataset, SWNB, LWL+MNB and MBSC achieve under 30 % 0–1 loss which indicates a better classification accuracy than the others. Interestingly, we can see that the weight-based and projection-based models, such as MBSC, SWNB, FWNB, KWNB, and LWL+MNB, perform better than NB, MapNB, SVMLIN and DLR+SGD on the two datasets, because the two classes of each dataset overlap each other and cannot be distinguished in the full space. NB, SVMLIN and DLR+SGD fail to classify the two classes, yielding close to 50 % 0–1 loss (absolute misclassification rate). In contrast, MapNB performs better, because it builds multiple decision boundaries to make classification easier, although these boundaries are built in the full space. In fact, the overlapping in LDMSS data is a common challenge to classifiers. One major characteristic of such data is that the distribution of values for one or more features may be the same for different classes; this results in difficulty in distinguishing them from each other in overlapping space. The weight-based and projection-based models can easily identify class boundaries in subspaces and thus achieve more effective classification.
5.4.2 Experimental time complexity
We were concerned with the training velocity of MBSC and other competing NB-based classifiers in the same context. As NB has exhibited high speed when it runs on large datasets, we graphically compare the training runtime ratios of classifiers with NB in Fig. 10. Keep in mind that the ratios here do not indicate the classifiers’ training speeds with respect to sample size and (feature) dimensionality, because each ratio partly depends on NB’s performance. As we can see, MBSC performs a training approximately 8 times slower than NB, while SWNB, FWNB and KWNB are 6 times slower, and SVMLIN and DLR+SGD 14 times slower. That is, the training efficiency of MBSC is within an acceptable range, better than that of SVMLIN, LWL+MNB and DLR+SGD, yet a little bit inferior to that of NB. From the results in the figure, we conclude that MBSC yields high classification accuracy at a moderate expense in terms of increase in training runtime. We omitted the corresponding comparisons in terms of competitors’ test runtimes because there was no significant difference between them.
5.4.3 Scalability for our approach
Since our approach is designed to tackle massive data, we need to examine the time efficiency of MBSC with respect to increasing numbers of samples and features. Hence, we ran MBSC on the Trojan-\(*\) dataset to test its scalability by varying the sample size (N) and dimensionality (V) of the input data. N ranges from 20 to 100 K with fixed \(V=\)100 K, and V from 20 to 100 K with \(N=\)100 K. All reported runtimes were obtained using a single-threaded version of the implementation. Figure 11 shows the results of the scalability test. Note that we did not perform the scalability test on the competing classifiers, as they have turned out to be linearly increasing with the growth of dimensionality and sample size, although they may perform slowly on high-dimensional data. We can see from Fig. 11 that both the training and test runtimes of MBSC increase linearly with respect to dimensionality and sample size; this indicates that our new approach can be a Bayesian classifier of choice for classifying HDMSS data.
6 Conclusions
To effectively address the class-dispersion problem arising from high-dimensional, massive sample-size data, we proposed a new representation of Bayesian discriminant functions, which allows multiple decision boundaries in subspaces per class, and developed a simple, effective Bayesian classifier. For this representation, the classifier makes full use of structural risk minimization to learn Bayesian discriminant functions and, simultaneously, to optimize decision boundaries. The case study demonstrates that the classifier’s characteristics include the following:
-
flexibility of feature selection for each class by means of Dirichlet prior and local feature weighting in conjunction;
-
effectiveness of NB learning in exploiting complex class distributions;
-
a tradeoff between flexibility and generalization ability.
Comparative experiments we have done show that, on data involving millions of samples and features, NB strengthened by the new representation is effective, time-efficient and suitable for tackling extremely complex data. One avenue of further investigation is to extend the multiple decision boundaries per class diagram beyond the NB cases to model-based classifiers.
Notes
They are available at: Macfee http://www.mcafee.com/ca/mcafee-labs.aspx; Symantec http://www.symantec.com/security_response/publications/threatreport.jsp; Panda http://www.pandasecurity.com/mediacenter/reports/.
The terminologies “feature”, “attribute” and “dimension” are used interchangeably throughout the paper.
We have borrowed the terminology “class-dispersion” from Vilalta and Rish (2003); the dispersion of our case is much more complicated, however, because it is subspace-related.
References
Aggarwal CC, Han J, Wang J, Yu PS (2004) A framework for projected clustering of high dimensional data streams. In: Proceedings of the thirtieth international conference on very large data bases (VLDB), pp 852–863
Aha D, Kibler D (1991) Instance-based learning algorithms. Mach Learn 6:37–66
Albert MK, Aha DW (1991) Analyses of instance-based learning algorithms. In: Proceedings of the ninth national conference on artificial intelligence (AAAI), pp 553–558
Atashpaz-Gargari E, Sima C, Braga-Neto UM, Dougherty ER (2013) Relationship between the accuracy of classifier error estimation and complexity of decision boundary. Pattern Recognit 46(5):1315–1322
Bengio Y, Bengio S (1999) Modeling high-dimensional discrete data with multi-layer neural networks. In: The twelfth advances in neural information processing systems (NIPS), pp 400–406
Boyd S, Vandenberghe L (2004) Convex optimization. Cambridge University Press, Cambridge
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
Chang CC, Lin CJ (2011) Libsvm: a library of the support vector machines. ACM Trans Intell Syst Technol 2:27
Chen L, Wang S (2012a) Automated feature weighting in naive Bayes for high-dimensional data classification. In: Proceedings of the twenty-first ACM international conference on information and knowledge management (CIKM), pp 1243–1252
Chen L, Wang S (2012b) Semi-naive Bayesian classification by weighted kernel density estimation. In: Proceedings of the eighth international conference on advanced data mining and applications (ADMA), pp 260–270
Chen L, Jiang Q, Wang S (2012) Model-based method for projective clustering. IEEE Trans Knowl Data Eng 24(7):1291–1305
Dahl GE, Stokes JW, Deng L, Yu D (2013) Large-scale malware classification using random projections and neural networks. In: Proceedings of the IEEE international conference on acoustics, speech and signal processing (ICASSP), pp 3422–3426
Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Ser B 39(1):1–38
Domeniconi C, Gunopulos D, Ma S, Yan B, Al-Razgan M, Papadopoulos D (2007) Locally adaptive metrics for clustering high dimensional data. Data Min Knowl Discov 14(1):63–97
Domingos P, Pazzani M (1997) On the optimality of the simple Bayesian classifier under zero-one loss. Mach Learn 29(2–3):103–130
Fan W, Bifet A (2013) Mining big data: current status, and forecast to the future. ACM SIGKDD Explor Newslett 14(2):1–5. doi:10.1145/2481244.2481246
Fawcett T (2006) An introduction to ROC analysis. Pattern Recognit Lett 27(8):861–874
Fayyad UM, Irani KB (1993) Multi-interval discretization of continuous-valued attributes for classification learning. In: Proceedings of the thirteenth international joint conference on artificial intelligence (IJCAI), pp 1022–1029
Fortuny EJ, Martens D, Provost F (2013) Predictive modeling with big data: is bigger really better? Big Data 1(4):215–226
Frank E, Hall M, Pfahringer B (2003) Locally weighted naive Bayes. In: Proceedings of the nineteenth annual conference on uncertainty in artificial intelligence (UAI), pp 249–256
Friedman JH (1997) On bias, variance, 0/1-loss, and the curse-of-dimensionality. Data Min Knowl Discov 1(1):55–77
Gopal S, Yang Y, Bai B, Mizil AN (2012) Bayesian models for large-scale hierarchical classification. In: Proceedings of the twenty-sixth annual conference on neural information processing systems (NIPS), pp 2420–2428
Han EHS, Karypis G (2000) Centroid-based document classification: analysis and experimental results. In: Proceedings of the fourth European conference on principles of knowledge discovery and data mining, pp 424–431
Hinneburg A, Aggarwal CC, Keim DA (2000) What is the nearest neighbor in high dimensional spaces? In: The twenty-sixth international conference on very large databases (VLDB), pp 506–515
Hsu CN, Huang HJ, Wong TT (2003) Implications of the dirichlet assumption for discretization of continuous variables in naive Bayesian classifiers. Mach Learn 53(3):235–263
Ifrim G, Bakır G, Weikum G (2008) Fast logistic regression for text categorization with variable-length n-grams. In: Proceedings of the fourteenth ACM SIGKDD international conference on knowledge discovery and data mining (KDD), pp 354–362
Indyk P, Motwani R (1998) Approximate nearest neighbors: towards removing the curse of dimensionality. In: Proceedings of the thirtieth annual ACM symposium on theory of computing (STOC), pp 604–613
Jing L, Ng MK, Huang JZ (2007) An entropy weighting k-means algorithm for subspace clustering of high-dimensional sparse data. IEEE Trans Knowl Data Eng 19(8):1026–1041
Joachims T (1996) A probabilistic analysis of the rocchio algorithm with tfidf for text categorization. Technical Reports, DTIC Document
Joachims T (1998) Text categorization with support vector machines: learning with many relevant features. Springer, Berlin
John GH, Langley P (1995) Estimating continuous distributions in Bayesian classifiers. In: Proceedings of the eleventh conference on uncertainty in artificial intelligence (UAI), pp 338–345
Kang DK, Silvescu A, Honavar V (2006) RNBL-MN: a recursive naive Bayes learner for sequence classification. In: Proceedings of the eighteenth Pacific-Asia conference on knowledge discovery and data mining (PAKDD), pp 45–54
Kim J, Le DX, Thoma GR (2008) Naive Bayes classifier for extracting bibliographic information from biomedical online articles. In: Proceedings of the international conference on data mining (DMIN), pp 373–378
Kohavi R, Langley P, Yun Y (1997) The utility of feature weighting in nearest-neighbor algorithms. In: Proceedings of the ninth European conference on machine learning (ECML), pp 85–92
Kooij AJ, et al (2007) Chapter 4: regularization with ridge penalties, the lasso, and the elastic net for regression with optimal scaling transformations. In: Prediction accuracy and stability of regression with optimal scaling transformations, pp 65–90
Lee CH, Gutierrez F, Dou D (2011) Calculating feature weights in naive Bayes with kullback–leibler measure. In: Proceedings of the eleventh IEEE international conference on data mining (ICDM), pp 1146–1151
Li P, Shrivastava A, Moore JL, König AC (2011) Hashing algorithms for large-scale learning. In: Proceedings of the twenty-fifth annual conference on neural information processing systems (NIPS), pp 2672–2680
Lin C, Weng RC, Keerthi SS (2007) Trust region Newton methods for large-scale logistic regression. In: Proceedings of the twenty-fourth international conference on machine learning (ICML), pp 561–568
Lin G, Shen C, Shi Q, van den Hengel A, Suter D (2014) Fast supervised hashing with decision trees for high-dimensional data. In: Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR), pp 1963–1970
Liu S, Trenkler G (2008) Hadamard, khatri-rao, kronecker and other matrix products. Int J Inf Syst Sci 4(1):160–177
Manevitz LM, Yousef M (2002) One-class svms for document classification. J Mach Learn Res 2:139–154
Marchiori E (2013) Class dependent feature weighting and k-nearest neighbor classification. In: Proceedings of the eighth IAPR international conference on pattern recognition in bioinformatics (PRIB), pp 69–78
Martens D, Provost F (2011) Pseudo-social network targeting from consumer transaction data. Faculty of Applied Economics, University of Antwerp, Belgium
Martínez AM, Kak AC (2001) PCA versus LDA. IEEE Trans Pattern Anal Mach Intell 23(2):228–233
Masud MM, Khan L, Thuraisingham B (2008) A scalable multi-level feature extraction technique to detect malicious executables. Inf Syst Front 10(1):33–45
Nakajima S, Watanabe S (2005) Generalization error of linear neural networks in an empirical Bayes approach. In: Proceedings of the nineteenth international joint conference on artificial intelligence (IJCAI), pp 804–810
Navon IM, Phua PK, Ramamurthy M (1988) Vectorization of conjugate-gradient methods for large-scale minimization. In: Proceedings of the second ACM/IEEE conference on supercomputing, pp 410–418
Parsons L, Haque E, Liu H (2004) Subspace clustering for high dimensional data: a review. ACM SIGKDD Explor Newslett 6(1):90–105
Rani P, Pudi V (2008) RBNBC: Repeat based naive Bayes classifier for biological sequences. In: Proceedings of the eighth IEEE international conference on data mining (ICDM), pp 989–994
Rao J, Wu C (2010) Bayesian pseudo-empirical-likelihood intervals for complex surveys. J R Stat Soc Ser B 72(4):533–544
Seeger M (2006) Bayesian modelling in machine learning: a tutorial review. Technical Reports
Shabtai A, Moskovitch R, Elovici Y, Glezer C (2009) Detection of malicious code by applying machine learning classifiers on static features: a state-of-the-art survey. Inf Secur Tech Rep 14(1):16–29
Straub WO (2009) A brief look at gaussian integrals. Article, Pasadena California. http://www.weylmann.com/gaussian.pdf
Su J, Shirab JS, Matwin S (2011) Large scale text classification using semisupervised multinomial naive Bayes. In: Proceedings of the twenty-eighth international conference on machine learning (ICML), pp 97–104
Tan M, Wang L, Tsang IW (2010) Learning sparse SVM for feature selection on very high dimensional datasets. In: Proceedings of the twenty-seventh international conference on machine learning (ICML), pp 1047–1054
Tan M, Tsang IW, Wang L (2014) Towards ultrahigh dimensional feature selection for big data. J Mach Learn Res 15(1):1371–1429
Tan S (2008) An improved centroid classifier for text categorization. Exp Syst Appl 35(1–2):279–285
Teh YW, Jordan MI, Beal MJ, Blei DM (2006) Hierarchical dirichlet processes. J Am Stat Assoc 101(476):1566–1581
Vapnik VN (1992) Principles of risk minimization for learning theory. In: Proceedings of the fifth annual conference on neural information processing systems (NIPS), pp 831–838
Vapnik VN (1999) An overview of statistical learning theory. IEEE Trans Neural Netw Learn Syst 10(5):988–999
Vapnik VN (2000) The nature of statistical learning theory. Springer, Berlin
Vapnik VN, Levin E, Le Cun Y (1994) Measuring the VC-dimension of a learning machine. Neural Comput 6(5):851–876
Verweij PJ, Van Houwelingen HC (1994) Penalized likelihood in cox regression. Stat Med 13(23–24):2427–2436
Vilalta R, Rish I (2003) A decomposition of classes via clustering to explain and improve naive Bayes. In: Proceedings of the fourteenth European conference on machine learning (ECML), pp 444–455
Vilalta R, Achari MK, Eick CF (2003) Class decomposition via clustering: a new framework for low-variance classifiers. In: Proceedings of the third IEEE international conference on data mining (ICDM), pp 673–676
Weinberger K, Dasgupta A, Langford J, Smola A, Attenberg J (2009) Feature hashing for large scale multitask learning. In: Proceedings of the twenty-sixth annual international conference on machine learning (ICML), pp 1113–1120
Xu B, Huang JZ, Williams G, Wang Q, Ye Y (2012) Classifying very high-dimensional data with random forests built from small subspaces. Int J Data Warehous Min 8(2):44–63
Yu S, Fung G, Rosales R, Krishnan S, Rao RB, Dehing-Oberije C, Lambin P (2008) Privacy-preserving Cox regression for survival analysis. In: Proceedings of the fourteenth ACM SIGKDD international conference on knowledge discovery and data mining (KDD), pp 1034–1042
Yuan YC (2010) Multiple imputation for missing data: concepts and new development (version 9.0). SAS Institute Inc, Rockville
Zaidi NA, Cerquides J, Carman MJ, Webb GI (2013) Alleviating naive Bayes attribute independence assumption by attribute weighting. J Mach Learn Res 14(1):1947–1988
Zhang J, Chen L, Guo G (2013) Projected-prototype based classifier for text categorization. Knowl Based Syst 49:179–189
Zhou Z, Chen Z (2002) Hybrid decision tree. Knowl Based Syst 15(8):515–528
Acknowledgments
We would like to thank Carol Harris for improving this paper significantly. This work has been supported by fundings from the Natural Sciences and Engineering Research Council of Canada (NSERC) to Shengrui Wang under Grant No. 396097-2010, and from the Natural Science Foundation of Fujian Province of China to Lifei Chen under Grant No. 2015J01238. Thanks UPMC—Université Pierre et Marie Curie for a financial support allowing the collaboration between Shengrui Wang and Patrick Gallinari. Shengrui Wang is also partly supported by Natural Science Foundation of China (NSFC) under Grant No. 61170130.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Kristian Kersting.
Rights and permissions
About this article
Cite this article
Zhang, J., Wang, S., Chen, L. et al. Multiple Bayesian discriminant functions for high-dimensional massive data classification. Data Min Knowl Disc 31, 465–501 (2017). https://doi.org/10.1007/s10618-016-0481-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10618-016-0481-y